Bovine Fibroblast-Derived Extracellular Matrix Promotes the Growth and Preserves the Stemness of Bovine Stromal Cells during In Vitro Expansion
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Culturing of Bovine Fibroblasts and Bovine Umbilical Cord Stem Cells
2.2. Evaluation of the Multipotency of BUSC
2.3. Preparation and Characterization of BF-ECM
2.4. Immunofluorescent Staining of BF-ECM
2.5. Quantification of Residual DNA in BF-ECM
2.6. In Vitro Expansion of BUSCs on BF-ECM and Tissue Culture Treated Polystyrene (TCP)
2.7. Monitoring of Cell Adhesion and Proliferation
2.8. Adipogenic Differentiation of BUSCs
2.9. Determination of Growth Factors in BF-ECM Using ELISA Assays
2.10. Statistical Analysis
3. Results and Discussion
3.1. Isolation and Characterization of Bovine Umbilical Cord Stem Cells (BUSC)
3.2. Preparation and Characterization of Decellularized ECM from BFs
3.3. The Growth of BUSC on BF-ECM and TCP
3.4. Expansion of BUSC on BF-ECM and TCP
3.5. The Presence of BF-ECM Reduced the Requirement of FBS in Cell Culture Medium
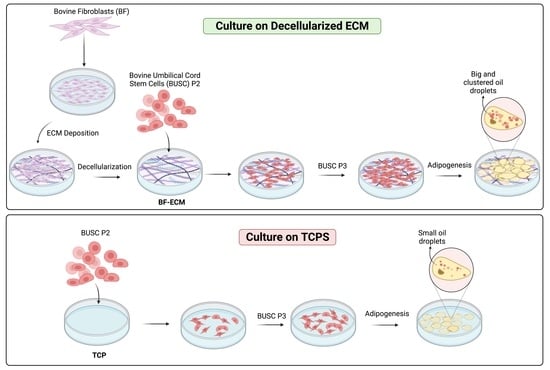
3.6. The Adipogenic Differentiation Potential of Busc Was Preserved Better in Cells Expanded on BF-ECM than on TCP
3.7. The Presence of Growth Factors in BF-ECM
4. Conclusions
- -
- retains the key proteins fibronectin and type I collagen after decellularization;
- -
- is a promising cell culture substrate to support bovine stem cell expansion in vitro faster and better;
- -
- reduces the requirement for FBS in the cell culture medium;
- -
- retains the stemness of bovine stem cells at later passages.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ben-Arye, T.; Levenberg, S. Tissue Engineering for Clean Meat Production. Front. Sustain. Food Syst. 2019, 3, 46. [Google Scholar] [CrossRef]
- Ozhava, D.; Bhatia, M.; Freman, J.; Mao, Y. Sustainable Cell Sources for Cultivated Meat. J. Biomed. Res. Environ. Sci. 2022, 3, 6. [Google Scholar] [CrossRef]
- Post, M.J.; Levenberg, S.; Kaplan, D.L.; Genovese, N.; Fu, J.A.; Bryant, C.J.; Negowetti, N.; Verzijden, K.; Moutsatsou, P. Scientific, sustainability and regulatory challenges of cultured meat. Nat. Food 2020, 1, 403–415. [Google Scholar] [CrossRef]
- Ritchie, H.; Rosado, P.; Roser, M. Meat and Dairy Production. Our World Data. 2017. Available online: https://ourworldindata.org/meat-production (accessed on 28 February 2023).
