SpAD Biofunctionalized Cellulose Acetate Scaffolds Inhibit Staphylococcus aureus Adherence in a Coordinating Function with the von Willebrand A1 Domain (vWF A1)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
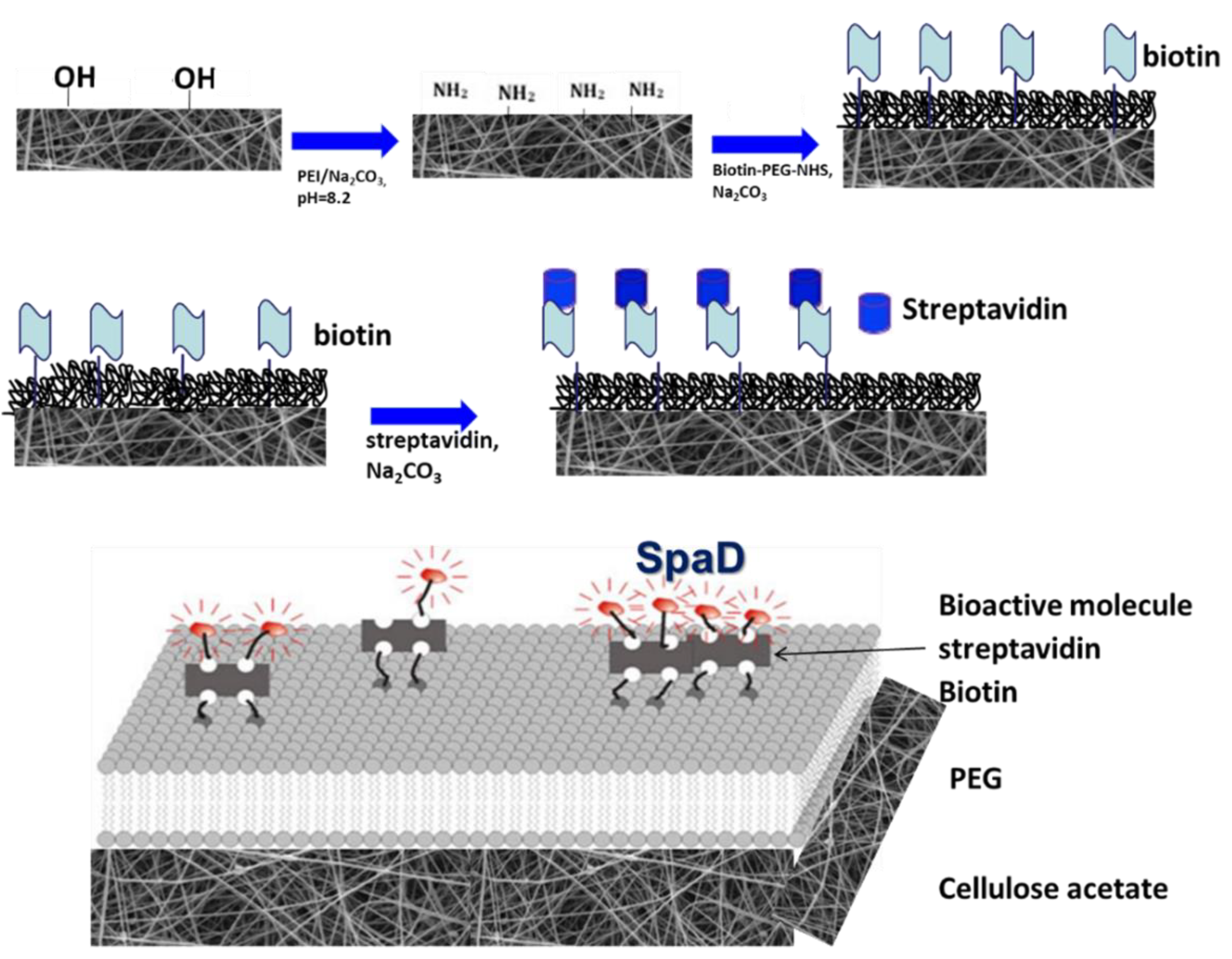
2.2.1. Scaffold Preparation
2.2.2. Determination of Amoxicillin Loading
2.2.3. In Vivo Biotinylation and Purification of SpAD and GFP
2.2.4. Purification of vWF A1
2.2.5. Incubation of S. aureus with CA Scaffolds in Human Plasma
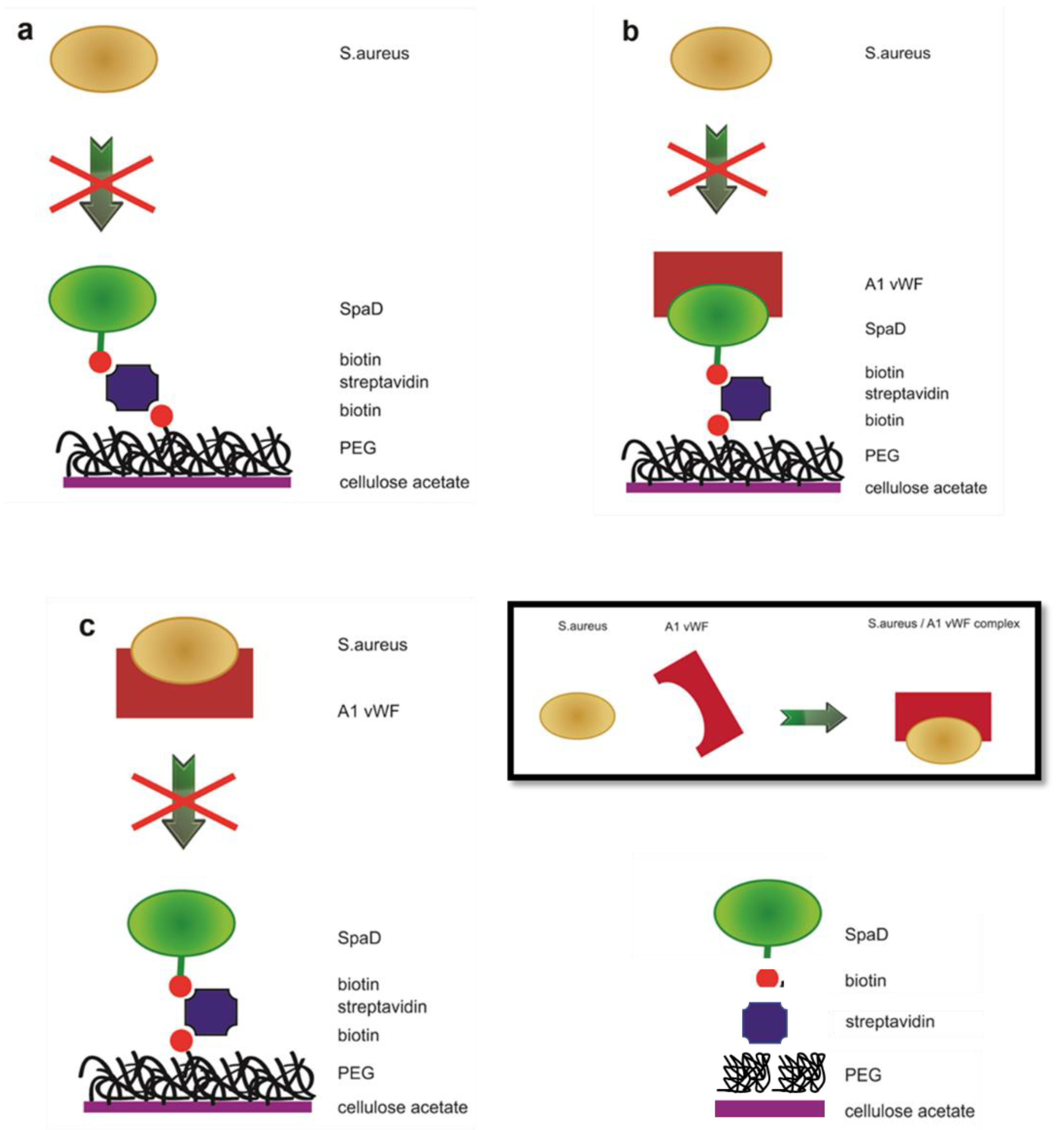
2.2.6. Treatment of Scaffolds for Specific Tethering of SpAD and Incubation of S. aureus with Biofunctionalized CA Scaffolds
2.2.7. Sample Treatment for Scanning Electron Microscopy (SEM)
2.2.8. Molecular Simulations
3. Results and Discussion
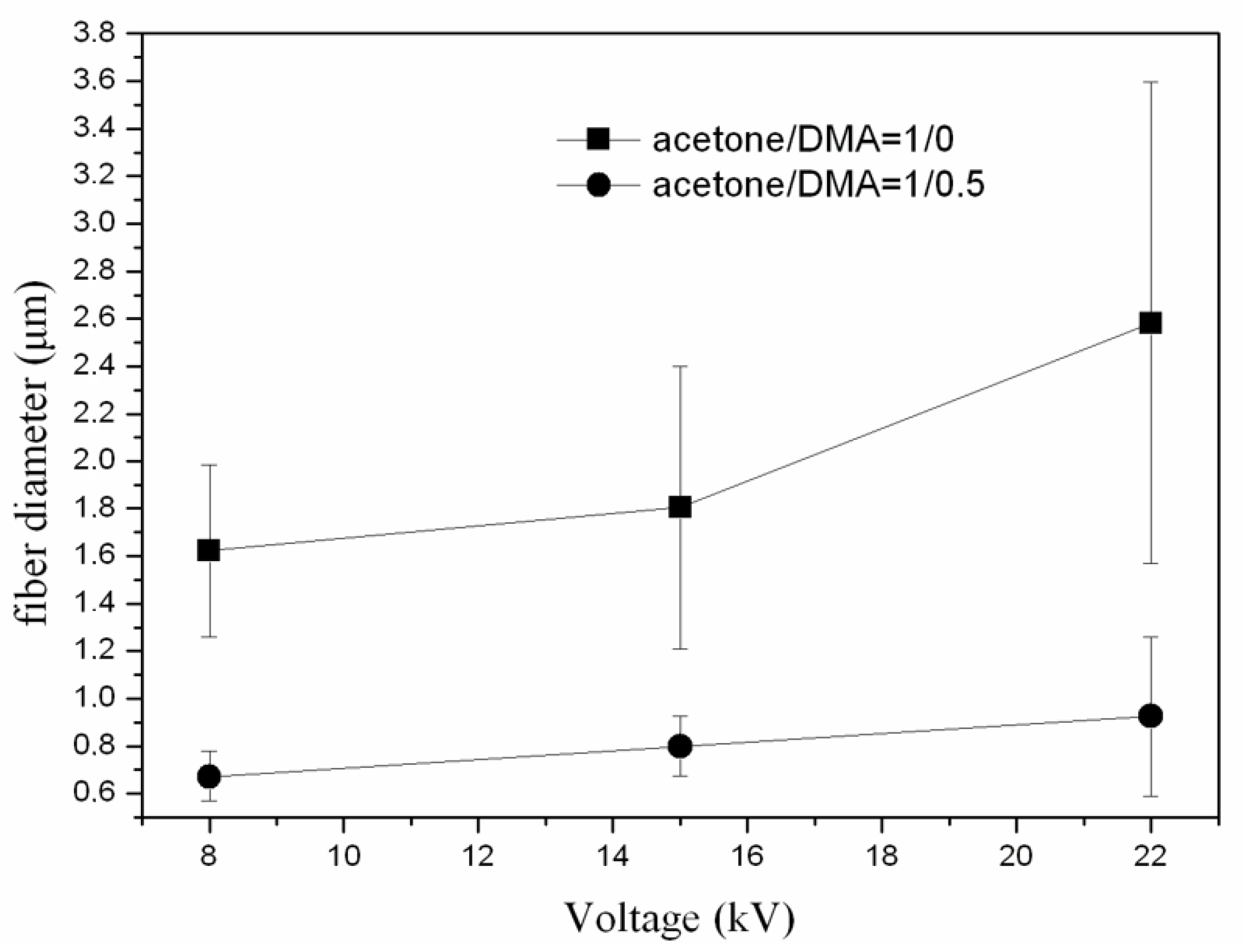
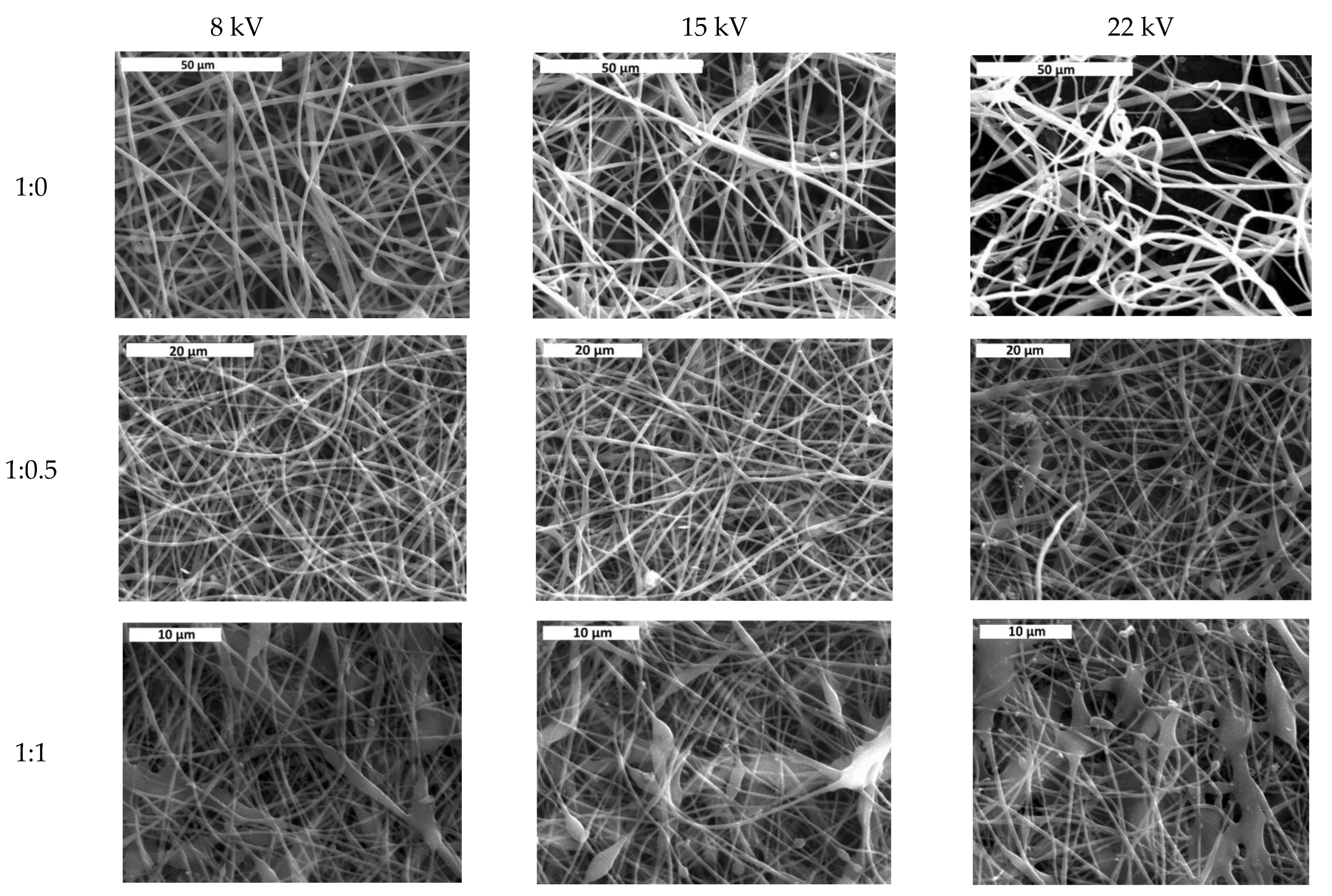
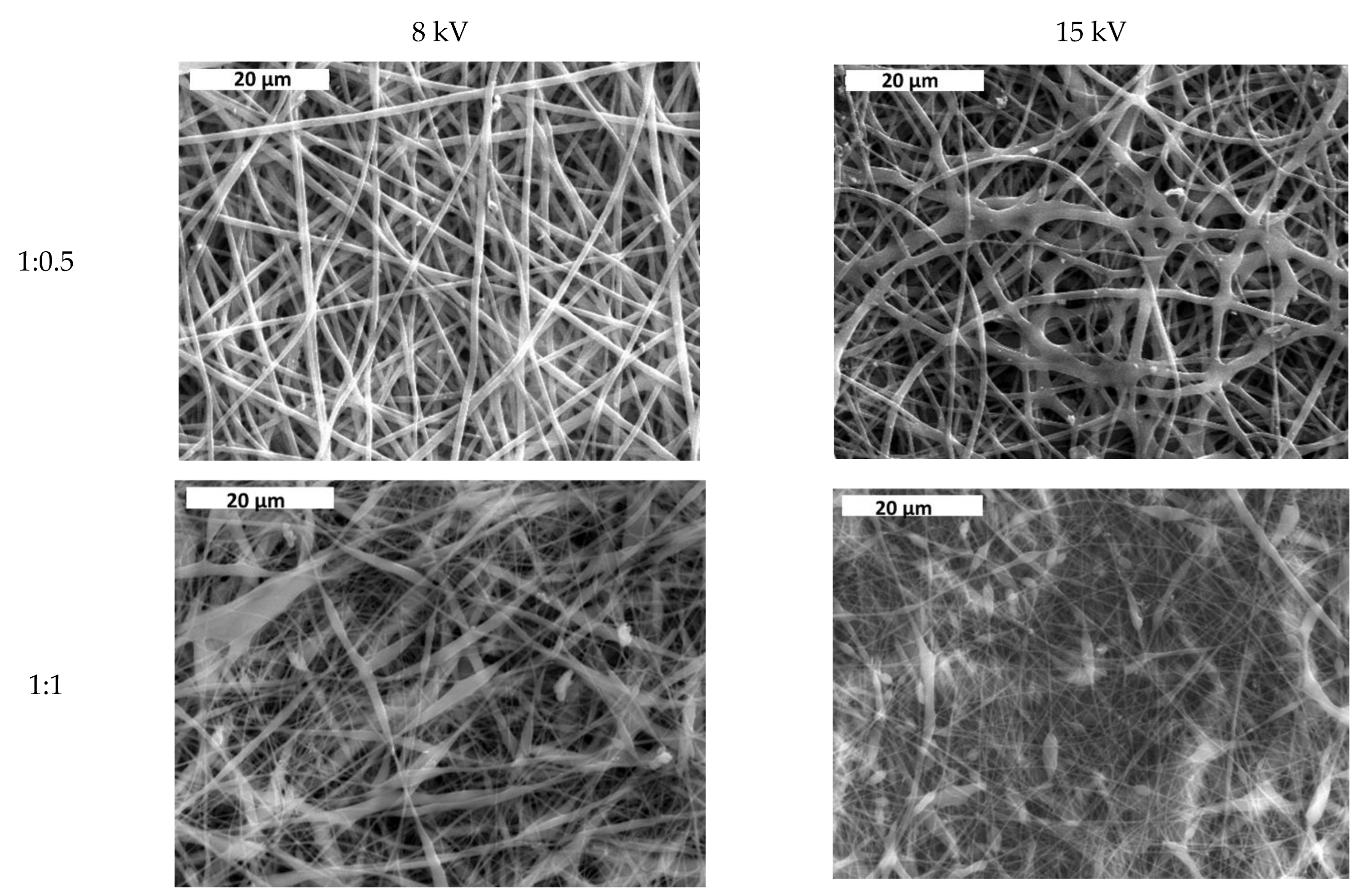
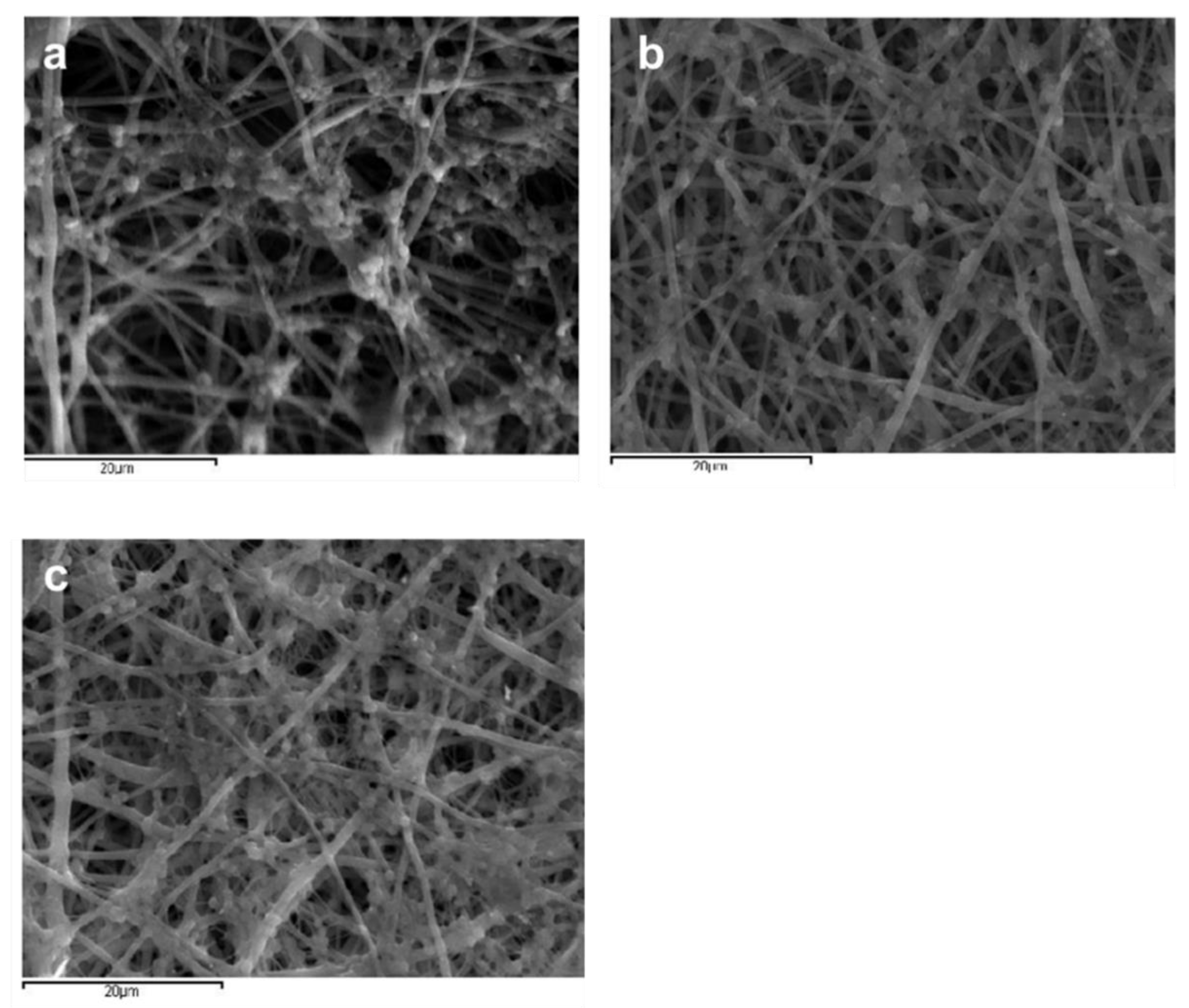
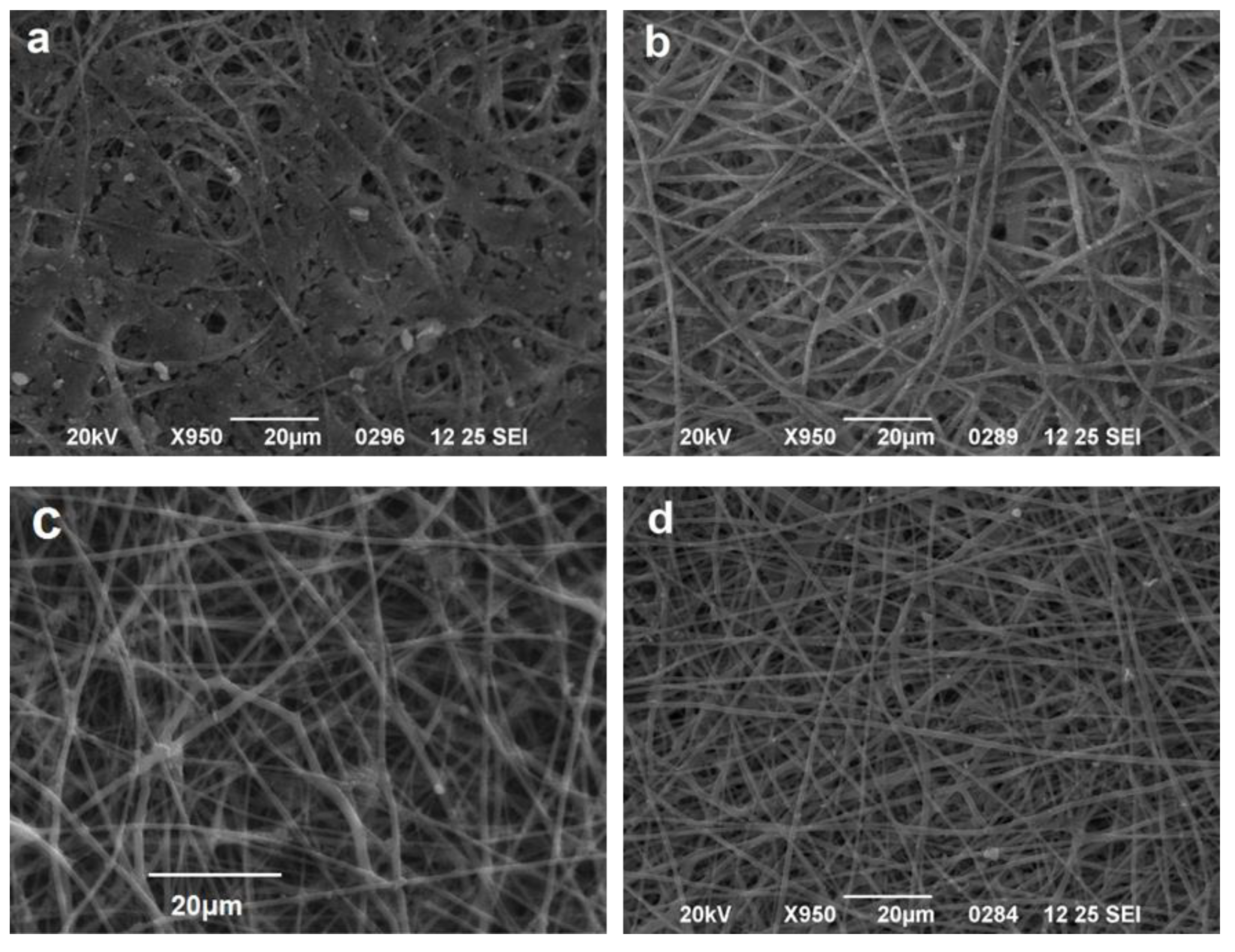
3.1. Effect of Solvent Ratio on Fiber Size and Morphology
3.2. Determination of Amoxicillin Loading and Effect of Amoxicillin on Fiber Size and Morphology
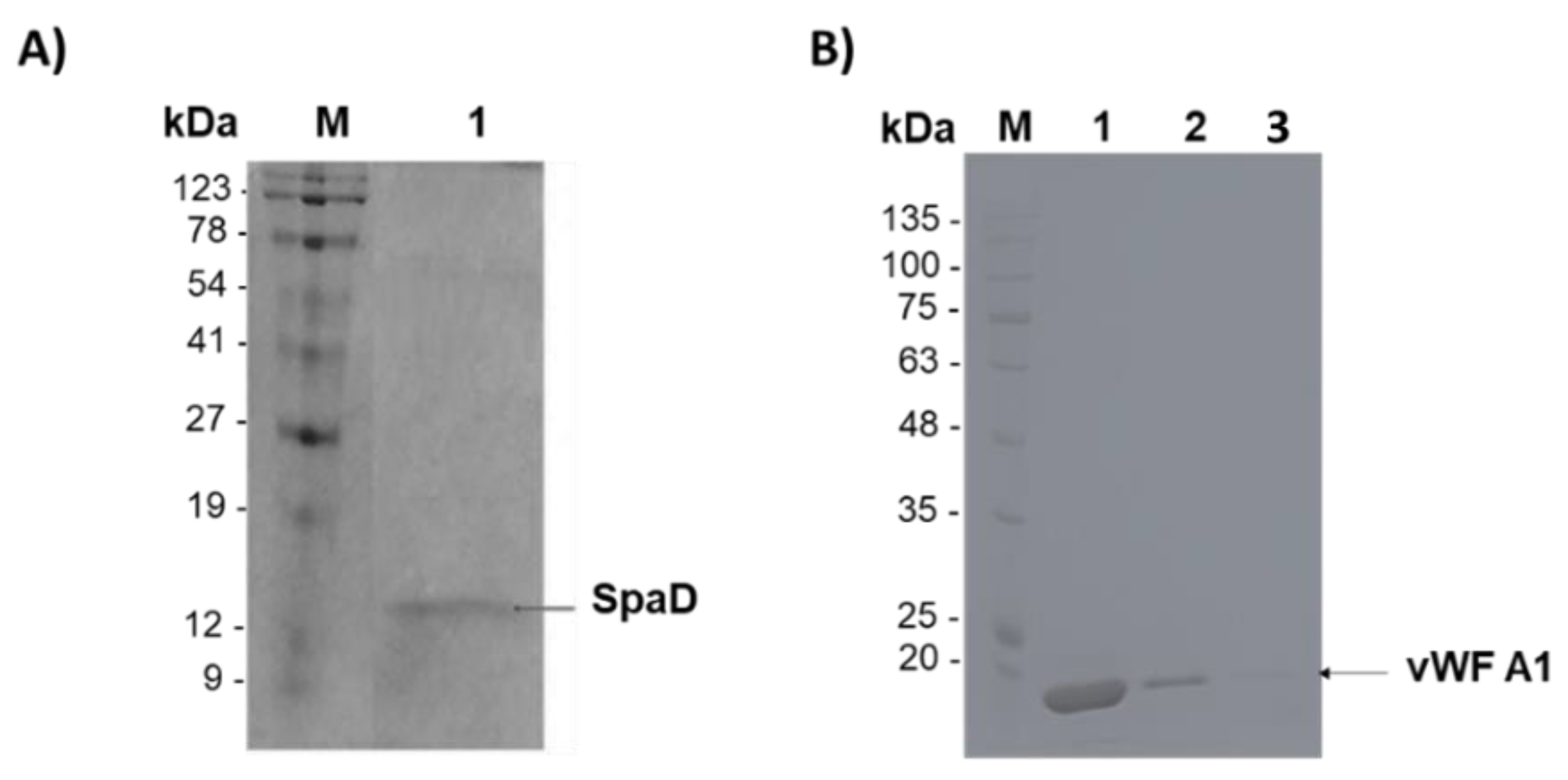
3.3. Purification of SpAD and vWF A1
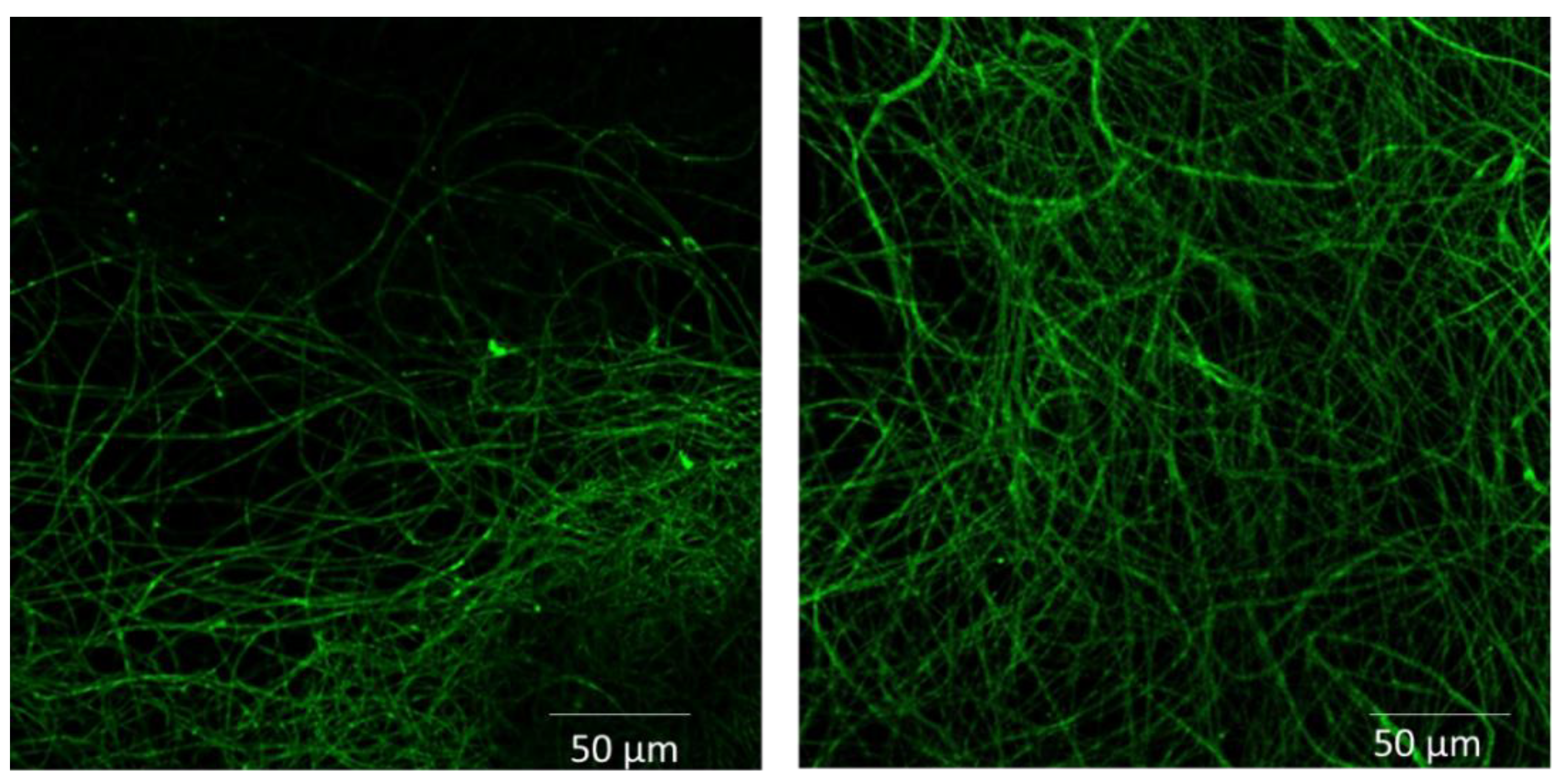
3.4. Functionalization of Cellulose Acetate Scaffolds with GFP or SpAD
3.5. Incubation of S. aureus or Ε. coli with CA Scaffolds
3.6. Incubation of S. aureus with CA Scaffolds in Human Plasma
3.7. Incubation of S. aureus with the Biofunctionalized CA Scaffolds
3.8. Incubation of S. aureus with Biofunctionalized CA Scaffolds in Human Plasma
