Immobilisation of Cellobiose Dehydrogenase and Laccase on Chitosan Particles as a Multi-Enzymatic System for the Synthesis of Lactobionic Acid
Abstract
1. Introduction
2. Materials and Method
2.1. Microorganisms
2.2. Materials
2.3. Enzyme Preparation
2.4. Analysis of Proteins and Enzyme Activity
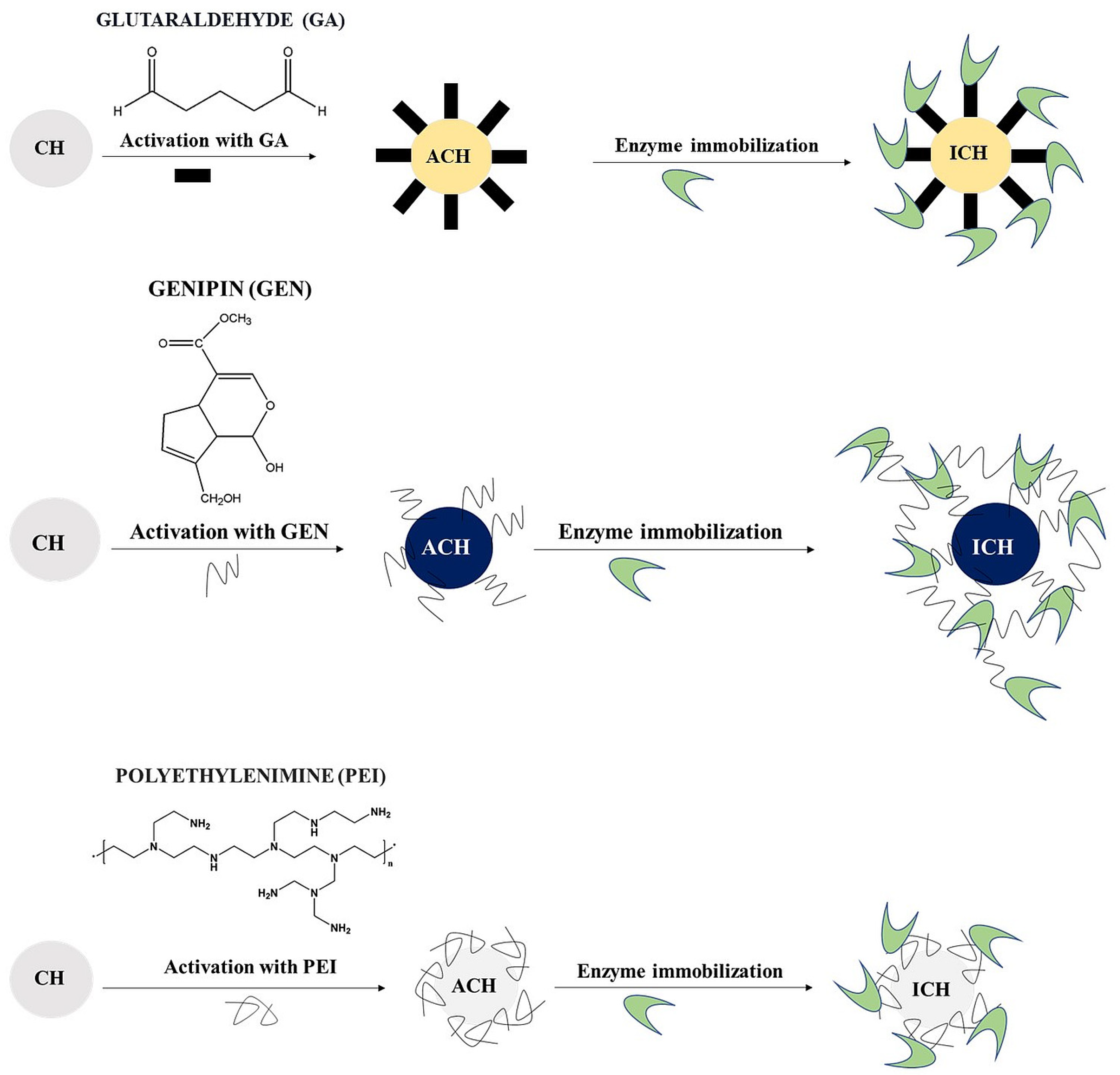
2.5. Immobilisation of Cellobiose Dehydrogenase on Chitosan Microspheres
2.5.1. Preparation of Chitosan Microspheres
2.5.2. Activation
2.5.3. Immobilisation
Activity of Immobilised Enzymes
Efficiency of Enzyme Immobilisation
2.6. Characterisation of Chitosan Microspheres
