HexA-Enzyme Coated Polymer Nanoparticles for the Development of a Drug-Delivery System in the Treatment of Sandhoff Lysosomal Storage Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of PLA Nanoparticles (NPs)
2.3. Immobilization of β-d-N-Acetyl-Hexosaminidase A
2.4. SEM and DLS Analysis
2.5. β-d-N-Acetyl-Hexosaminidase A Enzyme Assay
2.6. Biochemical Characterization of HexA-NPs
2.7. Sandhoff Cell Culture and HexA-NP Treatment
2.8. Immunofluorescence Assay
2.9. GM3 Analysis by Quadrupole Time-of-Flight Liquid Chromatography/Mass Spectrometry (Q-TOF LC/MS)
2.10. Statistical Analysis
3. Results and Discussion
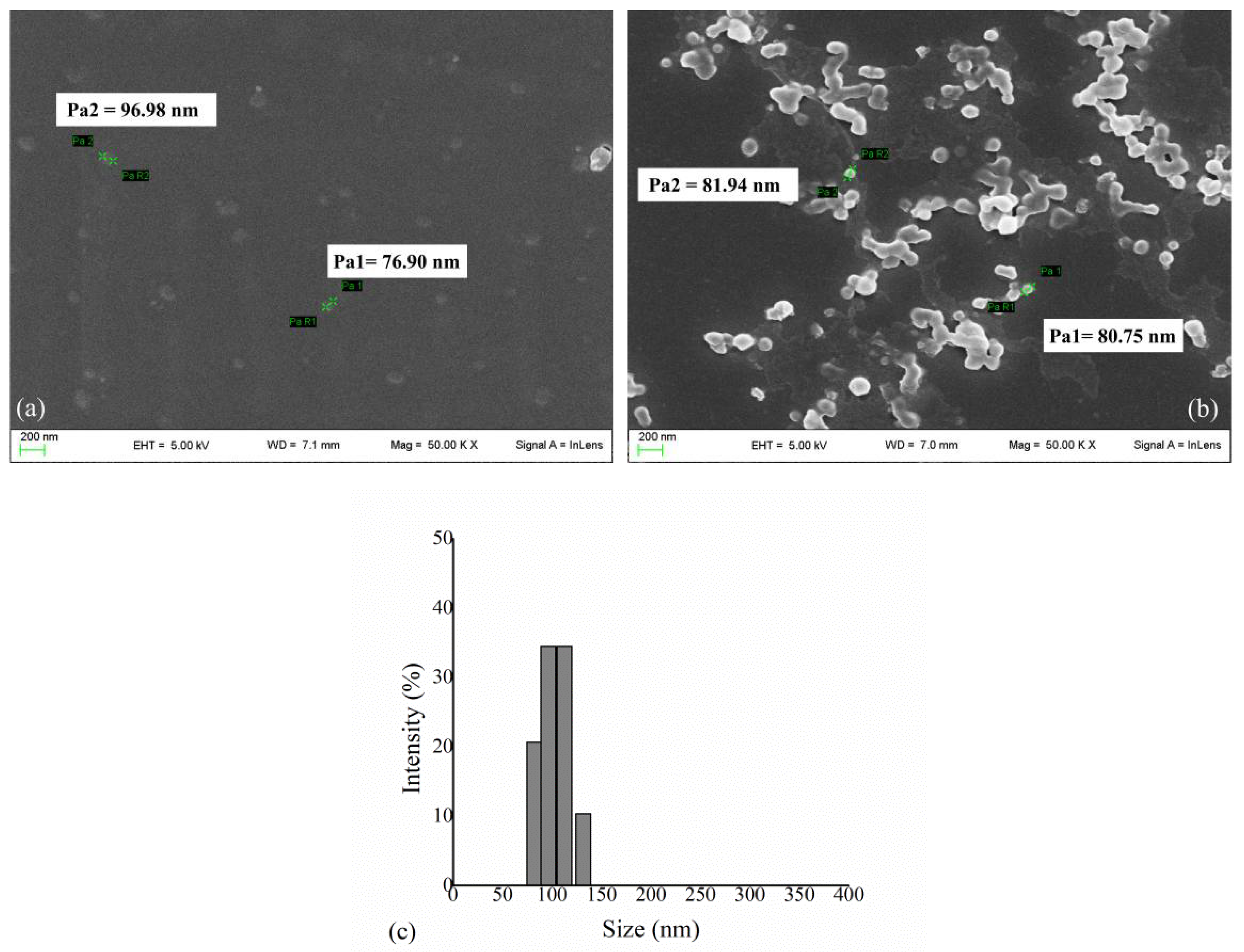
3.1. HexA Immobilization on PLA NPs and Morphological Analysis
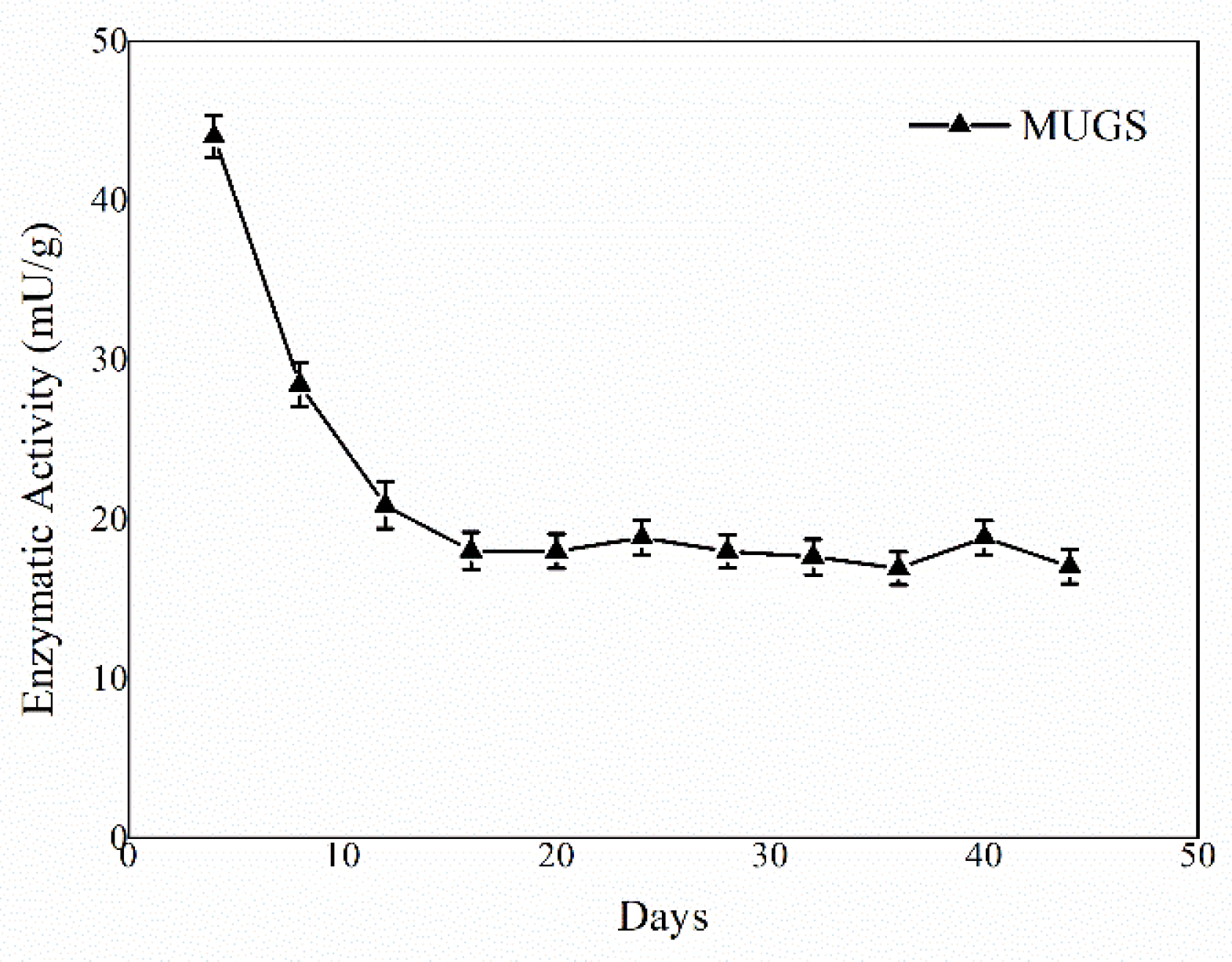
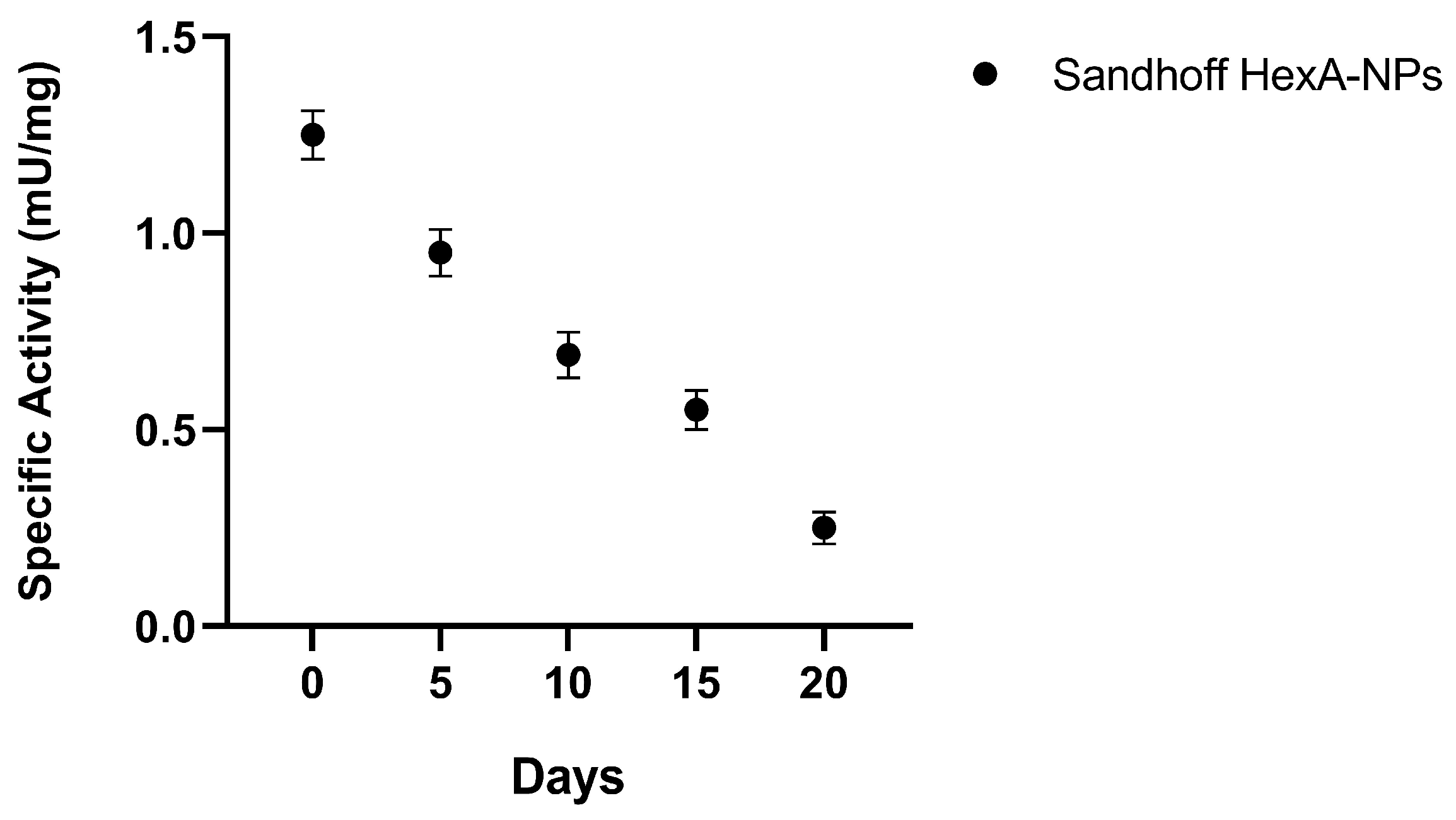
3.2. HexA-NP Stability
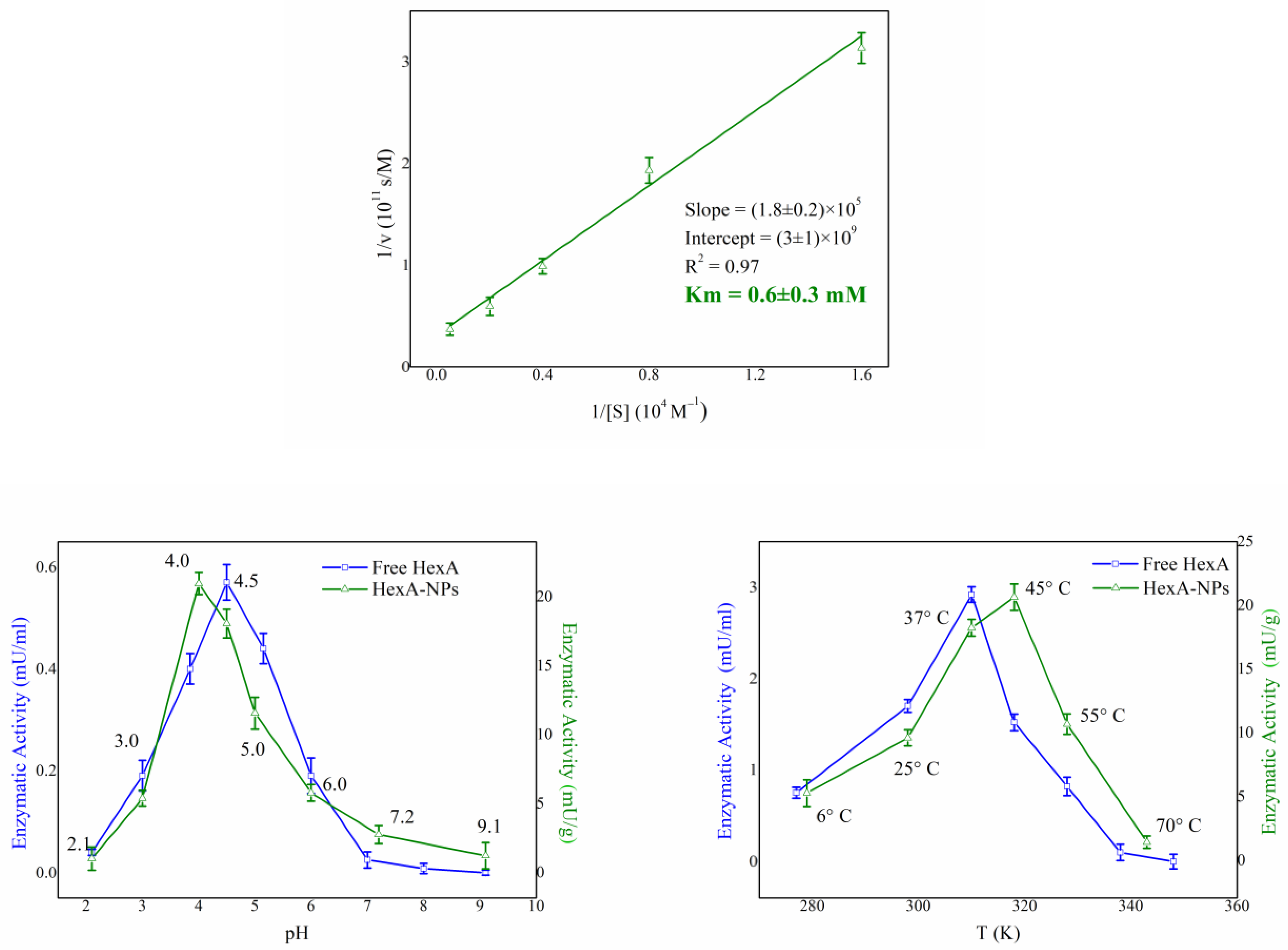
3.3. Biochemical Characterization of HexA-NPs
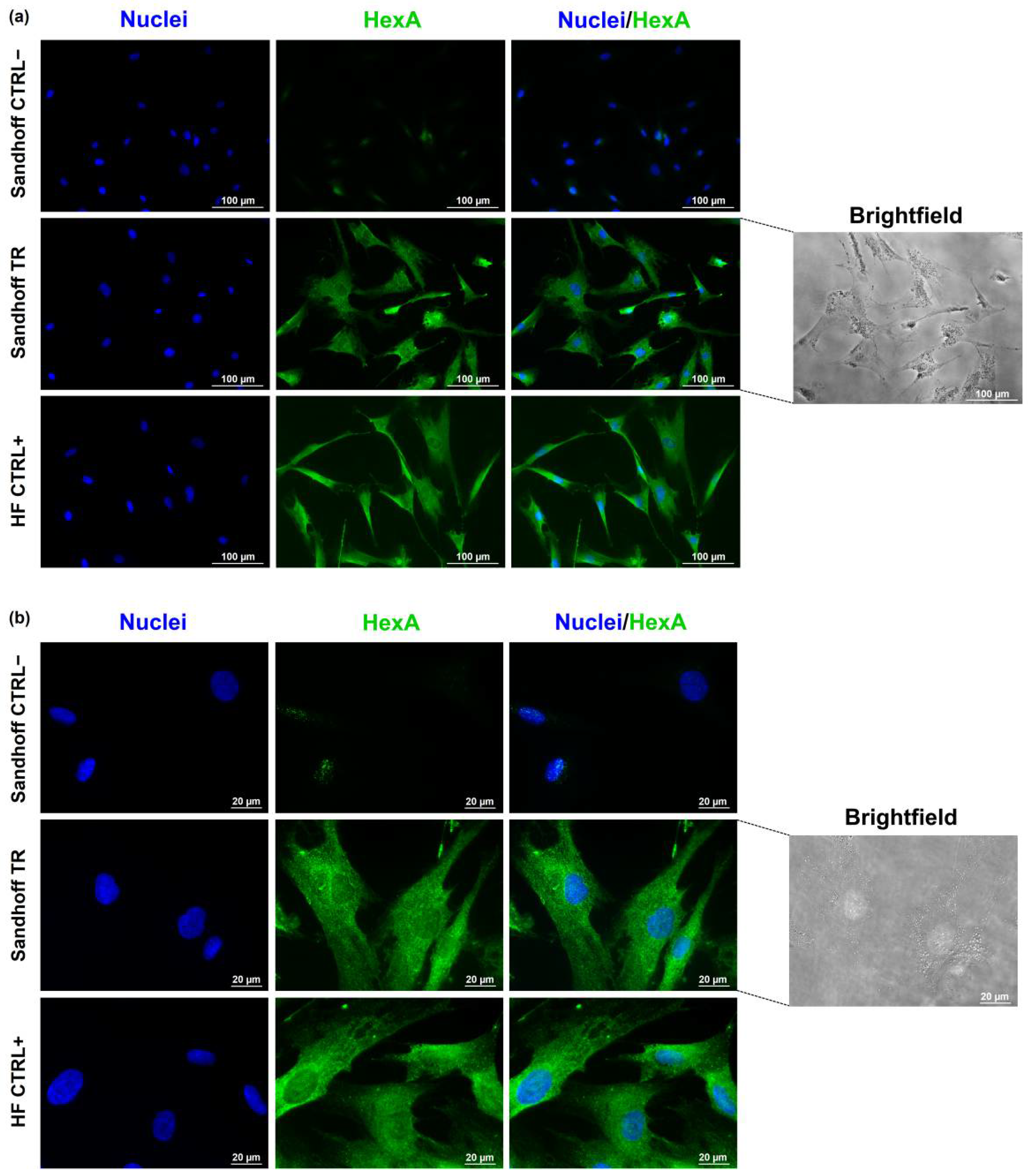
3.4. Immunofluorescence Assay
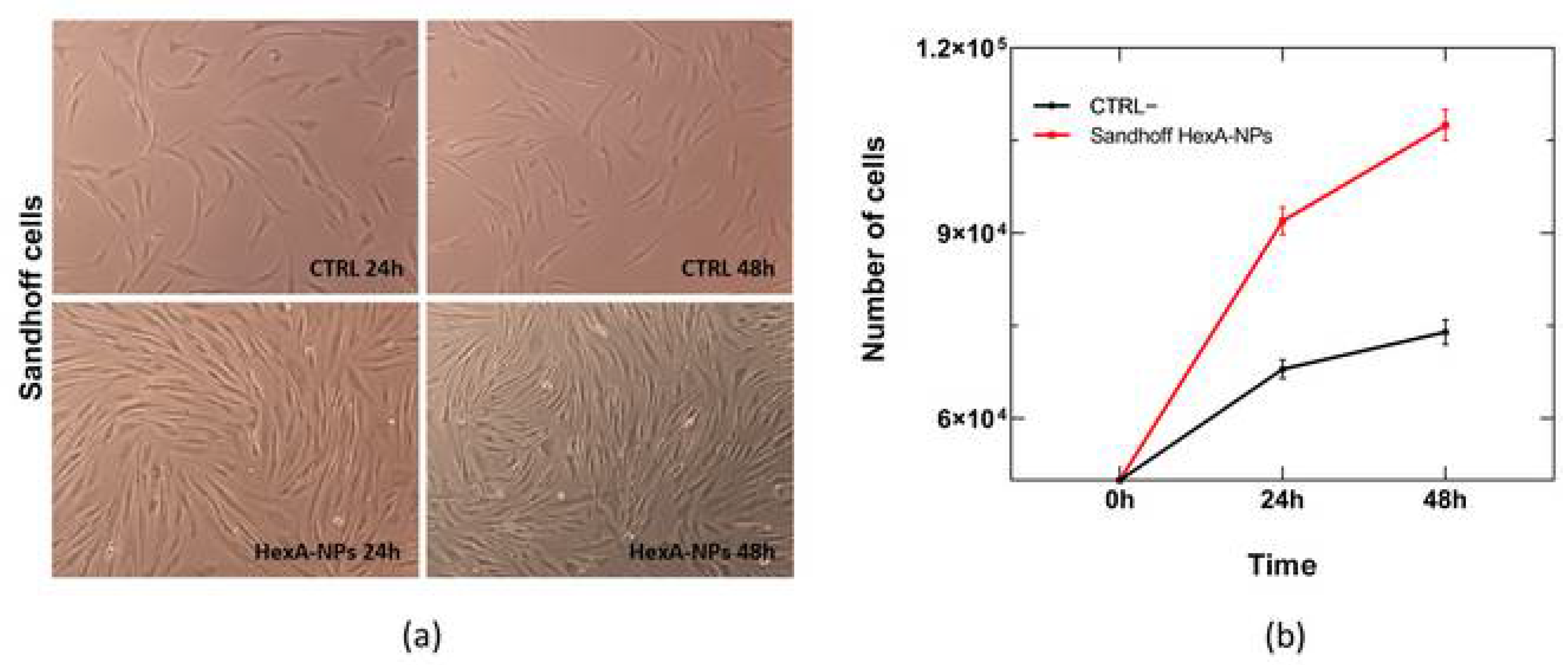
3.5. In Vitro Tests with HexA-NPs
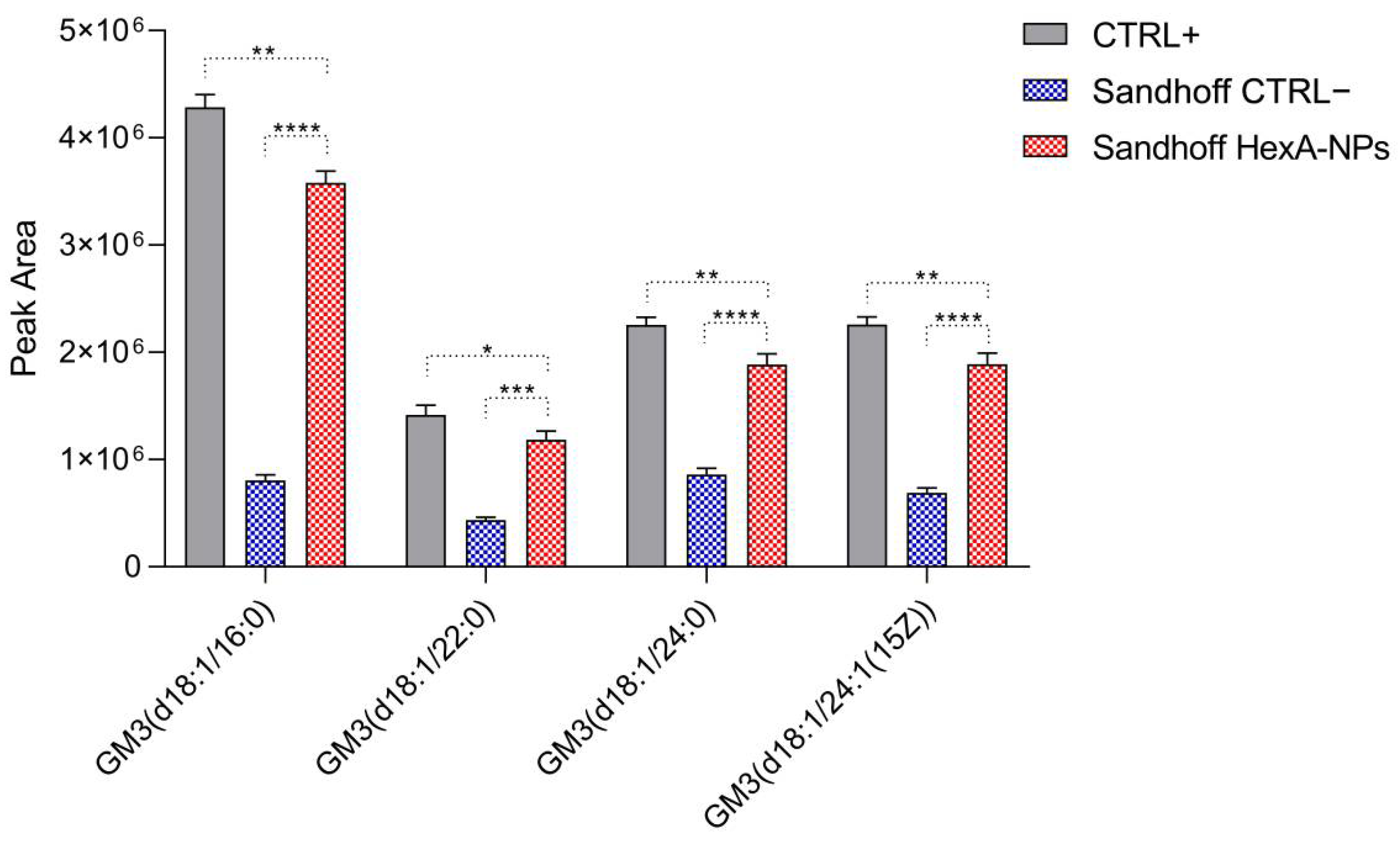
3.6. GM3 Analysis by Quadrupole Time-of-Flight Liquid Chromatography/Mass Spectrometry (Q-TOF LC/MS)
4. Conclusions
5. Patents
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Martino, S.; Marconi, P.C.R.; Tancini, B.; Dolcetta, D.; De Angelis, M.C.; Montanucci, P.; Bregola, G.; Sandhoff, K.; Bordignon, C.; Emiliani, C.; et al. A direct gene transfer strategy via brain internal capsule reverses the biochemical defect in Tay–Sachs disease. Hum. Mol. Genet. 2005, 14, 2113–2123. [Google Scholar] [CrossRef] [PubMed]
- Arfi, A.; Bourgoin, C.; Basso, L.; Emiliani, C.; Tancini, B.; Chigorno, V.; Li, Y.-T.; Orlacchio, A.; Poenaru, L.; Sonnino, S.; et al. Bicistronic lentiviral vector corrects β-hexosaminidase deficiency in transduced and cross-corrected human Sandhoff fibroblasts. Neurobiol. Dis. 2005, 20, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Li, Y.; Nikolaishvili-Feinberg, N.; Scesa, G.; Bi, Y.; Pan, D.; Moore, D.; Bongarzone, E.R.; Sands, M.; Miller, C.; et al. Hematopoietic Stem cell transplantation and lentiviral vector-based gene therapy for Krabbe’s disease: Present convictions and future prospects. J. Neurosci. Res. 2016, 94, 1152–1168. [Google Scholar] [CrossRef] [PubMed]