3.9. Interaction between SpAD and vWF A1
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Taj, Y.; Abdullah, F.E.; Kazmi, S.U. Current pattern of antibiotic resistance in Staphylococcus aureus clinical isolates and the emergence of vancomycin resistance. J. Coll. Physicians Surg. Pak. 2010, 20, 728–732. [Google Scholar] [PubMed]
- Akoglu, H.; Zarakolu, P.; Altun, B.; Unal, S. Epidemiological and molecular characteristics of hospital-acquired methicillin-resistant Staphylococcus aureus strains isolated in Hacettepe Universty Adult Hospital in 2004–2005. Mikrobiyol. Bul. 2010, 44, 343–355. [Google Scholar] [PubMed]
- Liang, D.; Hsiao, B.S.; Chu, B. Functional electrospun nanofibrous scaffolds for biomedical applications. Adv. Drug. Deliv. Rev. 2007, 59, 1392–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Decraene, V.; Pratten, J.; Wilson, M. Cellulose acetate containing toluidine blue and rose bengal is an effective antimicrobial coating when exposed to white light. Appl. Environ. Microb. 2006, 72, 4436–4439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heunis, T.D.; Dicks, L.M. Nanofibers offer alternative ways to the treatment of skin infections. J. Biomed. Biotechnol. 2010, 2010, 510682. [Google Scholar] [CrossRef]
- Zhou, M.; Green, T.B.; Joo, Y.L. The thermal effects on electrospinning of polylactic acid melts. Polymer 2006, 47, 7497–7505. [Google Scholar] [CrossRef]
- Lyons, J.; Ko, F. Melt Electrospinning of Polymers: A Review. Polym. News 2005, 30, 170–178. [Google Scholar] [CrossRef]