- Kang, D.H.; Louis, F.; Liu, H.; Shimoda, H.; Nishiyama, Y.; Nozawa, H.; Kakitani, M.; Takagi, D.; Kasa, D.; Nagamori, E.; et al. Engineered whole cut meat-like tissue by the assembly of cell fibers using tendon-gel integrated bioprinting. Nat. Commun. 2021, 12, 5059. [Google Scholar] [CrossRef]
- Zagury, Y.; Ianovici, I.; Landau, S.; Lavon, N.; Levenberg, S. Engineered marble-like bovine fat tissue for cultured meat. Commun. Biol. 2022, 5, 927. [Google Scholar] [CrossRef] [PubMed]
- Reiss, J.; Robertson, S.; Suzuki, M. Cell Sources for Cultivated Meat: Applications and Considerations throughout the Production Workflow. Int. J. Mol. Sci. 2021, 22, 7513. [Google Scholar] [CrossRef]
- Campisi, J.; di Fagagna, F.D. Cellular senescence: When bad things happen to good cells. Nat. Rev. Mol. Cell Bio. 2007, 8, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Rebollo, E.; Franzen, J.; Goetzke, R.; Hollmann, J.; Ostrowska, A.; Oliverio, M.; Sieben, T.; Rath, B.; Kornfeld, J.W.; Wagner, W. Senescence-Associated Metabolomic Phenotype in Primary and iPSC-Derived Mesenchymal Stromal Cells. Stem Cell Rep. 2020, 14, 201–209. [Google Scholar] [CrossRef] [Green Version]
- Allan, S.J.; De Bank, P.A.; Ellis, M.J. Bioprocess Design Considerations for Cultured Meat Production With a Focus on the Expansion Bioreactor. Front. Sustain. Food Syst. 2019, 3, 44. [Google Scholar] [CrossRef] [Green Version]
- Song, W.J.; Liu, P.P.; Li, H.X.; Ding, S.J. Large-Scale Expansion of Porcine Adipose-Derived Stem Cells Based on Microcarriers System for Cultured Meat Production. Foods 2022, 11, 3364. [Google Scholar] [CrossRef]
- Chia, W.K.; Cheah, F.C.; Aziz, N.H.A.; Kampan, N.C.; Shuib, S.; Khong, T.Y.; Tan, G.C.; Wong, Y.P. A Review of Placenta and Umbilical Cord-Derived Stem Cells and the Immunomodulatory Basis of Their Therapeutic Potential in Bronchopulmonary Dysplasia. Front. Pediatr. 2021, 9, 615508. [Google Scholar] [CrossRef]
- Gu, Y.J.; Li, T.; Ding, Y.L.; Sun, L.X.; Tu, T.; Zhu, W.; Hu, J.B.; Sun, X.C. Changes in mesenchymal stem cells following long-term culture in vitro. Mol. Med. Rep. 2016, 13, 5207–5215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Izadpanah, R.; Kaushal, D.; Kriedt, C.; Tsien, F.; Patel, B.; Dufour, J.; Bunnell, B.A. Long-term in vitro expansion alters the biology of adult mesenchymal stem cells. Cancer Res. 2008, 68, 4229–4238. [Google Scholar] [CrossRef] [Green Version]
- Jeske, R.; Yuan, X.G.; Fu, Q.; Bunnell, B.A.; Logan, T.M.; Li, Y. In Vitro Culture Expansion Shifts the Immune Phenotype of Human Adipose-Derived Mesenchymal Stem Cells. Front. Immunol. 2021, 12, 621744. [Google Scholar] [CrossRef]
- Jiang, T.; Xu, G.; Wang, Q.; Yang, L.; Zheng, L.; Zhao, J.; Zhang, X. In vitro expansion impaired the stemness of early passage mesenchymal stem cells for treatment of cartilage defects. Cell Death Dis. 2017, 8, e2851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, Y.; Sun, Y.; Skinner, C.M.; Son, E.L.; Lu, Z.; Tuan, R.S.; Jilka, R.L.; Ling, J.; Chen, X.D. Reconstitution of marrow-derived extracellular matrix ex vivo: A robust culture system for expanding large-scale highly functional human mesenchymal stem cells. Stem Cells Dev. 2010, 19, 1095–1107. [Google Scholar] [CrossRef] [Green Version]
- Witt, R.; Weigand, A.; Boos, A.M.; Cai, A.; Dippold, D.; Boccaccini, A.R.; Schubert, D.W.; Hardt, M.; Lange, C.; Arkudas, A.; et al. Mesenchymal stem cells and myoblast differentiation under HGF and IGF-1 stimulation for 3D skeletal muscle tissue engineering. BMC Cell Biol. 2017, 18, 15. [Google Scholar] [CrossRef] [Green Version]
- Eom, Y.W.; Oh, J.E.; Lee, J.I.; Baik, S.K.; Rhee, K.J.; Shin, H.C.; Kim, Y.M.; Ahn, C.M.; Kong, J.H.; Kim, H.S.; et al. The role of growth factors in maintenance of stemness in bone marrow-derived mesenchymal stem cells. Biochem. Biophys. Res. Commun. 2014, 445, 16–22. [Google Scholar] [CrossRef]