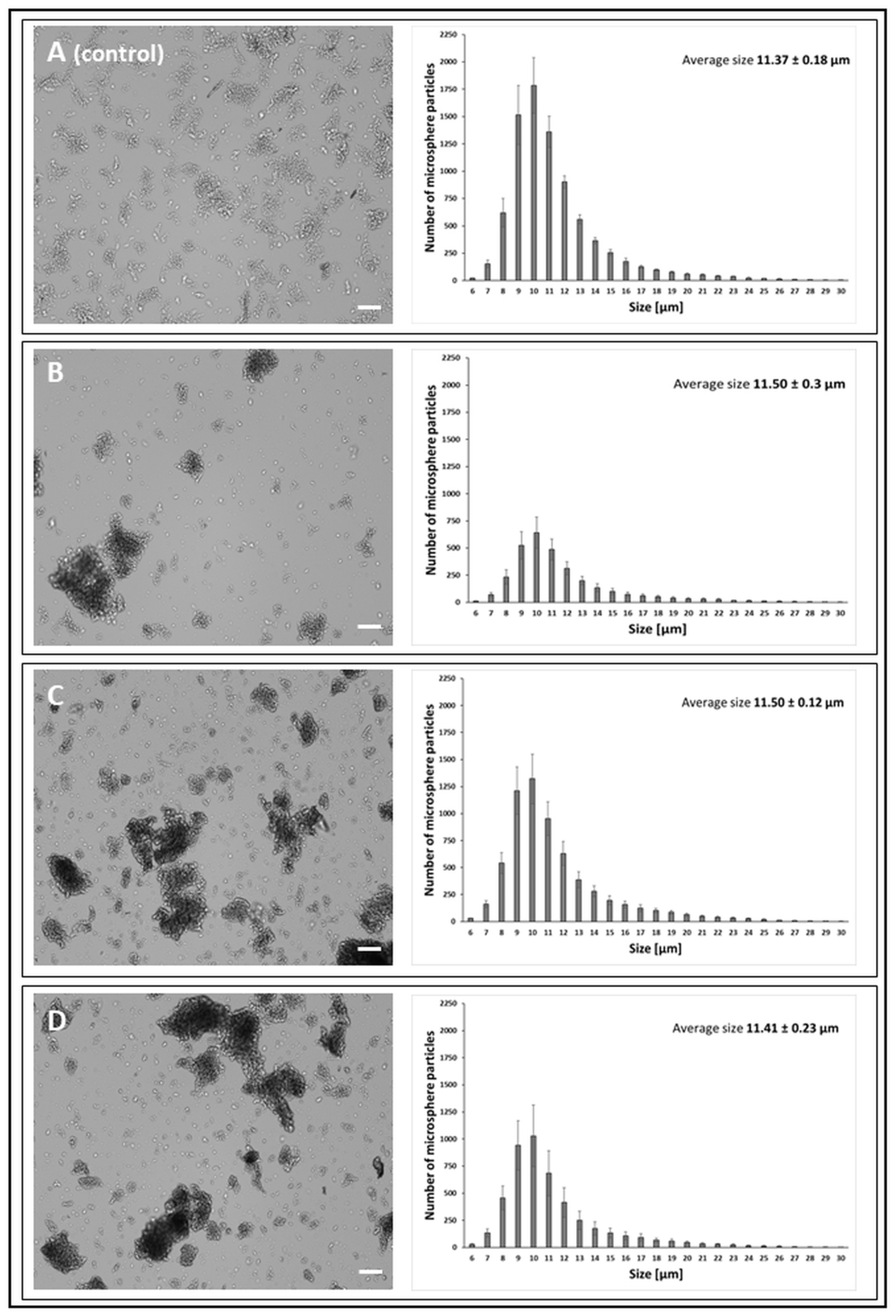
2.6.1. CellDrop
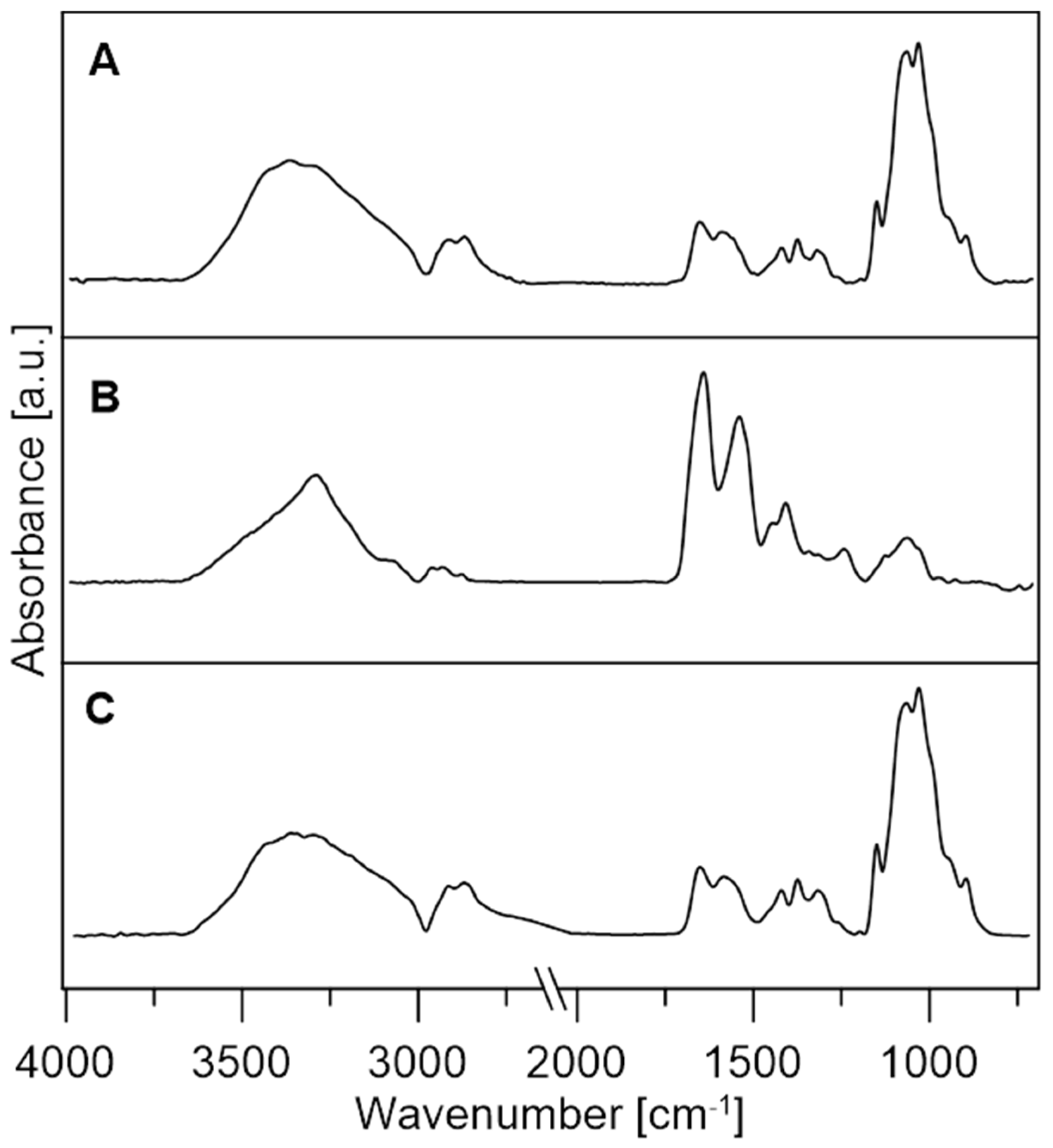
2.6.2. Fourier-Transform Infrared Spectroscopy (FTIR)
2.7. Enzymatic Oxidation of Lactose and Synthesis of Lactobionic Acid (LBA)
2.7.1. Use of Thin-Layer Chromatography to Determine LBA
2.7.2. Determination of LBA Using High-Performance Liquid Chromatography
2.8. Statistical Analysis
3. Results and Discussion
3.1. Immobilisation of Enzymes on Chitosan Microspheres
3.2. Characterisation of Chitosan Microspheres
3.2.1. CellDrop
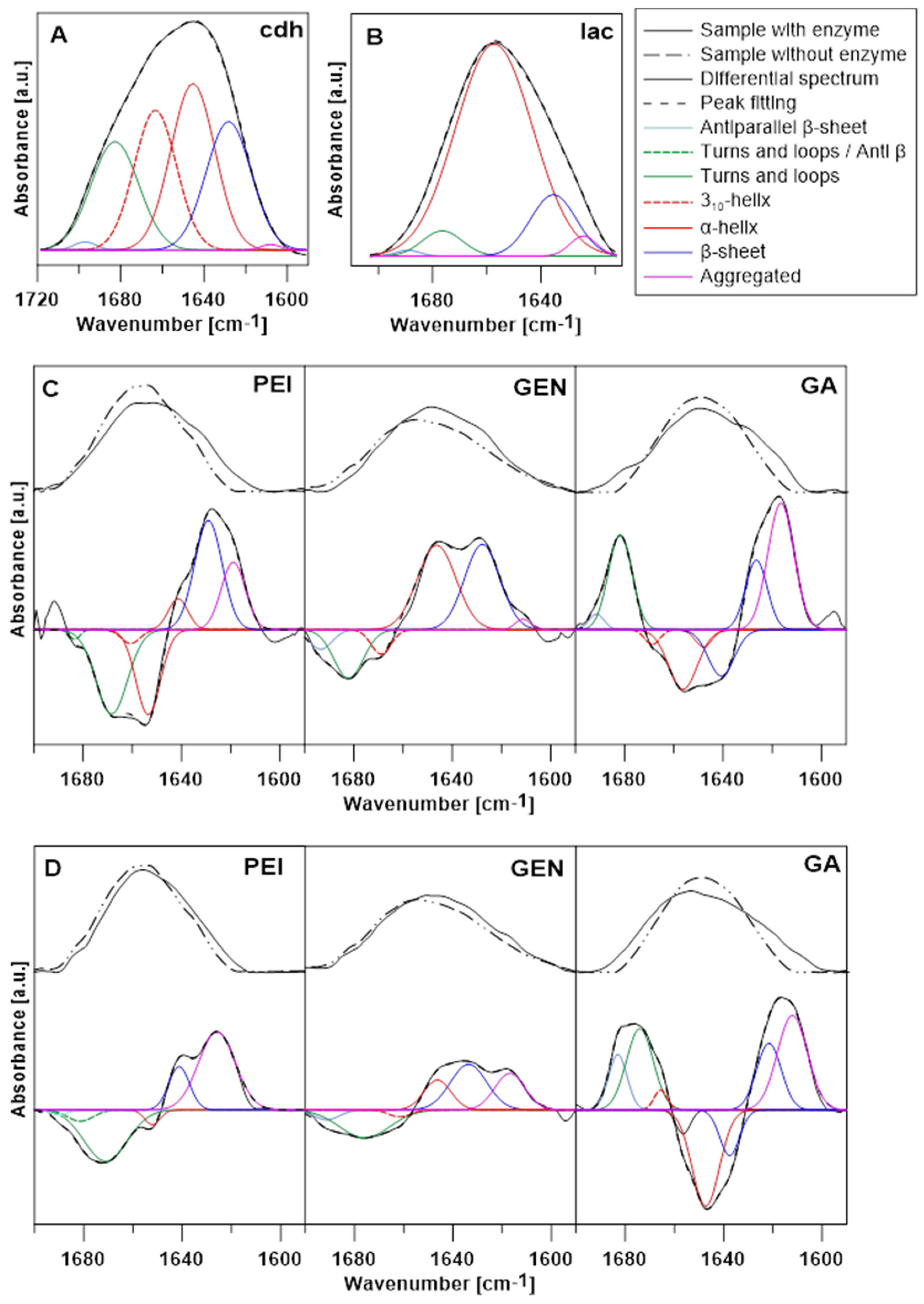
3.2.2. FTIR Analysis
3.3. Lactose Oxidation by Enzymes and Lactobionic Acid (LBA) Production
- ✓
- PchCDH/GEN/GA/PEI-chitosan activated with genipin, glutaraldehyde, or polyethyleneimine with immobilised CDH.
- ✓
- CuLAC/GEN/GA/PEI-chitosan activated with genipin, glutaraldehyde, or polyethyleneimine with immobilised LAC.
- PchCDH/GA + CuLAC,
- PchCDH/PEI + CuLAC,
- PchCDH/GEN + CuLAC,
- CuLAC/GA + PchCDH,
- CuLAC/PEI + PchCDH,
- CuLAC/GEN + PchCDH,
- PchCDH/GEN + CuLAC/GEN,
- PchCDH/GEN + CuLAC/PEI,
- PchCDH/GEN + CuLAC/GA,
- PchCDH/GA + CuLAC/GEN,
- PchCDH/GA + CuLAC/PEI,
- PchCDH/GA + CuLAC/GA,
- PchCDH/PEI + CuLAC/GEN,
- PchCDH/PEI + CuLAC/PEI,
- PchCDH/PEI + CuLAC/GA.