- Casper, C.L.; Stephens, J.S.; Tassi, N.G.; Chase, D.B.; Rabolt, J.F. Controlling Surface Morphology of Electrospun Polystyrene Fibers: Effect of Humidity and Molecular Weight in the Electrospinning Process. Macromolecules 2004, 37, 573–578. [Google Scholar] [CrossRef]
- Tungprapa, S.; Puangparn, T.; Weerasombut, M.; Jangchud, I.; Fakum, P.; Semongkhol, S.; Meechaisue, C.; Supaphol, P. Electrospun cellulose acetate fibers: Effect of solvent system on morphology and fiber diameter. Cellulose 2007, 14, 563–575. [Google Scholar] [CrossRef]
- Ito, Y.; Hasuda, H.; Kamitakahara, M.; Ohtsuki, C.; Tanihara, M.; Kang, I.K.; Kwon, O.H. A Composite of Hydroxyapatite with Electrospun Biodegradable Nanofibers as a Tissue Engineering Material. J. Biosci. Bioeng. 2005, 100, 43–49. [Google Scholar] [CrossRef]
- Fujihara, K.; Kotaki, M.; Ramakrishna, S. Guided bone regeneration membrane made of polycaprolactone/calcium carbonate composite nano-fibers. Biomaterials 2005, 26, 4139–4147. [Google Scholar] [CrossRef] [PubMed]
- Marras, S.I.; Kladi, K.P.; Tsivintzelis, I.; Zuburtikudis, I.; Panayiotou, C. Biodegradable polymer nanocomposites: The role of nanoclays on the thermomechanical characteristics and the electrospun fibrous structure. Acta Biomater. 2008, 4, 756–765. [Google Scholar] [CrossRef] [PubMed]
- Van, E.L.; Grondahl, L.; Chua, K.N.; Leong, K.W.; Nurcombe, V.; Cool, S.M. Controlled release of heparin from poly(ε-caprolactone) electrospun fibers. Biomaterials 2006, 27, 2042–2050. [Google Scholar]