- McKee, C.; Chaudhry, G.R. Advances and challenges in stem cell culture. Colloids Surf. B Biointerfaces 2017, 159, 62–77. [Google Scholar] [CrossRef]
- Mao, Y.; Block, T.; Singh-Varma, A.; Sheldrake, A.; Leeth, R.; Griffey, S.; Kohn, J. Extracellular matrix derived from chondrocytes promotes rapid expansion of human primary chondrocytes in vitro with reduced dedifferentiation. Acta Biomater. 2019, 85, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Hoffman, T.; Wu, A.; Goyal, R.; Kohn, J. Cell type-specific extracellular matrix guided the differentiation of human mesenchymal stem cells in 3D polymeric scaffolds. J. Mater. Sci. Mater. Med. 2017, 28, 100. [Google Scholar] [CrossRef] [Green Version]
- Rakian, R.; Block, T.J.; Johnson, S.M.; Marinkovic, M.; Wu, J.; Dai, Q.; Dean, D.D.; Chen, X.D. Native extracellular matrix preserves mesenchymal stem cell “stemness” and differentiation potential under serum-free culture conditions. Stem Cell Res. Ther. 2015, 6, 235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes, R.N.; Manuel, F.; Nascimento, D.S. The bright side of fibroblasts: Molecular signature and regenerative cues in major organs. NPJ Regen. Med. 2021, 6, 43. [Google Scholar] [CrossRef] [PubMed]
- Kendall, R.T.; Feghali-Bostwick, C.A. Fibroblasts in fibrosis: Novel roles and mediators. Front. Pharmacol. 2014, 5, 123. [Google Scholar] [CrossRef] [Green Version]
- Carrel, A.; Ebeling, A.H. Age and multiplication of fibroblasts. J. Exp. Med. 1921, 34, 24. [Google Scholar] [CrossRef] [Green Version]
- Jedrzejczak-Silicka, M. History of Cell Culture; BoD—Books on Demand: Paris, France, 2017. [Google Scholar]
- Denu, R.A.; Nemcek, S.; Bloom, D.D.; Goodrich, A.D.; Kim, J.; Mosher, D.F.; Hematti, P. Fibroblasts and Mesenchymal Stromal/Stem Cells Are Phenotypically Indistinguishable. Acta Haematol. 2016, 136, 85–97. [Google Scholar] [CrossRef] [Green Version]
- Unger, C.; Gao, S.; Cohen, M.; Jaconi, M.; Bergstrom, R.; Holm, F.; Galan, A.; Sanchez, E.; Irion, O.; Dubuisson, J.B.; et al. Immortalized human skin fibroblast feeder cells support growth and maintenance of both human embryonic and induced pluripotent stem cells. Hum. Reprod. 2009, 24, 2567–2581. [Google Scholar] [CrossRef] [Green Version]
- Cardoso, T.C.; Ferrari, H.F.; Garcia, A.F.; Novais, J.B.; Silva-Frade, C.; Ferrarezi, M.C.; Andrade, A.L.; Gameiro, R. Isolation and characterization of Wharton’s jelly-derived multipotent mesenchymal stromal cells obtained from bovine umbilical cord and maintained in a defined serum-free three-dimensional system. BMC Biotechnol. 2012, 12, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.S.; Hung, S.C.; Peng, S.T.; Huang, C.C.; Wei, H.M.; Guo, Y.J.; Fu, Y.S.; Lai, M.C.; Chen, C.C. Mesenchymal stem cells in the Wharton’s jelly of the human umbilical cord. Stem Cells 2004, 22, 1330–1337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, H.; Bai, C.Y.; Wu, S.; Gao, Y.H.; Lu, T.F.; Hu, Q.Y.; Guan, W.J.; Ma, Y.H. Biological characterization of mesenchymal stem cells from bovine umbilical cord. Anim. Cells Syst. 2014, 18, 59–67. [Google Scholar] [CrossRef] [Green Version]
- Mehta, F.; Theunissen, R.; Post, M.J. Adipogenesis from Bovine Precursors. In Myogenesis: Methods in Molecular Biology; Humana Press: New York, NY, USA, 2019. [Google Scholar]
- Martinez-Ibanez, M.; Murthy, N.S.; Mao, Y.; Suay, J.; Gurruchaga, M.; Goni, I.; Kohn, J. Enhancement of plasma protein adsorption and osteogenesis of hMSCs by functionalized siloxane coatings for titanium implants. J. Biomed. Mater. Res. B 2018, 106, 1138–1147. [Google Scholar] [CrossRef] [PubMed]
- Coriell. Protocols for Adipogenic Differentiation Assays for Characterization of Adipose Stromal Cells (ASC). Available online: https://www.coriell.org/0/pdf/1401_38.pdf (accessed on 12 September 2022).