- Biffi, A. Hematopoietic Stem Cell Gene Therapy for Storage Disease: Current and New Indications. Mol. Ther. 2017, 25, 1155–1162. [Google Scholar] [CrossRef] [Green Version]
- Ohashi, T. Enzyme replacement therapy for lysosomal storage diseases. Pediatr. Endocrinol. Rev. 2012, 10, 26–34. [Google Scholar]
- Muro, S. Strategies for delivery of therapeutics into the central nervous system for treatment of lysosomal storage disorders. Drug Deliv. Transl. Res. 2012, 2, 169–186. [Google Scholar] [CrossRef] [Green Version]
- de Garibay, A.P.R.; Solinís, M.; Rodríguez-Gascón, A. Gene Therapy for Fabry Disease: A Review of the Literature. BioDrugs 2013, 27, 237–246. [Google Scholar] [CrossRef]
- Calzoni, E.; Argentati, C.; Cesaretti, A.; Montegiove, N.; Tortorella, I.; Bazzucchi, M.; Morena, F.; Martino, S.; Emiliani, C. RNA Modifications in Neurodegenerations. In Epitranscriptomics; Jurga, S., Barciszewski, J., Eds.; RNA Technologies; Springer International Publishing: Cham, Switzerland, 2021; pp. 23–77. ISBN 978-3-030-71612-7. [Google Scholar]
- Pastores, G.M. Recombinant Glucocerebrosidase (Imiglucerase) as a Therapy for Gaucher Disease. BioDrugs 2010, 24, 41–47. [Google Scholar] [CrossRef]
- McCafferty, E.H.; Scott, L.J. Vestronidase Alfa: A Review in Mucopolysaccharidosis VII. BioDrugs 2019, 33, 233–240. [Google Scholar] [CrossRef] [Green Version]
- Cunha, A.; Gaubert, A.; Latxague, L.; Dehay, B. PLGA-Based Nanoparticles for Neuroprotective Drug Delivery in Neurodegenerative Diseases. Pharmaceutics 2021, 13, 1042. [Google Scholar] [CrossRef]
- Calzoni, E.; Cesaretti, A.; Polchi, A.; Di Michele, A.; Tancini, B.; Emiliani, C. Biocompatible Polymer Nanoparticles for Drug Delivery Applications in Cancer and Neurodegenerative Disorder Therapies. J. Funct. Biomater. 