- Tungprapa, S.; Jangchud, I.; Supaphol, P. Release characteristics of four model drugs from drug-loaded electrospun cellulose acetate fiber mats. Polymer 2007, 48, 5030–5041. [Google Scholar] [CrossRef]
- Chen, L.; Bromberg, L.; Hatton TA Rutledge, G.C. Electrospun cellulose acetate fibers containing chlorhexidine as a bactericide. Polymer 2008, 49, 1266–1275. [Google Scholar] [CrossRef]
- Suwantong, O.; Opanasopit, P.; Ruktanonchai, U.; Supaphol, P. Electrospun cellulose acetate fiber mats containing curcumin and release characteristic of the herbal substance. Polymer 2007, 48, 7546–7557. [Google Scholar] [CrossRef]
- Zhang, L.; Menkhausb, T.J.; Fong, H. Fabrication and bioseparation studies of adsorptive membranes/felts made from electrospun cellulose acetate nanofibers. J. Membr. Sci. 2008, 319, 176–184. [Google Scholar] [CrossRef]
- Han, S.O.; Youk, J.H.; Min, K.D.; Kang, Y.O.; Park, W.H. Electrospinning of cellulose acetate nanofibers using a mixed solvent of acetic acid/water: Effects of solvent composition on the fiber diameter. Mater. Lett. 2008, 62, 759–762. [Google Scholar] [CrossRef]
- Zhang, L.; Hsieh, Y.L. Ultra-fine cellulose acetate/poly(ethylene oxide) bicomponent fibers. Carbohydr. Polym. 2008, 71, 196–207. [Google Scholar] [CrossRef]
- Kalogiannis, C.G.; Michailof, C.M.; Panayiotou, C.G. Microencapsulation of Amoxicillin in Poly(L-lactic acid) by Supercritical Antisolvent Precipitation. Ind. Eng. Chem. Res. 2006, 45, 8738–8743. [Google Scholar] [CrossRef]