- Lee, M.J.; Fried, S.K. Optimal Protocol for the Differentiation and Metabolic Analysis of Human Adipose Stromal Cells. Method Enzymol. 2014, 538, 49–65. [Google Scholar] [CrossRef] [Green Version]
- Brigido, S.A.; Carrington, S.; Protzman, N.M.; Mao, Y.; Thomas, P.E.; Kohn, J.; Bhatia, M. The Use of an Acellular Connective Tissue Matrix in Hindfoot and Ankle Fusions: Understanding the Cellular Bench Top Data with a Consecutive Patient Series: A Pilot Study. Clin. Res. Foot Ankle 2018, 6, 276. [Google Scholar] [CrossRef]
- Chen, X.D.; Dusevich, V.; Feng, J.Q.; Manolagas, S.C.; Jilka, R.L. Extracellular matrix made by bone marrow cells facilitates expansion of marrow-derived mesenchymal progenitor cells and prevents their differentiation into osteoblasts. J. Bone Miner. Res. 2007, 22, 1943–1956. [Google Scholar] [CrossRef]
- Mao, Y.; John, N.; Protzman, N.M.; Kuehn, A.; Long, D.; Sivalenka, R.; Junka, R.A.; Gosiewska, A.; Hariri, R.J.; Brigido, S.A. A decellularized flowable placental connective tissue matrix supports cellular functions of human tenocytes in vitro. J. Exp. Orthop. 2022, 9, 69. [Google Scholar] [CrossRef]
- Mao, Y.; Schwarzbauer, J.E. Stimulatory effects of a three-dimensional microenvironment on cell-mediated fibronectin fibrillogenesis. J. Cell Sci. 2005, 118, 4427–4436. [Google Scholar] [CrossRef] [Green Version]
- Mao, Y.; Jacob, V.; Singal, A.; Lei, S.Y.; Park, M.S.; Lima, M.R.N.; Li, C.Y.; Dhall, S.; Sathyamoorthy, M.; Kohn, J. Exosomes Secreted from Amniotic Membrane Contribute to Its Anti-Fibrotic Activity. Int. J. Mol. Sci. 2021, 22, 2055. [Google Scholar] [CrossRef] [PubMed]
- Żuławińska, J. Cell Doubling Time Calculator. Available online: https://www.omnicalculator.com/biology/cell-doubling-time (accessed on 5 December 2022).
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, T.W.; Sellaro, T.L.; Badylak, S.F. Decellularization of tissues and organs. Biomaterials 2006, 27, 3675–3683. [Google Scholar] [CrossRef]
- Lopez-Martinez, M.; Gomez-Elizondo, D.; Guerrero-Ramirez, G.; Morales-Aseff, D.; Zavala, J.; Valdez-Garcia, J.E. Effect of different FBS concentrations on the proliferation of primary subculture of human pterygium fibroblasts. Invest. Ophth. Vis. Sci. 2019, 60, 6267. [Google Scholar]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef] [Green Version]
- Padhi, A.; Nain, A.S. ECM in Differentiation: A Review of Matrix Structure, Composition and Mechanical Properties. Ann. Biomed. Eng. 2020, 48, 1071–1089. [Google Scholar] [CrossRef] [PubMed]
- Pei, M.; Li, J.T.; Shoukry, M.; Zhang, Y. A review of decellularized stem cell matrix: A novel cell expansion system for cartilage tissue engineering. Eur. Cell Mater. 2011, 22, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Martino, M.M.; Brkic, S.; Bovo, E.; Burger, M.; Schaefer, D.J.; Wolff, T.; Gurke, L.; Briquez, P.S.; Larsson, H.M.; Gianni-Barrera, R.; et al. Extracellular matrix and growth factor engineering for controlled angiogenesis in regenerative medicine. Front. Bioeng. Biotechnol. 2015, 3, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taipale, J.; Keski-Oja, J. Growth factors in the extracellular matrix. FASEB J. 1997, 11, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Turnbull, J.; Guimond, S. Extracellular matrix and cell signalling: The dynamic cooperation of integrin, proteoglycan and growth factor receptor. J. Endocrinol. 2011, 209, 139–151. [Google Scholar] [CrossRef] [Green Version]
