Evaluation of the Cytotoxicity of Cationic Polymers on Glioblastoma Cancer Stem Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Storage of Compounds
2.2. Cell Culture and Treatment Conditions
2.3. Cell Viability Assay
2.4. Synthesis of 22 kDa L-PEI and Rhodamine–PEI Conjugate
2.5. Cationic Polymer Affinity Assay
2.5.1. U87 and NCH421K Cells
2.5.2. CHO-K1 and pgsA-745 Cells
2.6. Heparan Sulfate Expression
3. Results and Discussion
3.1. GSC Specific Toxicity Is Not a General Characteristic of Polycations
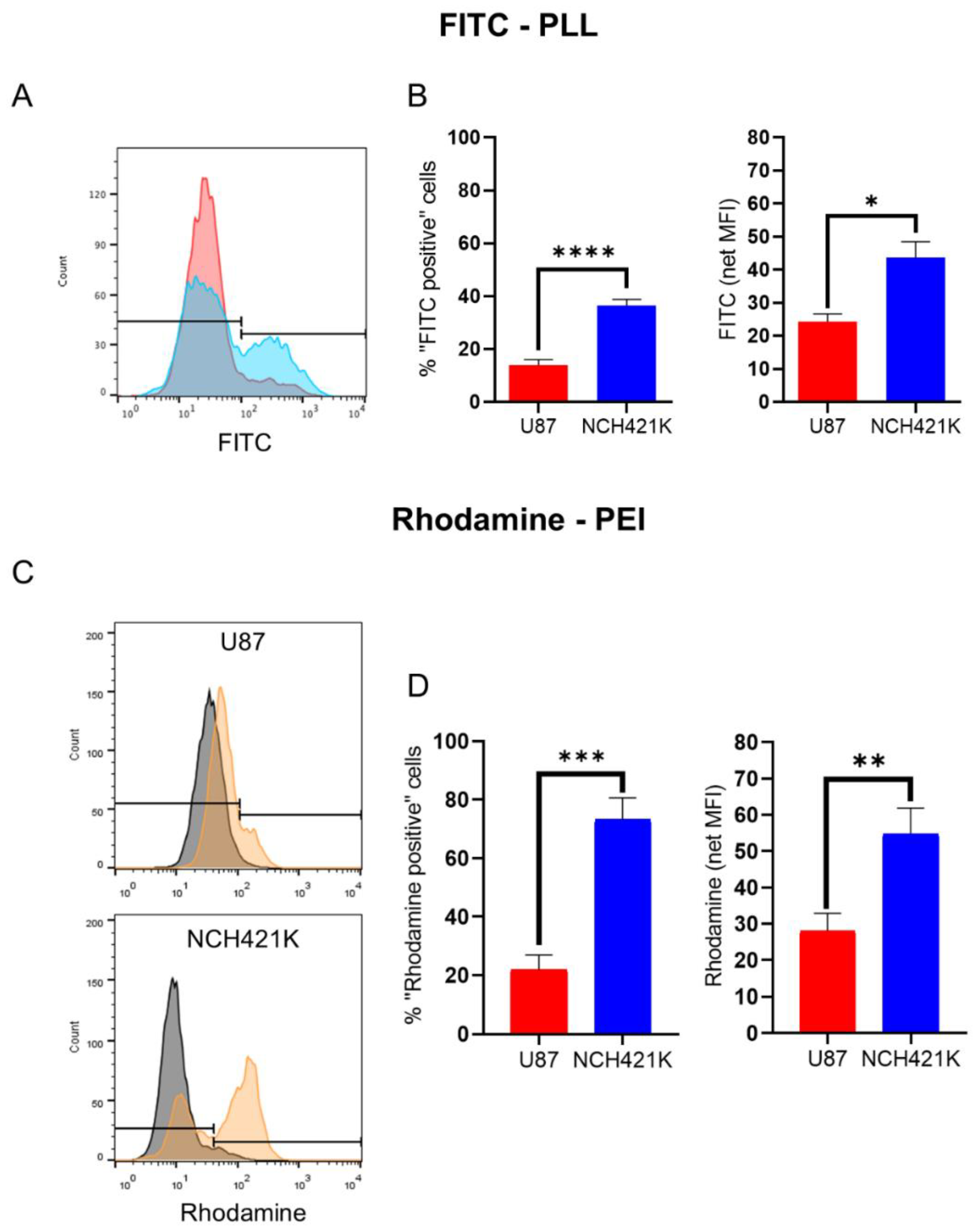
3.2. GSC Polycation Affinity Is Not Linked to Heparan Sulfate Expression
3.3. GSC Polycation Toxicity Is Size-Dependent
4. Conclusions and Outlook
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| B-PEI | Branched PEI |
| CSC | Cancer stem cell |
| DMEM | Dulbecco’s Modified Eagle Medium |
| DPBS | Dulbecco’s phosphate-buffered saline |
| FBS | Fetal bovine serum |
| GSC | Glioblastoma stem cell |
| L-PEI | Linear PEI |
| MFI | Median/Geometric mean fluorescence intensity |
| PDC | Polymer–drug conjugate |
| PEI | Polyethylenimine |
| PLL | Poly-L-lysine |
| RLU | Relative luminescence unit |
| RPMI | Roswell Park Memorial Institute |
References
- Moore, C.; Gao, W.; Fatehi, P. Cationic Lignin Polymers as Flocculant for Municipal Wastewater. Polymers 2021, 13, 3871. [Google Scholar] [CrossRef] [PubMed]
- Boussif, O.; Lezoualc’h, F.; Zanta, M.A.; Mergny, M.D.; Scherman, D.; Demeneix, B.; Behr, J.P. A Versatile Vector for Gene and Oligonucleotide Transfer into Cells in Culture and in Vivo: Polyethylenimine. Proc. Natl. Acad. Sci. USA 1995, 92, 7297–7301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samal, S.K.; Dash, M.; Van Vlierberghe, S.; Kaplan, D.L.; Chiellini, E.; van Blitterswijk, C.; Moroni, L.; Dubruel, P. Cationic Polymers and Their Therapeutic Potential. Chem. Soc. Rev. 2012, 41, 7147–7194. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Chen, W.C.W.; Heo, Y.; Wang, Y. Polycations and Their Biomedical Applications. Prog. Polym. Sci. 2016, 60, 18–50. [Google Scholar] [CrossRef]