3.3.1. Thin-Layer Liquid Chromatography (TLC) Analysis
3.3.2. High-Performance Liquid Chromatography Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Alonso, S.; Rendueles, M.; Díaz, M. Bio-production of lactobionic acid: Current status, applications and future prospects. Biotechnol. Adv. 2013, 31, 1275–1291. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, T.; Marques, C.; Dagostin, J.L.A.; Masson, M.L. Lactobionic acid as a potential food ingredient: Recent studies and applications. J. Food Sci. 2019, 84, 1672–1681. [Google Scholar] [CrossRef] [PubMed]
- Southard, M.; James, H.; Belzer, M.; Folkert, O. Organ preservation. Annu. Rev. Med. 1995, 46, 235–247. [Google Scholar] [CrossRef]
- Gutiérrez, L.-F.; Hamoudi, S.; Belkacemi, K. Lactobionic acid: A high value-added lactose derivative for food and pharmaceutical applications. Int. Dairy J. 2012, 26, 103–111. [Google Scholar] [CrossRef]
- Isaacson, Y.; Chensny, L.; Shepherd, R.E.; Zhang, S.; Kortes, R.; John, K. Lactobionic and gluconic acid complexes of FeII and FeIII; control of oxidation pathways by an organ transplantation preservant. J. Inorg. Biochem. 1993, 49, 23–48. [Google Scholar] [CrossRef]
- Green, B.A.; Ruey, J.Y.; Van Scott, E.J. Clinical and cosmeceutical uses of hydroxyacids. Clin. Dermatol. 2009, 27, 495–501. [Google Scholar] [CrossRef]
- Goderska, K.; Kozłowski, P. Evaluation of microencapsulated synbiotic preparations containing lactobionic acid. Appl. Biochem. Biotechnol. 2021, 193, 3483–3495. [Google Scholar] [CrossRef]
- Henriksson, G.; Sild, V.; Szabó, I.J.; Pettersson, G.; Johansson, G. Substrate specificity of cellobiose dehydrogenase from Phanerochaete chrysosporium. Biochim. Biophys. Acta (BBA)-Protein Struct. Mol. Enzymol. 1998, 1383, 48–54. [Google Scholar] [CrossRef]
- Baminger, U.; Ludwig, R.; Galhaup, C.; Leitner, C.; Kulbe, K.D.; Haltrich, D. Continuous enzymatic regeneration of redox mediators used in biotransformation reactions employing flavoproteins. J. Mol. Catal. B Enzym. 2001, 11, 541–550. [Google Scholar] [CrossRef]
- Van Hecke, W.; Bhagwat, A.; Ludwig, R.; Dewulf, J.; Haltrich, D.; Van Langenhove, H. Kinetic modeling of a bi-enzymatic system for efficient conversion of lactose to lactobionic acid. Biotechnol. Bioeng. 2009, 102, 1475–1482. [Google Scholar] [CrossRef]
- Gangwar, R.; Rasool, S.; Mishra, S. Evaluation of cellobiose dehydrogenase and laccase containing culture fluids of Termitomyces sp. OE147 for degradation of Reactive blue 21. Biotechnol. Rep. 2016, 12, 52–61. [Google Scholar] [CrossRef]
- Kaczmarek, M.B.; Kwiatos, N.; Szczęsna-Antczak, M.; Bielecki, S. Laccases–enzymes with an unlimited potential. Biotechnol. Food Sci. 2017, 81, 41–70. [Google Scholar]
- Abdolahinia, E.D.; Nadri, S.; Rahbarghazi, R.; Barar, J.; Aghanejad, A.; Omidi, Y. Enhanced penetration and cytotoxicity of metformin and collagenase conjugated gold nanoparticles in breast cancer spheroids. Life Sci. 2019, 231, 116545. [Google Scholar] [CrossRef] [PubMed]
- Ajoolabady, A.; Aghanejad, A.; Bi, Y.; Zhang, Y.; Aslkhodapasandhukmabad, H.; Abhari, A.; Ren, J. Enzyme-based autophagy in anti-neoplastic management: From molecular mechanisms to clinical therapeutics. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2020, 1874, 188366. [Google Scholar] [CrossRef]
- Liu, S.; Bilal, M.; Rizwan, K.; Gul, I.; Rasheed, T.; Iqbal, H.M. Smart chemistry of enzyme immobilization using various support matrices—A review. Int. J. Biol. Macromol. 2021, 190, 396–408. [Google Scholar] [CrossRef] [PubMed]
- Lindeque, R.M.; Woodley, J.M. Reactor selection for effective continuous biocatalytic production of pharmaceuticals. Catalysts 2019, 9, 262. [Google Scholar] [CrossRef]
- Basso, A.; Serban, S. Industrial applications of immobilized enzymes—A review. Mol. Catal. 2019, 479, 110607. [Google Scholar] [CrossRef]
- Biró, E.; Németh, Á.S.; Sisak, C.; Feczkó, T.; Gyenis, J. Preparation of chitosan particles suitable for enzyme immobilization. J. Biochem. Biophys. Methods 2008, 70, 1240–1246. [Google Scholar] [CrossRef]
- Homaei, A.A.; Sariri, R.; Vianello, F.; Stevanato, R. Enzyme immobilization: An update. J. Chem. Biol. 2013, 6, 185–205. [Google Scholar] [CrossRef]
- Pinheiro, B.B.; Rios, N.S.; Aguado, E.R.; Fernandez-Lafuente, R.; Freire, T.M.; Fechine, P.B.; Dos Santos, J.C.; Goncalves, L.R. Chitosan activated with divinyl sulfone: A new heterofunctional support for enzyme immobilization. Application in the immobilization of lipase B from Candida antarctica. Int. J. Biol. Macromol. 2019, 130, 798–809. [Google Scholar] [CrossRef]