- Basilico, C.; Moscatelli, D. The Fgf Family of Growth-Factors and Oncogenes. Adv. Cancer Res. 1992, 59, 115–165. [Google Scholar] [CrossRef]
- Macri, L.; Silverstein, D.; Clark, R.A.F. Growth factor binding to the pericellular matrix and its importance in tissue engineering. Adv. Drug Deliv. Rev. 2007, 59, 1366–1381. [Google Scholar] [CrossRef]
- Hoch, R.V.; Soriano, P. Roles of PDGF in animal development. Development 2003, 130, 4769–4784. [Google Scholar] [CrossRef] [Green Version]
- Smith, E.M.; Mitsi, M.; Nugent, M.A.; Symes, K. PDGF-A interactions with fibronectin reveal a critical role for heparan sulfate in directed cell migration during Xenopus gastrulation. Proc. Natl. Acad. Sci. USA 2009, 106, 21683–21688. [Google Scholar] [CrossRef] [Green Version]
- Cebinelli, G.C.; Trugilo, K.P.; Garcia, S.B.; de Oliveira, K.B. TGF-beta 1 functional polymorphisms: A review. Eur. Cytokine Netw. 2016, 27, 81–89. [Google Scholar] [CrossRef] [Green Version]
- Discher, D.E.; Mooney, D.J.; Zandstra, P.W. Growth Factors, Matrices, and Forces Combine and Control Stem Cells. Science 2009, 324, 1673–1677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horiguchi, M.; Ota, M.; Rifkin, D.B. Matrix control of transforming growth factor-beta function. J. Biochem. 2012, 152, 321–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doorn, J.; Moll, G.; Le Blanc, K.; van Blitterswijk, C.; de Boer, J. Therapeutic Applications of Mesenchymal Stromal Cells: Paracrine Effects and Potential Improvements. Tissue Eng. Part B Rev. 2012, 18, 101–115. [Google Scholar] [CrossRef] [PubMed]







| Batch 1 | P3→P4 | P4→P5 | P5→P6 | Total |
| Amplification (fold) on ECM | 2.2 ± 0.09 | 25 ± 0.3 | 9.6 ± 1.3 | 539 |
| Doubling time (h) on ECM | 185 | 41 | 51 | |
| Amplification (fold) on TCP | 0.6 ± 0.06 | 4.7 ± 0.1 | 1.9 ± 0.5 | 5.2 |
| Doubling time (h) on TCP | N/A | 86 | 179 | |
| Statistical difference (p value) | 0.005 | 0.0017 | 0.0006 | |
| Batch 2 | P3→P4 | P4→P5 | P5→P6 | Total |
| Amplification (fold) on ECM | 6.4 ± 0.6 | 9.8 ± 0.3 | 7.8 ± 0.7 | 486 |
| Doubling time (h) on ECM | 81 | 58 | 57 | |
| Amplification (fold) on TCP | 2.5 ± 0.15 | 2.8 ± 0.8 | 1.3 ± 0.4 | 8.9 |
| Doubling time (h) on TCP | 162 | 129 | 485 | |
| Statistical difference (p value) | 0.0043 | 0.0048 | 0.0006 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, K.; Jackson, A.; John, N.; Zhang, R.; Ozhava, D.; Bhatia, M.; Mao, Y. Bovine Fibroblast-Derived Extracellular Matrix Promotes the Growth and Preserves the Stemness of Bovine Stromal Cells during In Vitro Expansion. J. Funct. Biomater. 2023, 14, 218. https://doi.org/10.3390/jfb14040218
Lee K, Jackson A, John N, Zhang R, Ozhava D, Bhatia M, Mao Y. Bovine Fibroblast-Derived Extracellular Matrix Promotes the Growth and Preserves the Stemness of Bovine Stromal Cells during In Vitro Expansion. Journal of Functional Biomaterials. 2023; 14(4):218. https://doi.org/10.3390/jfb14040218
Chicago/Turabian StyleLee, Kathleen, Anisha Jackson, Nikita John, Ryan Zhang, Derya Ozhava, Mohit Bhatia, and Yong Mao. 2023. "Bovine Fibroblast-Derived Extracellular Matrix Promotes the Growth and Preserves the Stemness of Bovine Stromal Cells during In Vitro Expansion" Journal of Functional Biomaterials 14, no. 4: 218. https://doi.org/10.3390/jfb14040218