- Moreau, E.; Domurado, M.; Chapon, P.; Vert, M.; Domurado, D. Biocompatibility of Polycations: In Vitro Agglutination and Lysis of Red Blood Cells and in Vivo Toxicity. J. Drug Target. 2002, 10, 161–173. [Google Scholar] [CrossRef]
- Richter, F.; Leer, K.; Martin, L.; Mapfumo, P.; Solomun, J.I.; Kuchenbrod, M.T.; Hoeppener, S.; Brendel, J.C.; Traeger, A. The Impact of Anionic Polymers on Gene Delivery: How Composition and Assembly Help Evading the Toxicity-Efficiency Dilemma. J. Nanobiotechnol. 2021, 19, 292. [Google Scholar] [CrossRef]
- Zhang, Q.F.; Wang, B.; Yin, D.X.; Zhang, J.; Wu, W.X.; Yu, Q.Y.; Yu, X.Q. Linear TACN-Based Cationic Polymers as Non-Viral Gene Vectors. RSC Adv. 2014, 4, 59164–59174. [Google Scholar] [CrossRef]
- Kheraldine, H.; Rachid, O.; Habib, A.M.; Al Moustafa, A.E.; Benter, I.F.; Akhtar, S. Emerging Innate Biological Properties of Nano-Drug Delivery Systems: A Focus on PAMAM Dendrimers and Their Clinical Potential. Adv. Drug Deliv. Rev. 2021, 178, 113908. [Google Scholar] [CrossRef]
- Jevprasesphant, R.; Penny, J.; Jalal, R.; Attwood, D.; McKeown, N.B.; D’Emanuele, A. The Influence of Surface Modification on the Cytotoxicity of PAMAM Dendrimers. Int. J. Pharm. 2003, 252, 263–266. [Google Scholar] [CrossRef]
- Chekkat, N.; Dahm, G.; Chardon, E.; Wantz, M.; Sitz, J.; Decossas, M.; Lambert, O.; Frisch, B.; Rubbiani, R.; Gasser, G.; et al. N-Heterocyclic Carbene-Polyethylenimine Platinum Complexes with Potent in Vitro and in Vivo Antitumor Efficacy. Bioconjug. Chem. 2016, 27, 1942–1948. [Google Scholar] [CrossRef]
- Wantz, M.; Bouché, M.; Dahm, G.; Chekkat, N.; Fournel, S.; Bellemin-Laponnaz, S. N-Heterocyclic Carbene-Polyethyleneimine (PEI) Platinum Complexes Inducing Human Cancer Cell Death: Polymer Carrier Impact. Int. J. Mol. Sci. 2018, 19, 3472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cherng, J.Y.; Van De Wetering, P.; Talsma, H.; Crommelin, D.J.A.; Hennink, W.E. Effect of Size and Serum Proteins on Transfection Efficiency of Poly ((2-Dimethylamino)Ethyl Methacrylate)-Plasmid Nanoparticles. Pharm. Res. 1996, 13, 1038–1042. [Google Scholar] [CrossRef]
- Ekladious, I.; Colson, Y.L.; Grinstaff, M.W. Polymer–Drug Conjugate Therapeutics: Advances, Insights and Prospects. Nat. Rev. Drug Discov. 2019, 18, 273–294. [Google Scholar] [CrossRef] [PubMed]