2019, 10, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuster, T.; Mühlstein, A.; Yaghootfam, C.; Maksimenko, O.; Shipulo, E.; Gelperina, S.; Kreuter, J.; Gieselmann, V.; Matzner, U. Potential of surfactant-coated nanoparticles to improve brain delivery of arylsulfatase A. J. Control. Release 2017, 253, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Edelmann, M.J.; Maegawa, G.H.B. CNS-Targeting Therapies for Lysosomal Storage Diseases: Current Advances and Challenges. Front. Mol. Biosci. 2020, 7, 559804. [Google Scholar] [CrossRef] [PubMed]
- Del Grosso, A.; Galliani, M.; Angella, L.; Santi, M.; Tonazzini, I.; Parlanti, G.; Signore, G.; Cecchini, M. Brain-targeted enzyme-loaded nanoparticles: A breach through the blood-brain barrier for enzyme replacement therapy in Krabbe disease. Sci. Adv. 2019, 5, eaax7462. [Google Scholar] [CrossRef] [Green Version]
- Naqvi, S.; Panghal, A.; Flora, S.J.S. Nanotechnology: A Promising Approach for Delivery of Neuroprotective Drugs. Front. Neurosci. 2020, 14, 494. [Google Scholar] [CrossRef]
- Brambilla, D.; Le Droumaguet, B.; Nicolas, J.; Hashemi, S.H.; Wu, L.; Moghimi, S.M.; Couvreur, P.; Andrieux, K. Nanotechnologies for Alzheimer’s disease: Diagnosis, therapy, and safety issues. Nanomed. Nanotechnol. Biol. Med. 2011, 7, 521–540. [Google Scholar] [CrossRef]
- Stanimirovic, D.B.; Sandhu, J.K.; Costain, W.J. Emerging Technologies for Delivery of Biotherapeutics and Gene Therapy Across the Blood–Brain Barrier. BioDrugs 2018, 32, 547–559. [Google Scholar] [CrossRef] [Green Version]
- Ferrari, M.; Downing, G. Medical Nanotechnology. BioDrugs 2005, 19, 203–210. [Google Scholar] [CrossRef]
- Wells, C.M.; Harris, M.; Choi, L.; Murali, V.P.; Guerra, F.D.; Jennings, J.A. Stimuli-responsive drug release from smart polymers. J. Funct. Biomater. 2019, 10, 34. [Google Scholar] [CrossRef] [Green Version]
- Torchilin, V.P. Drug targeting. Eur. J. Pharm. Sci. 2000, 11, S81–S91. [Google Scholar] [CrossRef]
- Brunella, T.; Giovanni, T.; Barbara, B.; Diego, D.; Alessandro, M.; Eleonora, D.M.; Lorena, U.; Barbara, R.; Flavio, F.; Carla, E.; et al. Use of Polylactide-Co-Glycolide-Nanoparticles for Lysosomal Delivery of a Therapeutic Enzyme in Glycogenosis Type II Fibroblasts. J. Nanosci. Nanotechnol. 2015, 15, 2657–2666. [Google Scholar] [CrossRef] [PubMed]