- Xu, Q.; Czernuszka, J.T. Controlled release of amoxicillin from hydroxyapatite-coated poly(lactic-co-glycolic acid) microspheres. J. Control. Release 2008, 127, 146–153. [Google Scholar] [CrossRef]
- Uhlen, M.; Guss, B.; Nilsson, B.; Gatenbeck, S.; Philipson, L.; Lindberg, M. Complete sequence of the staphylococcal gene encoding protein A. A gene evolved through multiple duplications. J. Biol. Chem. 1984, 259, 1695–1702. [Google Scholar] [CrossRef]
- Moks, T.; Abrahmsen, L.; Nilsson, B.; Hellman, U.; Sjoquist, J.; Uhlen, M. Staphylococcal protein A consists of five IgG-binding domains. Eur. J. Biochem. 1986, 156, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Tserki, V.; Philippou, J.; Panayiotou, C. Preparation and characterization of electrospun poly(butylene succinate-co-butylene adipate) nanofibrous nonwoven mats. Proc. Inst. Mech. Eng. Part N J. Nanoeng. Nanosyst. 2006, 220, 71–79. [Google Scholar] [CrossRef]
- Beckett, D.; Kovaleva, E.; Schatz, P.J. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein. Sci. 1999, 8, 921–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metzger, S.W.; Natesan, M.; Yanavich, C.; Schneider, J.; Lee, G.U. Development and characterization of surface chemistries for microfabricated biosensors. J. Vac. Sci. Technol. A 1999, 17, 2623–2628. [Google Scholar] [CrossRef]
- Katranidis, A.; Melachroinos, A.; Karagiannidis, P.G.; Lousinian, S.; Papadopoulos, G.; Logothetidis, S.; Choli-Papadopoulou, T. Biofunctionalization of PET/SiO2 surfaces for single molecule experiments and medical applications. Nano 2011, 6, 271–277. [Google Scholar] [CrossRef]