- Fahira, A.I.; Abdulah, R.; Barliana, M.I.; Gatera, V.A.; Amalia, R. Polyethyleneimine (PEI) as a Polymer-Based Co-Delivery System for Breast Cancer Therapy. Breast Cancer Targets Ther. 2022, 14, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Dufès, C.; Keith, W.N.; Bilsland, A.; Proutski, I.; Uchegbu, I.F.; Schätzlein, A.G. Synthetic Anticancer Gene Medicine Exploits Intrinsic Antitumor Activity of Cationic Vector to Cure Established Tumors. Cancer Res. 2005, 65, 8079–8084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, K.; Mikami, T.; Okawa, Y.; Tokoro, A.; Suzuki, S.; Suzuki, M. Antitumor Effect of Hexa-N-Acetylchitohexaose and Chitohexaose. Carbohydr. Res. 1986, 151, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Zou, P.; Yang, X.; Wang, J.; Li, Y.; Yu, H.; Zhang, Y.; Liu, G. Advances in Characterisation and Biological Activities of Chitosan and Chitosan Oligosaccharides. Food Chem. 2016, 190, 1174–1181. [Google Scholar] [CrossRef]
- He, W.; Liang, P.; Guo, G.; Huang, Z.; Niu, Y.; Dong, L.; Wang, C.; Zhang, J. Re-Polarizing Myeloid-Derived Suppressor Cells (MDSCs) with Cationic Polymers for Cancer Immunotherapy. Sci. Rep. 2016, 6, 24506. [Google Scholar] [CrossRef]
- Chen, B.; Le, W.; Wang, Y.; Li, Z.; Wang, D.; Ren, L.; Lin, L.; Cui, S.; Hu, J.J.; Hu, Y.; et al. Targeting Negative Surface Charges of Cancer Cells by Multifunctional Nanoprobes. Theranostics 2016, 6, 1887–1898. [Google Scholar] [CrossRef]
- Motomura, M.; Ichihara, H.; Matsumoto, Y. Nano-Chemotherapy Using Cationic Liposome That Strategically Targets the Cell Membrane Potential of Pancreatic Cancer Cells with Negative Charge. Bioorganic Med. Chem. Lett. 2018, 28, 1161–1165. [Google Scholar] [CrossRef]
- Li, Z.; Ruan, J.; Zhuang, X. Effective Capture of Circulating Tumor Cells from an S180-Bearing Mouse Model Using Electrically Charged Magnetic Nanoparticles. J. Nanobiotechnol. 2019, 17, 59. [Google Scholar] [CrossRef] [PubMed]
- Beyerle, A.; Irmler, M.; Beckers, J.; Kissel, T.; Stoeger, T. Toxicity Pathway Focused Gene Expression Profiling of PEI-Based Polymers for Pulmonary Applications. Mol. Pharm. 2010, 7, 727–737. [Google Scholar] [CrossRef]