- Boddu, S.H.S.; Bhagav, P.; Karla, P.K.; Jacob, S.; Adatiya, M.D.; Dhameliya, T.M.; Ranch, K.M.; Tiwari, A.K. Polyamide/Poly(Amino Acid) Polymers for Drug Delivery. J. Funct. Biomater. 2021, 12, 58. [Google Scholar] [CrossRef] [PubMed]
- Farahani, A.; Zarei-Hanzaki, A.; Abedi, H.R.; Tayebi, L.; Mostafavi, E. Polylactic Acid Piezo-Biopolymers: Chemistry, Structural Evolution, Fabrication Methods, and Tissue Engineering Applications. J. Funct. Biomater. 2021, 12, 71. [Google Scholar] [CrossRef] [PubMed]
- Hill, M.; Cunningham, R.N.; Hathout, R.M.; Johnston, C.; Hardy, J.G.; Migaud, M.E. Formulation of Antimicrobial Tobramycin Loaded PLGA Nanoparticles via Complexation with AOT. J. Funct. Biomater. 2019, 10, 26. [Google Scholar] [CrossRef] [Green Version]
- Kost, B.; Gonciarz, W.; Krupa, A.; Socka, M.; Rogala, M.; Biela, T.; Brzeziński, M. pH-tunable nanoparticles composed of copolymers of lactide and allyl-glycidyl ether with various functionalities for the efficient delivery of anti-cancer drugs. Colloids Surfaces B Biointerfaces 2021, 204, 111801. [Google Scholar] [CrossRef]
- Revdekar, A.; Shende, P. Block copolymers in Alzheimer’s disease therapy: A perceptive to revolutionize biomaterials. J. Control. Release 2021, 340, 271–281. [Google Scholar] [CrossRef]
- da Silva, D.; Kaduri, M.; Poley, M.; Adir, O.; Krinsky, N.; Shainsky-Roitman, J.; Schroeder, A. Biocompatibility, biodegradation and excretion of polylactic acid (PLA) in medical implants and theranostic systems. Chem. Eng. J. 2018, 340, 9–14. [Google Scholar] [CrossRef]
- Gaumet, M.; Vargas, A.; Gurny, R.; Delie, F. Nanoparticles for drug delivery: The need for precision in reporting particle size parameters. Eur. J. Pharm. Biopharm. 2008, 69, 1–9. [Google Scholar] [CrossRef]
- Khan, S.; Mansoor, S.; Rafi, Z.; Kumari, B.; Shoaib, A.; Saeed, M.; Alshehri, S.; Ghoneim, M.M.; Rahamathulla, M.; Hani, U.; et al. A review on nanotechnology: Properties, applications, and mechanistic insights of cellular uptake mechanisms. J. Mol. Liq. 2021, 348, 118008. [Google Scholar] [CrossRef]