- Nuraliyah, A.; Perdani, M.S.; Putri, D.N.; Sahlan, M.; Wijanarko, A.; Hermansyah, H. Effect of Additional Amino Group to Improve the Performance of Immobilized Lipase From Aspergillus niger by Adsorption-Crosslinking Method. Front. Energy Res. 2021, 9, 616945. [Google Scholar] [CrossRef]
- Apriceno, A.; Bucci, R.; Girelli, A.M. Immobilization of laccase from Trametes versicolor on chitosan macrobeads for anthracene degradation. Anal. Lett. 2017, 50, 2308–2322. [Google Scholar] [CrossRef]
- Samprasit, W.; Akkaramongkolporn, P.; Jaewjira, S.; Opanasopit, P. Design of alpha mangostin-loaded chitosan/alginate controlled-release nanoparticles using genipin as crosslinker. J. Drug Deliv. Sci. Technol. 2018, 46, 312–321. [Google Scholar] [CrossRef]
- Chronopoulou, L.; Daniele, M.; Perez, V.; Gentili, A.; Gasperi, T.; Lupi, S.; Palocci, C. A physico-chemical approach to the study of genipin crosslinking of biofabricated peptide hydrogels. Process Biochem. 2018, 70, 110–116. [Google Scholar] [CrossRef]
- Somers, P.; De Somer, F.; Cornelissen, M.; Bouchez, S.; Gasthuys, F.; Narine, K.; Cox, E.; Van Nooten, G. Genipin blues: An alternative non-toxic crosslinker for heart valves? J. Heart Valve Dis. 2008, 17, 682. [Google Scholar]
- Tsai, C.C.; Huang, R.N.; Sung, H.W.; Liang, H.C. In vitro evaluation of the genotoxicity of a naturally occurring crosslinking agent (genipin) for biologic tissue fixation. J. Biomed. Mater. Res. 2000, 52, 58–65. [Google Scholar] [CrossRef]
- Roether, J.; Oelschlaeger, C.; Willenbacher, N. Hyaluronic acid cryogels with non-cytotoxic crosslinker genipin. Mater. Lett. X 2019, 4, 100027. [Google Scholar] [CrossRef]
- Manickam, B.; Sreedharan, R.; Elumalai, M. ‘Genipin’–the natural water soluble cross-linking agent and its importance in the modified drug delivery systems: An overview. Curr. Drug Deliv. 2014, 11, 139–145. [Google Scholar] [CrossRef]
- Tacias-Pascacio, V.G.; García-Parra, E.; Vela-Gutiérrez, G.; Virgen-Ortiz, J.J.; Berenguer-Murcia, Á.; Alcántara, A.R.; Fernandez-Lafuente, R. Genipin as an emergent tool in the design of biocatalysts: Mechanism of reaction and applications. Catalysts 2019, 9, 1035. [Google Scholar] [CrossRef]
- Saralidze, K.; Koole, L.H.; Knetsch, M.L. Polymeric microspheres for medical applications. Materials 2010, 3, 3537–3564. [Google Scholar] [CrossRef]
- Wang, Z.-Y.; Zhang, X.-W.; Ding, Y.-W.; Ren, Z.-W.; Wei, D.-X. Natural biopolyester microspheres with diverse structures and surface topologies as micro-devices for biomedical applications. Smart Mater. Med. 2022, 4, 15–36. [Google Scholar] [CrossRef]
- Mitra, A.; Dey, B. Chitosan microspheres in novel drug delivery systems. Indian J. Pharm. Sci. 2011, 73, 355. [Google Scholar]
- Alencastre, J.B.; Bentley, M.V.L.B.; Garcia, F.S.; Moragas, M.d.; Viladot, J.L.; Marchetti, J.M. A study of the characteristics and in vitro permeation properties of CMC/chitosan microparticles as a skin delivery system for vitamin E. Rev. Bras. Ciênc. Farm. 2006, 42, 69–76. [Google Scholar] [CrossRef]
- Piątek-Gołda, W.; Sulej, J.; Grąz, M.; Waśko, P.; Janik-Zabrotowicz, E.; Osińska-Jaroszuk, M. Multi-Enzymatic Synthesis of Lactobionic Acid Using Wood-Degrading Enzymes Produced by White Rot Fungi. Metabolites 2023, 13, 469. [Google Scholar] [CrossRef]
- Sulej, J.; Osińska-Jaroszuk, M.; Jaszek, M.; Olszewska, A.; Belcarz, A.; Piątek-Gołda, W. Chitosan as a Promising Support of a CDH Activity Preservation System for Biomedical and Industrial Applications. Int. J. Mol. Sci. 2023, 24, 4535. [Google Scholar] [CrossRef]
- Sulej, J.; Osińska-Jaroszuk, M.; Jaszek, M.; Grąz, M.; Kutkowska, J.; Pawlik, A.; Chudzik, A.; Bancerz, R. Antimicrobial and antioxidative potential of free and immobilised cellobiose dehydrogenase isolated from wood degrading fungi. Fungal Biol. 2019, 123, 875–886. [Google Scholar] [CrossRef]
- Baminger, U.; Nidetzky, B.; Kulbe, K.D.; Haltrich, D. A simple assay for measuring cellobiose dehydrogenase activity in the presence of laccase. J. Microbiol. Methods 1999, 35, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Wlizło, K.; Polak, J.; Kapral-Piotrowska, J.; Grąz, M.; Paduch, R.; Jarosz-Wilkołazka, A. Influence of carrier structure and physicochemical factors on immobilisation of fungal laccase in terms of Bisphenol a removal. Catalysts 2020, 10, 951. [Google Scholar] [CrossRef]