- Fischer, D.; Li, Y.; Ahlemeyer, B.; Krieglstein, J.; Kissel, T. In Vitro Cytotoxicity Testing of Polycations: Influence of Polymer Structure on Cell Viability and Hemolysis. Biomaterials 2003, 24, 1121–1131. [Google Scholar] [CrossRef]
- Monnery, B.D.; Wright, M.; Cavill, R.; Hoogenboom, R.; Shaunak, S.; Steinke, J.H.G.; Thanou, M. Cytotoxicity of Polycations: Relationship of Molecular Weight and the Hydrolytic Theory of the Mechanism of Toxicity. Int. J. Pharm. 2017, 521, 249–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prabhakar, N.; Merisaari, J.; Le Joncour, V.; Peurla, M.; Karaman, D.Ş.; Casals, E.; Laakkonen, P.; Westermarck, J.; Rosenholm, J.M. Circumventing Drug Treatment? Intrinsic Lethal Effects of Polyethyleneimine (PEI)-Functionalized Nanoparticles on Glioblastoma Cells Cultured in Stem Cell Conditions. Cancers 2021, 13, 2631. [Google Scholar] [CrossRef]
- Knauer, N.; Arkhipova, V.; Li, G.; Hewera, M.; Pashkina, E.; Nguyen, P.-H.; Meschaninova, M.; Kozlov, V.; Zhang, W.; Croner, R.; et al. In Vitro Validation of the Therapeutic Potential of Dendrimer-Based Nanoformulations against Tumor Stem Cells. Int. J. Mol. Sci. 2022, 23, 5691. [Google Scholar] [CrossRef] [PubMed]
- Brown, Y.; Hua, S.; Tanwar, P.S. Extracellular Matrix-Mediated Regulation of Cancer Stem Cells and Chemoresistance. Int. J. Biochem. Cell Biol. 2019, 109, 90–104. [Google Scholar] [CrossRef] [PubMed]
- Hermann, P.C.; Huber, S.L.; Herrler, T.; Aicher, A.; Ellwart, J.W.; Guba, M.; Bruns, C.J.; Heeschen, C. Distinct Populations of Cancer Stem Cells Determine Tumor Growth and Metastatic Activity in Human Pancreatic Cancer. Cell Stem Cell 2007, 1, 313–323. [Google Scholar] [CrossRef] [Green Version]
- Najafi, M.; Mortezaee, K.; Majidpoor, J. Cancer Stem Cell (CSC) Resistance Drivers. Life Sci. 2019, 234, 116781. [Google Scholar] [CrossRef]
- Lathia, J.D.; Mack, S.C.; Mulkearns-Hubert, E.E.; Valentim, C.L.L.; Rich, J.N. Cancer Stem Cells in Glioblastoma. Genes Dev. 2015, 29, 1203–1217. [Google Scholar] [CrossRef]