- Kim, D.H.; Lee, H.S.; Kwon, T.-W.; Han, Y.-M.; Kang, N.-W.; Lee, M.Y.; Kim, D.-D.; Kim, M.G.; Lee, J.-Y. Single enzyme nanoparticle, an effective tool for enzyme replacement therapy. Arch. Pharmacal Res. 2020, 43, 1–21. [Google Scholar] [CrossRef]
- Rigon, L.; Salvalaio, M.; Pederzoli, F.; Legnini, E.; Duskey, J.T.; D’Avanzo, F.; De Filippis, C.; Ruozi, B.; Marin, O.; Vandelli, M.A.; et al. Targeting Brain Disease in MPSII: Preclinical Evaluation of IDS-Loaded PLGA Nanoparticles. Int. J. Mol. Sci. 2019, 20, 2014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tosi, G.; Duskey, J.T.; Kreuter, J. Nanoparticles as carriers for drug delivery of macromolecules across the blood-brain barrier. Expert Opin. Drug Deliv. 2019, 17, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Duskey, J.T.; Da Ros, F.; Ottonelli, I.; Zambelli, B.; Vandelli, M.A.; Tosi, G.; Ruozi, B. Enzyme Stability in Nanoparticle Preparations Part 1: Bovine Serum Albumin Improves Enzyme Function. Molecules 2020, 25, 4593. [Google Scholar] [CrossRef] [PubMed]
- Calzoni, E.; Cesaretti, A.; Montegiove, N.; Di Michele, A.; Emiliani, C. Enhanced Stability of Long-Living Immobilized Recombinant β-d-N-Acetyl-Hexosaminidase A on Polylactic Acid (PLA) Films for Potential Biomedical Applications. J. Funct. Biomater. 2021, 12, 32. [Google Scholar] [CrossRef] [PubMed]
- Sharifi, M.; Sohrabi, M.J.; Hosseinali, S.H.; Hasan, A.; Kani, P.H.; Talaei, A.J.; Karim, A.; Nanakali, N.M.Q.; Salihi, A.; Aziz, F.M.; et al. Enzyme immobilization onto the nanomaterials: Application in enzyme stability and prodrug-activated cancer therapy. Int. J. Biol. Macromol. 2019, 143, 665–676. [Google Scholar] [CrossRef]
- Sandhoff, K.; Harzer, K. Gangliosides and Gangliosidoses: Principles of Molecular and Metabolic Pathogenesis. J. Neurosci. 2013, 33, 10195–10208. [Google Scholar] [CrossRef]
- Ferreira, C.R.; Gahl, W.A. Lysosomal storage diseases. Transl. Sci. Rare Dis. 2017, 2, 1–71. [Google Scholar] [CrossRef] [Green Version]