- Groll, J.; Amirgoulova, E.V.; Ameringer, T.; Heyes, C.D.; Röcker, C.; Nienhaus, G.U.; Möller, M. Biofunctionalized, ultrathin coatings of cross-linked star-shaped poly(ethylene oxide) allow reversible folding of immobilized proteins. J. Am. Chem. Soc. 2004, 126, 4234–4239. [Google Scholar] [CrossRef]
- Katranidis, A.; Atta, D.; Schlesinger, R.; Nierhaus, K.H.; Choli-Papadopoulou, T.; Gregor, I.; Gerrits, M.; Büldt, G.; Fitter, J. Fast Biosynthesis of GFP Molecules: A Single-Molecule Fluorescence Study. Angew. Chem. Int. Ed. 2009, 48, 1758–1761. [Google Scholar] [CrossRef]
- Kroger, D.; Hucho, F.; Vogel, H. Ligand binding to nicotinic acetylcholine receptor investigated by surface plasmon resonance. Anal. Chem. 1999, 71, 3157–3165. [Google Scholar] [CrossRef]
- Kroger, D.; Katerkamp, A.; Renneberg, R.; Cammann, K. Surface investigations on the development of a direct optical immunosensor. Biosens. Bioelectron. 1998, 13, 1141–1147. [Google Scholar] [PubMed]
- Mansfeld, J.; Ulbrich-Hofmann, R. Site-specific and random immobilization of thermolysin-like proteases reflected in the thermal inactivation kinetics. Biotechnol. Appl. Biochem. 2000, 32, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Gooding, J.J.; Hibbert, D.B.; Gooding, J.J.; Hibbert, D.B. The application of alkanethiol self-assembled monolayers to enzyme electrodes. Trends Anal. Chem. 1999, 18, 525–533. [Google Scholar] [CrossRef]
- Butterfield, D.A.; Bhattacharyya, D.; Daunert, S.; Bachas, L. Catalytic biofunctional membranes containing site-specifically immobilized enzyme arrays: A review. J. Membr. Sci. 2001, 181, 29–37. [Google Scholar] [CrossRef]