- Szałapata, K.; Osińska-Jaroszuk, M.; Bryjak, J.; Jaszek, M.; Jarosz-Wilkołazka, A. Novel application of porous and cellular materials for covalent immobilization of pepsin. Braz. J. Chem. Eng. 2016, 33, 251–260. [Google Scholar] [CrossRef]
- Yang, J.; Xu, P.; Long, L.; Ding, S. Production of lactobionic acid using an immobilized cellobiose dehydrogenase/laccase system on magnetic chitosan spheres. Process Biochem. 2021, 100, 1–9. [Google Scholar] [CrossRef]
- Kiryu, T.; Kiso, T.; Nakano, H.; Ooe, K.; Kimura, T.; Murakami, H. Involvement of Acetobacter orientalis in the production of lactobionic acid in Caucasian yogurt (“Caspian Sea yogurt”) in Japan. J. Dairy Sci. 2009, 92, 25–34. [Google Scholar] [CrossRef]
- Hanefeld, U.; Cao, L.; Magner, E. Enzyme immobilisation: Fundamentals and application. Chem. Soc. Rev. 2013, 42, 6211–6212. [Google Scholar] [CrossRef] [PubMed]
- Federsel, H.-J.; Moody, T.S.; Taylor, S.J. Recent trends in enzyme immobilization—Concepts for expanding the biocatalysis toolbox. Molecules 2021, 26, 2822. [Google Scholar] [CrossRef] [PubMed]
- de Andrades, D.; Graebin, N.G.; Kadowaki, M.K.; Ayub, M.A.; Fernandez-Lafuente, R.; Rodrigues, R.C. Immobilization and stabilization of different β-glucosidases using the glutaraldehyde chemistry: Optimal protocol depends on the enzyme. Int. J. Biol. Macromol. 2019, 129, 672–678. [Google Scholar] [CrossRef] [PubMed]
- Bilal, M.; Jing, Z.; Zhao, Y.; Iqbal, H.M. Immobilization of fungal laccase on glutaraldehyde cross-linked chitosan beads and its bio-catalytic potential to degrade bisphenol A. Biocatal. Agric. Biotechnol. 2019, 19, 101174. [Google Scholar] [CrossRef]
- Mo, H.; Qiu, J.; Yang, C.; Zang, L.; Sakai, E.; Chen, J. Porous biochar/chitosan composites for high performance cellulase immobilization by glutaraldehyde. Enzym. Microb. Technol. 2020, 138, 109561. [Google Scholar] [CrossRef] [PubMed]
- Sano, L.L.; Krueger, A.M.; Landrum, P.F. Chronic toxicity of glutaraldehyde: Differential sensitivity of three freshwater organisms. Aquat. Toxicol. 2005, 71, 283–296. [Google Scholar] [CrossRef]
- Pereira, S.P.; Oliveira, R.; Coelho, S.n.; Musso, C.; Soares, A.M.; Domingues, I.; Nogueira, A.n.J. From sub cellular to community level: Toxicity of glutaraldehyde to several aquatic organisms. Sci. Total Environ. 2014, 470, 147–158. [Google Scholar] [CrossRef]
- Ehrmann, A. Non-toxic crosslinking of electrospun gelatin nanofibers for tissue engineering and biomedicine—A review. Polymers 2021, 13, 1973. [Google Scholar] [CrossRef]
- Priddy-Arrington, T.R.; Edwards, R.E.; Colley, C.E.; Nguyen, M.M.; Hamilton-Adaire, T.; Caldorera-Moore, M.E. Characterization and Optimization of Injectable in situ Crosslinked Chitosan-Genipin Hydrogels. Macromol. Biosci. 2023, 23, 2200505. [Google Scholar] [CrossRef]
- Wang, W.; Jiang, Y.; Zhou, L.; Gao, J. Comparison of the properties of lipase immobilized onto mesoporous resins by different methods. Appl. Biochem. Biotechnol. 2011, 164, 561–572. [Google Scholar] [CrossRef] [PubMed]
- Cavello, I.A.; Contreras-Esquivel, J.C.; Cavalitto, S.F. Immobilization of a keratinolytic protease from Purpureocillium lilacinum on genipin activated-chitosan beads. Process Biochem. 2014, 49, 1332–1336. [Google Scholar] [CrossRef]
- Phadungcharoen, N.; Winotapun, W.; Khomniyawanit, A.; Krataichan, F.; Rojanarata, T. Facile and green fabrication of biocatalytic chitosan beads by one-step genipin-mediated β-glucosidase immobilization for production of bioactive genistein. Sustain. Chem. Pharm. 2019, 14, 100187. [Google Scholar] [CrossRef]
- Zabner, J.; Fasbender, A.J.; Moninger, T.; Poellinger, K.A.; Welsh, M.J. Cellular and molecular barriers to gene transfer by a cationic lipid. J. Biol. Chem. 1995, 270, 18997–19007. [Google Scholar] [CrossRef] [PubMed]
- Xiong, M.P.; Forrest, M.L.; Ton, G.; Zhao, A.; Davies, N.M.; Kwon, G.S. Poly (aspartate-g-PEI800), a polyethylenimine analogue of low toxicity and high transfection efficiency for gene delivery. Biomaterials 2007, 28, 4889–4900. [Google Scholar] [CrossRef]
- Liu, C.; Liu, F.; Feng, L.; Li, M.; Zhang, J.; Zhang, N. The targeted co-delivery of DNA and doxorubicin to tumor cells via multifunctional PEI-PEG based nanoparticles. Biomaterials 2013, 34, 2547–2564. [Google Scholar] [CrossRef]
- Vasiliu, T.; Craciun, B.F.; Neamtu, A.; Clima, L.; Isac, D.L.; Maier, S.S.; Pinteala, M.; Mocci, F.; Laaksonen, A. In silico study of PEI-PEG-squalene-dsDNA polyplex formation: The delicate role of the PEG length in the binding of PEI to DNA. Biomater. Sci. 2021, 9, 6623–6640. [Google Scholar] [CrossRef]