- Sandhoff, K.; Andreae, U.; Jatzkewitz, H. Deficient hexosaminidase activity in an exceptional case of Tay-Sachs disease with additional storage of kidney globoside in visceral organs. Life Sci. 1968, 7, 283–288. [Google Scholar] [CrossRef]
- Emiliani, C.; Beccari, T.; Tabilio, A.; Orlacchio, A.; Hosseini, R.; Stirling, J.L. An enzyme with properties similar to those of β-N-acetylhexosaminidase S is expressed in the promyelocytic cell line HL-60. Biochem. J. 1990, 267, 111–117. [Google Scholar] [CrossRef]
- Lemieux, M.J.; Mark, B.; Cherney, M.M.; Withers, S.G.; Mahuran, D.J.; James, M.N. Crystallographic Structure of Human β-Hexosaminidase A: Interpretation of Tay-Sachs Mutations and Loss of GM2 Ganglioside Hydrolysis. J. Mol. Biol. 2006, 359, 913–929. [Google Scholar] [CrossRef] [Green Version]
- Solovyeva, V.V.; Shaimardanova, A.A.; Chulpanova, D.S.; Kitaeva, K.V.; Chakrabarti, L.; Rizvanov, A.A. New Approaches to Tay-Sachs Disease Therapy. Front. Physiol. 2018, 9, 1663. [Google Scholar] [CrossRef] [PubMed]
- Beck, M. New therapeutic options for lysosomal storage disorders: Enzyme replacement, small molecules and gene therapy. Qual. Life Res. 2006, 121, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Leal, A.F.; Espejo-Mojica, A.J.; Sánchez, O.F.; Ramírez, C.M.; Reyes, L.H.; Cruz, J.C.; Alméciga-Díaz, C.J. Lysosomal storage diseases: Current therapies and future alternatives. Klin. Wochenschr. 2020, 98, 931–946. [Google Scholar] [CrossRef] [PubMed]
- Calzoni, E.; Cesaretti, A.; Tacchi, S.; Caponi, S.; Pellegrino, R.M.; Luzi, F.; Cottone, F.; Fioretto, D.; Emiliani, C.; Di Michele, A. Covalent Immobilization of Proteases on Polylactic Acid for Proteins Hydrolysis and Waste Biomass Protein Content Valorization. Catalysts 2021, 11, 167. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Beccari, T.; Emiliani, C.; Hosseini, R.; Orlacchio, A.; Stirling, J.L. Intermediate forms of human β-N-acetylhexosaminidase lack activity towards 4-methylumbelliferyl β-N-acetylglucosaminide 6-sulphate. Biochem. J. 1987, 244, 801–804. [Google Scholar] [CrossRef]
- Tancini, B.; Magini, A.; Latterini, L.; Urbanelli, L.; Ciccarone, V.; Elisei, F.; Emiliani, C. Occurrence of an Anomalous Endocytic Compartment in Fibroblast from Sandhoff Disease Patients. Mol. Cell. Biochem. 2010, 335, 273–282. [Google Scholar] [CrossRef]