- McCartin, C.; Dussouillez, C.; Bernhard, C.; Mathieu, E.; Blumberger, J.; Dontenwill, M.; Herold-Mende, C.; Idbaih, A.; Lavalle, P.; Bellemin-Laponnaz, S.; et al. Polyethylenimine, an Autophagy-Inducing Platinum-Carbene-Based Drug Carrier with Potent Toxicity towards Glioblastoma Cancer Stem Cells. Cancers 2022, 14, 5057. [Google Scholar] [CrossRef]
- Smith, A.G.; Macleod, K.F. Autophagy, Cancer Stem Cells and Drug Resistance. J. Pathol. 2019, 247, 708–718. [Google Scholar] [CrossRef] [Green Version]
- Esko, J.D.; Elgavish, A.; Prasthofer, T.; Taylor, W.H.; Weinke, J.L. Sulfate Transport-Deficient Mutants of Chinese Hamster Ovary Cells. Sulfation of Glycosaminoglycans Dependent on Cysteine. J. Biol. Chem. 1986, 261, 15725–15733. [Google Scholar] [CrossRef]
- Brissault, B.; Kichler, A.; Guis, C.; Leborgne, C.; Danos, O.; Cheradame, H. Synthesis of Linear Polyethylenimine Derivatives for DNA Transfection. Bioconjug. Chem. 2003, 14, 581–587. [Google Scholar] [CrossRef]
- Günay, K.A.; Ceccato, T.L.; Silver, J.S.; Bannister, K.L.; Bednarski, O.J.; Leinwand, L.A.; Anseth, K.S. PEG–Anthracene Hydrogels as an On-Demand Stiffening Matrix to Study Mechanobiology. Angew. Chem. Int. Ed. 2019, 58, 9912–9916. [Google Scholar] [CrossRef]
- Campos, B.; Wan, F.; Farhadi, M.; Ernst, A.; Zeppernick, F.; Tagscherer, K.E.; Ahmadi, R.; Lohr, J.; Dictus, C.; Gdynia, G.; et al. Differentiation Therapy Exerts Antitumor Effects on Stem-like Glioma Cells. Clin. Cancer Res. 2010, 16, 2715–2728. [Google Scholar] [CrossRef] [Green Version]
- Podergajs, N.; Brekka, N.; Radlwimmer, B.; Herold-Mende, C.; Talasila, K.M.; Tiemann, K.; Rajcevic, U.; Lah, T.T.; Bjerkvig, R.; Miletic, H. Expansive Growth of Two Glioblastoma Stem-like Cell Lines Is Mediated by BFGF and Not by EGF. Radiol. Oncol. 2013, 47, 330–337. [Google Scholar] [CrossRef]
- Francoia, J.-P.; Vial, L. Everything You Always Wanted to Know about Poly-l-Lysine Dendrigrafts (But Were Afraid to Ask). Chem. A Eur. J. 2018, 24, 2806–2814. [Google Scholar] [CrossRef] [Green Version]
- McCartin, C.; Mathieu, E.; Dontenwill, M.; Herold-Mende, C.; Idbaih, A.; Bonfiglio, A.; Mauro, M.; Fournel, S.; Kichler, A. An N-Heterocyclic Carbene Iridium(III) Complex as a Potent Anti-Cancer Stem Cell Therapeutic. Chem. Biol. Interact. 2022, 367, 110167. [Google Scholar] [CrossRef]