- Bahulekar, R.; Ayyangar, N.; Ponrathnam, S. Polyethyleneimine in immobilization of biocatalysts. Enzym. Microb. Technol. 1991, 13, 858–868. [Google Scholar] [CrossRef]
- Jia, H.; Zhao, J.-Z.; Jin, F.-Y.; Pu, W.-F.; Li, Y.-M.; Li, K.-X.; Li, J.-M. New insights into the gelation behavior of polyethyleneimine cross-linking partially hydrolyzed polyacrylamide gels. Ind. Eng. Chem. Res. 2012, 51, 12155–12166. [Google Scholar] [CrossRef]
- Feng, C.; Xu, J.; Li, M.; Tang, Y.; Gao, C. Studies on a novel nanofiltration membrane prepared by cross-linking of polyethyleneimine on polyacrylonitrile substrate. J. Membr. Sci. 2014, 451, 103–110. [Google Scholar] [CrossRef]
- Jung, H.; Jeon, S.; Jo, D.H.; Huh, J.; Kim, S.H. Effect of crosslinking on the CO2 adsorption of polyethyleneimine-impregnated sorbents. Chem. Eng. J. 2017, 307, 836–844. [Google Scholar] [CrossRef]
- Maia, M.d.M.D.; de Vasconcelos, E.A.; de Mascena Diniz, P.F.C.; da Costa Maciel, J.; Cajueiro, K.R.R.; da Silva, M.d.P.C.; da Silva Jr, E.F.; Dutra, R.A.F.; Freire, V.N.; de Lima Filho, J.L. Immobilization of urease on vapour phase stain etched porous silicon. Process Biochem. 2007, 42, 429–433. [Google Scholar] [CrossRef]
- Sima, V.; Cristea, C.; Bodoki, E.; Duţu, G.; Sāndulescu, R. Screen-printed electrodes modified with HRP-zirconium alcoxide film for the development of a biosensor for acetaminophen detection. Open Chem. 2010, 8, 1034–1040. [Google Scholar] [CrossRef]
- Patel, K.S.; Patel, M.B. Preparation and evaluation of chitosan microspheres containing nicorandil. Int. J. Pharm. Investig. 2014, 4, 32. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, K.; Schelfhout, C.; Bracke, M.; De Wever, O.; Van Bockstal, M.; Ceelen, W.; Remon, J.P.; Vervaet, C. Genipin-crosslinked gelatin microspheres as a strategy to prevent postsurgical peritoneal adhesions: In vitro and in vivo characterization. Biomaterials 2016, 96, 33–46. [Google Scholar] [CrossRef]
- Jha, R.; Mayanovic, R.A. A Review of the Preparation, Characterization, and Applications of Chitosan Nanoparticles in Nanomedicine. Nanomaterials 2023, 13, 1302. [Google Scholar] [CrossRef]
- Solorio, L.; Zwolinski, C.; Lund, A.W.; Farrell, M.J.; Stegemann, J.P. Gelatin microspheres crosslinked with genipin for local delivery of growth factors. J. Tissue Eng. Regen. Med. 2010, 4, 514–523. [Google Scholar] [CrossRef]
- Takahashi, T. Potential of an Automated-and Image-Based Cell Counter to Accelerate Microalgal Research and Applications. Energies 2020, 13, 6019. [Google Scholar] [CrossRef]
- Aijaz, A.; Trawinski, D.; McKirgan, S.; Parekkadan, B. Non-invasive cell counting of adherent, suspended and encapsulated mammalian cells using optical density. BioTechniques 2020, 68, 35–40. [Google Scholar] [CrossRef]
- Takahashi, T. Applicability of automated cell counter with a chlorophyll detector in routine management of microalgae. Sci. Rep. 2018, 8, 4967. [Google Scholar] [CrossRef] [PubMed]
- Lojk, J.; Čibej, U.; Karlaš, D.; Šajn, L.; Pavlin, M. Comparison of two automatic cell-counting solutions for fluorescent microscopic images. J. Microsc. 2015, 260, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.C.; Berenguer-Murcia, Á.; Carballares, D.; Morellon-Sterling, R.; Fernandez-Lafuente, R. Stabilization of enzymes via immobilization: Multipoint covalent attachment and other stabilization strategies. Biotechnol. Adv. 2021, 52, 107821. [Google Scholar] [CrossRef]
- Cuesta-Garrote, N.; Escoto-Palacios, M.J.; Arán-Ais, F.; Orgiles-Barcelo, C. Structural changes in the crosslinking process of a protein bioadhesive. Proc. Inst. Mech. Eng. Part L J. Mater. Des. Appl. 2014, 228, 115–124. [Google Scholar] [CrossRef]
- Andrade, J.; Pereira, C.G.; de Almeida Junior, J.C.; Viana, C.C.R.; de Oliveira Neves, L.N.; da Silva, P.H.F.; Bell, M.J.V.; dos Anjos, V.d.C. FTIR-ATR determination of protein content to evaluate whey protein concentrate adulteration. LWT 2019, 99, 166–172. [Google Scholar] [CrossRef]
- El-Batal, A.I.; ElKenawy, N.M.; Yassin, A.S.; Amin, M.A. Laccase production by Pleurotus ostreatus and its application in synthesis of gold nanoparticles. Biotechnol. Rep. 2015, 5, 31–39. [Google Scholar] [CrossRef]
- Kong, J.; Yu, S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim. Biophys. Sin. 2007, 39, 549–559. [Google Scholar] [CrossRef]
- Surewicz, W.K.; Mantsch, H.H.; Chapman, D. Determination of protein secondary structure by Fourier transform infrared spectroscopy: A critical assessment. Biochemistry 1993, 32, 389–394. [Google Scholar] [CrossRef]