- Montalvo, A.L.E.; Filocamo, M.; Vlahoviček, K.; Dardis, A.; Lualdi, S.; Corsolini, F.; Bembi, B.; Pittis, M.G. Molecular analysis of theHEXAgene in Italian patients with infantile and late Onset Tay-Sachs disease: Detection of fourteen novel alleles. Hum. Mutat. 2005, 26, 282. [Google Scholar] [CrossRef]
- Zampieri, S.; Filocamo, M.; Buratti, E.; Stroppiano, M.; Vlahovicek, K.; Rosso, N.; Bignulin, E.; Regis, S.; Carnevale, F.; Bembi, B.; et al. Molecular and functional analysis of the HEXB gene in Italian patients affected with Sandhoff disease: Identification of six novel alleles. Neurogenetics 2008, 10, 49–58. [Google Scholar] [CrossRef]
- Morena, F.; Argentati, C.; Calzoni, E.; Cordellini, M.; Emiliani, C.; D’Angelo, F.A.; Martino, S. Ex-Vivo tissues engineering modeling for reconstructive surgery using human adult adipose stem cells and polymeric nanostructured matrix. Nanomaterials 2016, 6, 57. [Google Scholar] [CrossRef]
- Koelmel, J.P.; Li, X.; Stow, S.M.; Sartain, M.J.; Murali, A.; Kemperman, R.; Tsugawa, H.; Takahashi, M.; Vasiliou, V.; Bowden, J.A.; et al. Lipid Annotator: Towards Accurate Annotation in Non-Targeted Liquid Chromatography High-Resolution Tandem Mass Spectrometry (LC-HRMS/MS) Lipidomics Using a Rapid and User-Friendly Software. Metabolites 2020, 10, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pellegrino, R.M.; Di Veroli, A.; Valeri, A.; Goracci, L.; Cruciani, G. LC/MS lipid profiling from human serum: A new method for global lipid extraction. Anal. Bioanal. Chem. 2014, 406, 7937–7948. [Google Scholar] [CrossRef] [PubMed]
- Fleige, E.; Quadir, M.A.; Haag, R. Stimuli-responsive polymeric nanocarriers for the controlled transport of active compounds: Concepts and applications. Adv. Drug Deliv. Rev. 2012, 64, 866–884. [Google Scholar] [CrossRef] [PubMed]
- James, H.P.; John, R.; Alex, A.; Anoop, K. Smart polymers for the controlled delivery of drugs—A concise overview. Acta Pharm. Sin. B 2014, 4, 120–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parveen, S.; Misra, R.; Sahoo, S.K. Nanoparticles: A boon to drug delivery, therapeutics, diagnostics and imaging. Nanomed. Nanotechnol. Biol. Med. 2012, 8, 147–166. [Google Scholar] [CrossRef]