- Santos Franco, S.; Raveh-Amit, H.; Kobolák, J.; Alqahtani, M.H.; Mobasheri, A.; Dinnyes, A. The Crossroads between Cancer Stem Cells and Aging. BMC Cancer 2015, 15 (Suppl. 1), S1. [Google Scholar] [CrossRef]
- Weiswald, L.-B.; Bellet, D.; Dangles-Marie, V. Spherical Cancer Models in Tumor Biology. Neoplasia 2015, 17, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.K.; Clarke, I.D.; Terasaki, M.; Bonn, V.E.; Hawkins, C.; Squire, J.; Dirks, P.B. Identification of a Cancer Stem Cell in Human Brain Tumors. Cancer Res. 2003, 63, 5821–5828. [Google Scholar]
- Bishop, J.R.; Schuksz, M.; Esko, J.D. Heparan Sulphate Proteoglycans Fine-Tune Mammalian Physiology. Nature 2007, 446, 1030–1037. [Google Scholar] [CrossRef]
- Mislick, K.A.; Baldeschwieler, J.D. Evidence for the Role of Proteoglycans in Cation-Mediated Gene Transfer. Proc. Natl. Acad. Sci. USA 1996, 93, 12349–12354. [Google Scholar] [CrossRef] [Green Version]
- Poon, G.M.K.; Gariépy, J. Cell-Surface Proteoglycans as Molecular Portals for Cationic Peptide and Polymer Entry into Cells. Biochem. Soc. Trans. 2007, 35, 788–793. [Google Scholar] [CrossRef]
- Kopatz, I.; Remy, J.-S.; Behr, J.-P. A Model for Non-Viral Gene Delivery: Through Syndecan Adhesion Molecules and Powered by Actin. J. Gene Med. 2004, 6, 769–776. [Google Scholar] [CrossRef] [Green Version]
- Xiong, A.; Kundu, S.; Forsberg-Nilsson, K. Heparan Sulfate in the Regulation of Neural Differentiation and Glioma Development. FEBS J. 2014, 281, 4993–5008. [Google Scholar] [CrossRef]
- Steck, P.A.; Moser, R.P.; Bruner, J.M.; Liang, L.; Freidman, A.N.; Hwang, T.L.; Yung, W.K. Altered Expression and Distribution of Heparan Sulfate Proteoglycans in Human Gliomas. Cancer Res. 1989, 49, 2096–2103. [Google Scholar]
- Bertolotto, A.; Magrassi, M.L.; Orsi, L.; Sitia, C.; Schiffer, D. Glycosaminoglycan Changes in Human Gliomas. A Biochemical Study. J. Neurooncol. 1986, 4, 43–48. [Google Scholar] [CrossRef]
- Holley, R.J.; Meade, K.A.; Merry, C.L.R. Using Embryonic Stem Cells to Understand How Glycosaminoglycans Regulate Differentiation. Biochem. Soc. Trans. 2014, 42, 689–695. [Google Scholar] [CrossRef]