| Sample | Applied Protein [mg/g Carrier] | Immobilised Protein [mg/g Carrier] | Protein Yield [%] | Activity of Enzyme Bound with Chitosan [U/g Carrier] | Activity Yield [%] |
|---|---|---|---|---|---|
| CDH + GA | 0.56 | 0.47 | 83.21 | 1.08 | 60.00 |
| CDH + PEI | 0.56 | 0.53 | 94.29 | 0.79 | 44.89 |
| CDH + GEN | 0.56 | 0.55 | 97.86 | 1.77 | 100.00 |
| LAC + GA | 0.03 | 0.00 | 0.00 | 0.00 | 0.00 |
| LAC + PEI | 0.03 | 0.00 | 14.29 | 23.42 | 100.00 |
| LAC + GEN | 0.03 | 0.01 | 35.71 | 0.00 | 0.00 |
| Enzyme | Secondary Structure | Position [cm−1] | % of Area |
|---|---|---|---|
| CDH | Antiparallel β-sheet | 1697 | 0.75 |
| Turns and loops | 1682 | 21.02 | |
| 310-helix | 1663 | 23.95 | |
| α-helix | 1645 | 29.78 | |
| β-sheet | 1628 | 23.60 | |
| Aggregated | 1608 | 0.47 | |
| laccase | Antiparallel β-sheet | 1689 | 0.68 |
| Turns and loops | 1676 | 4.83 | |
| α-helix | 1657 | 77.96 | |
| β-sheet | 1635 | 13.35 | |
| Aggregated | 1624 | 2.59 |
| Enzyme | Activating Substance | Structure | Position [cm−1] | Rise [+]/Loss [34] |
|---|---|---|---|---|
| CDH | PEI | Turns and loops/Anti β | 1683 | - |
| Turns and loops | 1668 | - | ||
| 310-helix | 1660 | - | ||
| α-helix | 1653 | - | ||
| α-helix | 1641 | + | ||
| β-sheet | 1629 | + | ||
| Aggregated | 1618 | + | ||
| GEN | Antiparallel β-sheet | 1693 | - | |
| Turns and loops | 1682 | - | ||
| 310-helix | 1668 | - | ||
| α-helix | 1646 | + | ||
| β-sheet | 1627 | + | ||
| Aggregated | 1611 | + | ||
| GA | Antiparallel β-sheet | 1691 | + | |
| Turns and loops | 1681 | + | ||
| 310-helix | 1668 | - | ||
| Unordered | 1656 | - | ||
| α-helix | 1647 | - | ||
| α-helix | 1640 | - | ||
| β-sheet | 1626 | + | ||
| Aggregated | 1616 | + | ||
| laccase | PEI | Antiparallel β-sheet | 1689 | - |
| Turns and loops/Anti β | 1681 | - | ||
| Turns and loops | 1671 | - | ||
| α-helix | 1652 | - | ||
| β-sheet | 1641 | + | ||
| Aggregated | 1625 | + | ||
| GEN | Antiparallel β-sheet | 1691 | - | |
| Turns and loops | 1675 | - | ||
| 310-helix | 1661 | - | ||
| α-helix | 1646 | + | ||
| β-sheet | 1633 | + | ||
| Aggregated | 1616 | + | ||
| GA | Turns and loops/Anti β | 1683 | + | |
| Turns and loops | 1674 | + | ||
| 310-helix | 1665 | + | ||
| Unordered | 1656 | - | ||
| α-helix | 1647 | - | ||
| β-sheet | 1637 | - | ||
| β-sheet | 1621 | + | ||
| Aggregated | 1612 | + |
| Enzymatic System | Lactobionic Acid Concentration [mM] | Conversion Efficiencies [%] |
|---|---|---|
| Control PchCDH + CuLAC | 22.66 | 45.32 |
| PchCDH/GA + CuLAC | 8.32 | 14.07 |
| PchCDH/PEI + CuLAC | 43.01 | 93.34 |
| PchCDH/GEN + CuLAC | 14.80 | 26.14 |
| CuLAC/GA + PchCDH | 4.35 | 8.38 |
| CuLAC/PEI + PchCDH | 1.71 | 3.57 |
| CuLAC/GEN + PchCDH | 5.21 | 10.34 |
| PchCDH/GEN + CuLAC/GEN | 0.00 | 0.00 |
| PchCDH/GEN+ CuLAC/PEI | 4.76 | 10.23 |
| PchCDH/GEN + CuLAC/GA | 0.24 | 0.51 |
| PchCDH/GA+ CuLAC/GEN | 0.00 | 0.00 |
| PchCDH/GA + CuLAC/PEI | 0.84 | 1.81 |
| PchCDH/GA+ CuLAC/GA | 0.00 | 0.00 |
| PchCDH/PEI + CuLAC/GEN | 0.74 | 1.41 |
| PchCDH/PEI + CuLAC/PEI | 9.81 | 22.34 |
| PchCDH/PEI + CuLAC/GA | 0.00 | 0.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sulej, J.; Piątek-Gołda, W.; Grąz, M.; Szałapata, K.; Waśko, P.; Janik-Zabrotowicz, E.; Osińska-Jaroszuk, M. Immobilisation of Cellobiose Dehydrogenase and Laccase on Chitosan Particles as a Multi-Enzymatic System for the Synthesis of Lactobionic Acid. J. Funct. Biomater. 2023, 14, 383. https://doi.org/10.3390/jfb14070383
Sulej J, Piątek-Gołda W, Grąz M, Szałapata K, Waśko P, Janik-Zabrotowicz E, Osińska-Jaroszuk M. Immobilisation of Cellobiose Dehydrogenase and Laccase on Chitosan Particles as a Multi-Enzymatic System for the Synthesis of Lactobionic Acid. Journal of Functional Biomaterials. 2023; 14(7):383. https://doi.org/10.3390/jfb14070383
Chicago/Turabian StyleSulej, Justyna, Wiktoria Piątek-Gołda, Marcin Grąz, Katarzyna Szałapata, Piotr Waśko, Ewa Janik-Zabrotowicz, and Monika Osińska-Jaroszuk. 2023. "Immobilisation of Cellobiose Dehydrogenase and Laccase on Chitosan Particles as a Multi-Enzymatic System for the Synthesis of Lactobionic Acid" Journal of Functional Biomaterials 14, no. 7: 383. https://doi.org/10.3390/jfb14070383
APA StyleSulej, J., Piątek-Gołda, W., Grąz, M., Szałapata, K., Waśko, P., Janik-Zabrotowicz, E., & Osińska-Jaroszuk, M. (2023). Immobilisation of Cellobiose Dehydrogenase and Laccase on Chitosan Particles as a Multi-Enzymatic System for the Synthesis of Lactobionic Acid. Journal of Functional Biomaterials, 14(7), 383. https://doi.org/10.3390/jfb14070383