- Zhu, H.; Bilgin, M.; Bangham, R.; Hall, D.; Casamayor, A.; Bertone, P.; Lan, N.; Jansen, R.; Bidlingmaier, S.; Houfek, T.; et al. Global analysis of protein activities using proteome chips. Science 2001, 293, 2101–2105. [Google Scholar] [CrossRef]
- Mitchell, P. A perspective on protein microarrays. Nat. Biotechnol. 2002, 20, 225–229. [Google Scholar] [CrossRef]
- Wilson, D.S.; Nock, S. Functional protein microarrays. Curr. Opin. Chem. Biol. 2002, 6, 81–85. [Google Scholar] [CrossRef]
- Norde, W. Adsorption of proteins from solution at the solid-liquid interface. Adv. Colloid Interface Sci. 1986, 25, 267–340. [Google Scholar] [CrossRef]
- Hogt, A.H.; Dankert, J.; de Vries, J.A.; Feijen, J. Adhesion of coagulase-negative staphylococci to biomaterials. J. Gen. Microbiol. 1983, 129, 2959–2968. [Google Scholar] [CrossRef] [Green Version]
- Cardile, A.P.; Sanchez, C.J., Jr.; Samberg, M.E.; Romano, D.R.; Hardy, S.K.; Wenke, J.C.; Murray, C.K.; Akers, K.S. Human plasma enhances the expression of Staphylococcal microbial surface components recognizing adhesive matrix molecules promoting biofilm formation and increases antimicrobial tolerance In Vitro. BMC Res. Notes 2004, 7, 457. [Google Scholar] [CrossRef] [Green Version]
- Whimbey, E.; Kiehn, T.E.; Brannon, P.; Benezra, D.; Armstrong, D. Clinical significance of colony counts in immunocompromised patients with Staphylococcus aureus bacteremia. J. Infect. Dis. 1987, 155, 1328–1330. [Google Scholar] [CrossRef] [PubMed]