- Sasaki, N.; Hirano, T.; Kobayashi, K.; Toyoda, M.; Miyakawa, Y.; Okita, H.; Kiyokawa, N.; Akutsu, H.; Umezawa, A.; Nishihara, S. Chemical Inhibition of Sulfation Accelerates Neural Differentiation of Mouse Embryonic Stem Cells and Human Induced Pluripotent Stem Cells. Biochem. Biophys. Res. Commun. 2010, 401, 480–486. [Google Scholar] [CrossRef]
- Vitale, D.; Kumar Katakam, S.; Greve, B.; Jang, B.; Oh, E.S.; Alaniz, L.; Götte, M. Proteoglycans and Glycosaminoglycans as Regulators of Cancer Stem Cell Function and Therapeutic Resistance. FEBS J. 2019, 286, 2870–2882. [Google Scholar] [CrossRef]
- Esko, J.D.; Stewart, T.E.; Taylor, W.H. Animal Cell Mutants Defective in Glycosaminoglycan Biosynthesis. Proc. Natl. Acad. Sci. USA 1985, 82, 3197–3201. [Google Scholar] [CrossRef] [Green Version]
- Kunath, K.; von Harpe, A.; Fischer, D.; Petersen, H.; Bickel, U.; Voigt, K.; Kissel, T. Low-Molecular-Weight Polyethylenimine as a Non-Viral Vector for DNA Delivery: Comparison of Physicochemical Properties, Transfection Efficiency and in Vivo Distribution with High-Molecular-Weight Polyethylenimine. J. Control. Release 2003, 89, 113–125. [Google Scholar] [CrossRef]
- Fischer, D.; Bieber, T.; Li, Y.; Elsässer, H.P.; Kissel, T. A Novel Non-Viral Vector for DNA Delivery Based on Low Molecular Weight, Branched Polyethylenimine: Effect of Molecular Weight on Transfection Efficiency and Cytotoxicity. Pharm. Res. 1999, 16, 1273–1279. [Google Scholar] [CrossRef]
- Choksakulnimitr, S.; Masuda, S.; Tokuda, H.; Takakura, Y.; Hashida, M. In Vitro Cytotoxicity of Macromolecules in Different Cell Culture Systems. J. Control. Release 1995, 34, 233–241. [Google Scholar] [CrossRef]
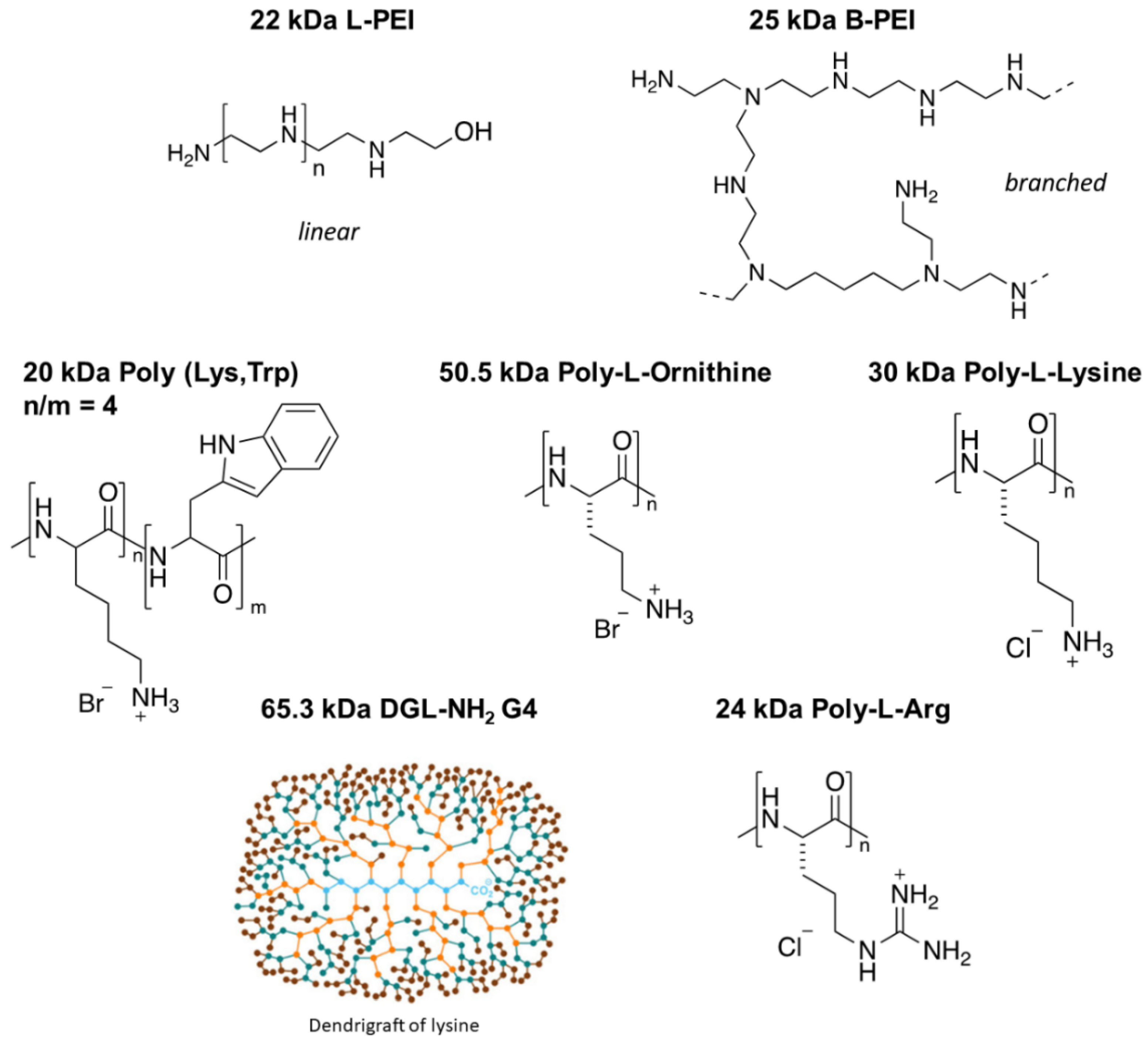


| POLYMERS | IC50 ± SEM (nM Polymer) | ||
|---|---|---|---|
| [Approx. Number of Monomers/Molecule] | U87 | NCH421K | |
| 22 kDa L-PEI [523] | 1547 ± 206 | **** | 194 ± 23.5 |
| 25 kDa B-PEI [595] | 508.8 ± 12.3 | *** | 146.2 ± 12.9 |
| 30 kDa PLL [182] | 583.4 ± 17.1 | ** | 280.5 ± 18.2 |
| 20 kDa P-(Lys,Trp) [164] | 514.9 ± 9.7 | ** | 257.1 ± 17.3 |
| 50.5 kDa PL-Orn [259] | 344.2 ± 26.7 | * | 133.3 ± 5.1 |