- Ulbrich, K.; Hola, K.; Šubr, V.; Bakandritsos, A.; Tuček, J.; Zbořil, R. Targeted Drug Delivery with Polymers and Magnetic Nanoparticles: Covalent and Noncovalent Approaches, Release Control, and Clinical Studies. Chem. Rev. 2016, 116, 5338–5431. [Google Scholar] [CrossRef]
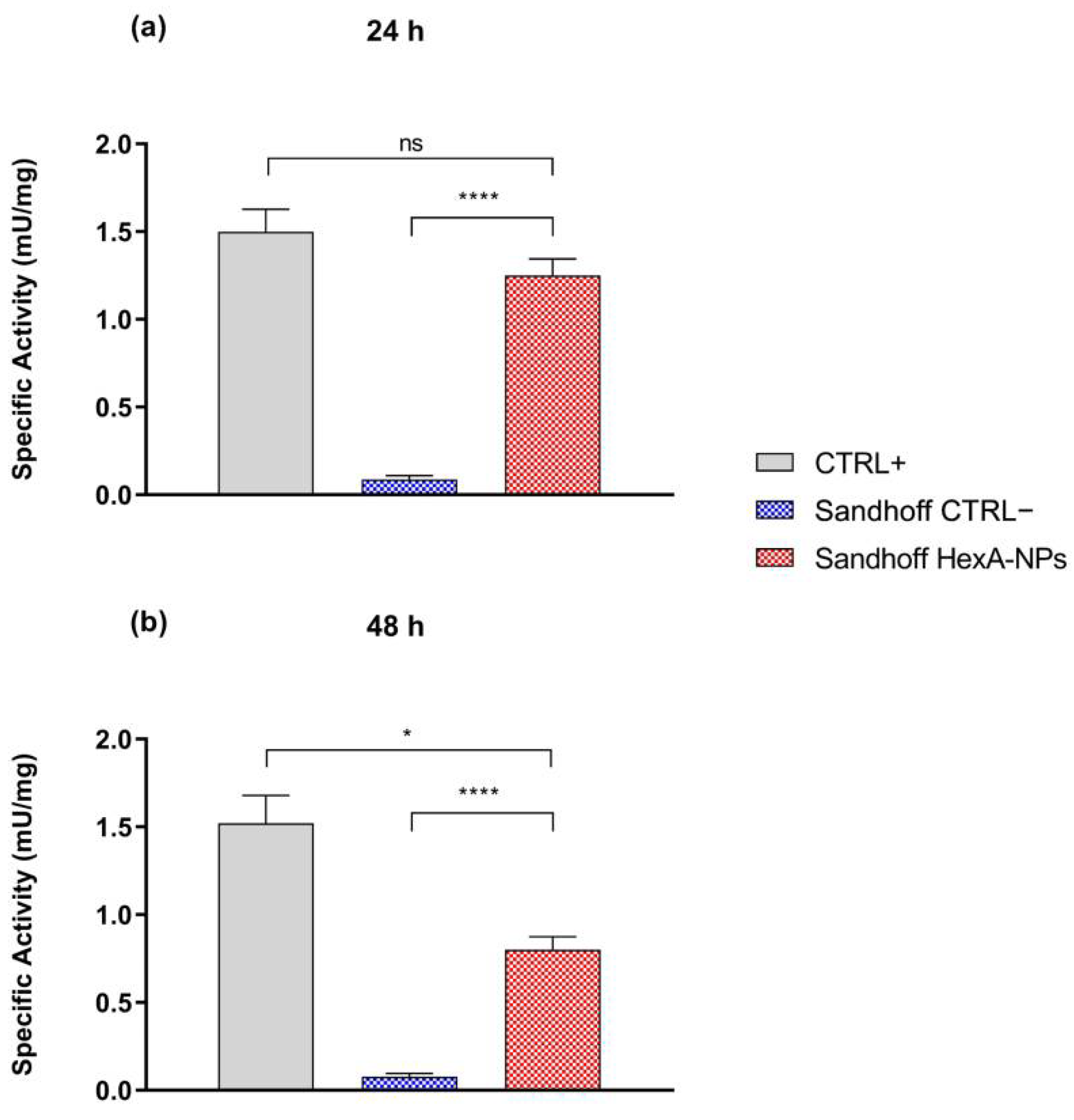
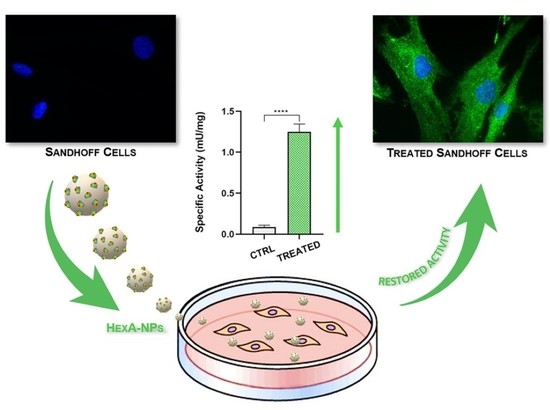
| Specific Activity (mU/mg) | CTRL+ | CTRL− | Sandhoff HexA-NPs |
|---|---|---|---|
| 24 h | 1.5 ± 0.1 | 0.09 ± 0.02 | 1.25 ± 0.09 |
| 48 h | 1.5 ± 0.2 | 0.08 ± 0.02 | 0.85 ± 0.07 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calzoni, E.; Cesaretti, A.; Montegiove, N.; Di Michele, A.; Pellegrino, R.M.; Emiliani, C. HexA-Enzyme Coated Polymer Nanoparticles for the Development of a Drug-Delivery System in the Treatment of Sandhoff Lysosomal Storage Disease. J. Funct. Biomater. 2022, 13, 37. https://doi.org/10.3390/jfb13020037
Calzoni E, Cesaretti A, Montegiove N, Di Michele A, Pellegrino RM, Emiliani C. HexA-Enzyme Coated Polymer Nanoparticles for the Development of a Drug-Delivery System in the Treatment of Sandhoff Lysosomal Storage Disease. Journal of Functional Biomaterials. 2022; 13(2):37. https://doi.org/10.3390/jfb13020037
Chicago/Turabian StyleCalzoni, Eleonora, Alessio Cesaretti, Nicolò Montegiove, Alessandro Di Michele, Roberto Maria Pellegrino, and Carla Emiliani. 2022. "HexA-Enzyme Coated Polymer Nanoparticles for the Development of a Drug-Delivery System in the Treatment of Sandhoff Lysosomal Storage Disease" Journal of Functional Biomaterials 13, no. 2: 37. https://doi.org/10.3390/jfb13020037
APA StyleCalzoni, E., Cesaretti, A., Montegiove, N., Di Michele, A., Pellegrino, R. M., & Emiliani, C. (2022). HexA-Enzyme Coated Polymer Nanoparticles for the Development of a Drug-Delivery System in the Treatment of Sandhoff Lysosomal Storage Disease. Journal of Functional Biomaterials, 13(2), 37. https://doi.org/10.3390/jfb13020037