- O’Seaghdha, M.; Van Schooten, C.J.; Kerrigan, S.W.; Emsley, J.; Silverman, G.; Cox, D.; Lenting, P.; Foster, T.J. Staphylococcus aureus protein A binding to von Willebrand factor A1 domain is mediated by conserved IgG binding regions. FEBS J. 2006, 273, 4831–4841. [Google Scholar] [CrossRef] [PubMed]




| 7 kV | 15 kV | 22 kV | |
|---|---|---|---|
| 1:0.5 | 0.07 | 0.06 | - |
| 1:1 | 0.06 | 0.16 | 0.03 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pendas, S.; Asiminas, A.; Katranidis, A.; Tsioptsias, C.; Pitou, M.; Papadopoulos, G.; Choli-Papadopoulou, T. SpAD Biofunctionalized Cellulose Acetate Scaffolds Inhibit Staphylococcus aureus Adherence in a Coordinating Function with the von Willebrand A1 Domain (vWF A1). J. Funct. Biomater. 2022, 13, 21. https://doi.org/10.3390/jfb13010021
Pendas S, Asiminas A, Katranidis A, Tsioptsias C, Pitou M, Papadopoulos G, Choli-Papadopoulou T. SpAD Biofunctionalized Cellulose Acetate Scaffolds Inhibit Staphylococcus aureus Adherence in a Coordinating Function with the von Willebrand A1 Domain (vWF A1). Journal of Functional Biomaterials. 2022; 13(1):21. https://doi.org/10.3390/jfb13010021
Chicago/Turabian StylePendas, Stefanos, Antonis Asiminas, Alexandros Katranidis, Costas Tsioptsias, Maria Pitou, Georgios Papadopoulos, and Theodora Choli-Papadopoulou. 2022. "SpAD Biofunctionalized Cellulose Acetate Scaffolds Inhibit Staphylococcus aureus Adherence in a Coordinating Function with the von Willebrand A1 Domain (vWF A1)" Journal of Functional Biomaterials 13, no. 1: 21. https://doi.org/10.3390/jfb13010021