| 65.3 kDa DGL-NH2G4 [365] | 558.8 ± 16.5 | * | 292.2 ± 35.9 |
| 24 kDa PL-Arg [120] | 1570 ± 85 | Ns | 620 ± 55 |
| Poly-Arginine [Monomers/Molecule] | IC50 ± SEM (nM Polymer) |
|---|---|
| 1.9 kDa PL-Arg [10] | >8000 |
| 5.8 kDa PL-Arg [30] | 2380 ± 64 |
| 13 kDa PL-Arg [70] | 890 ± 98 **** |
| 24 kDa PL-Arg [120] | 620 ± 64 * |
| 38.5 kDa PL-Arg [200] | 460 ± 75 ns |
| Polyethylenimine | IC50 ± SEM (nM PEI) | ||
|---|---|---|---|
| [Monomers/Molecule] | U87 | NCH421K | |
| 0.73 kDa L-PEI [17] | ND | / | >32,000 |
| 4 kDa L-PEI [60] | 3337.2 ± 178.8 | **** | 1285 ± 188.7 |
| 22 kDa L-PEI [523] | 1547 ± 205.9 **** | **** | 194 ± 23.5 **** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McCartin, C.; Blumberger, J.; Dussouillez, C.; Fernandez de Larrinoa, P.; Dontenwill, M.; Herold-Mende, C.; Lavalle, P.; Heurtault, B.; Bellemin-Laponnaz, S.; Fournel, S.; et al. Evaluation of the Cytotoxicity of Cationic Polymers on Glioblastoma Cancer Stem Cells. J. Funct. Biomater. 2023, 14, 17. https://doi.org/10.3390/jfb14010017
McCartin C, Blumberger J, Dussouillez C, Fernandez de Larrinoa P, Dontenwill M, Herold-Mende C, Lavalle P, Heurtault B, Bellemin-Laponnaz S, Fournel S, et al. Evaluation of the Cytotoxicity of Cationic Polymers on Glioblastoma Cancer Stem Cells. Journal of Functional Biomaterials. 2023; 14(1):17. https://doi.org/10.3390/jfb14010017
Chicago/Turabian StyleMcCartin, Conor, Juliette Blumberger, Candice Dussouillez, Patricia Fernandez de Larrinoa, Monique Dontenwill, Christel Herold-Mende, Philippe Lavalle, Béatrice Heurtault, Stéphane Bellemin-Laponnaz, Sylvie Fournel, and et al. 2023. "Evaluation of the Cytotoxicity of Cationic Polymers on Glioblastoma Cancer Stem Cells" Journal of Functional Biomaterials 14, no. 1: 17. https://doi.org/10.3390/jfb14010017
APA StyleMcCartin, C., Blumberger, J., Dussouillez, C., Fernandez de Larrinoa, P., Dontenwill, M., Herold-Mende, C., Lavalle, P., Heurtault, B., Bellemin-Laponnaz, S., Fournel, S., & Kichler, A. (2023). Evaluation of the Cytotoxicity of Cationic Polymers on Glioblastoma Cancer Stem Cells. Journal of Functional Biomaterials, 14(1), 17. https://doi.org/10.3390/jfb14010017







