DNA-Based Single-Molecule Electronics: From Concept to Function
Abstract
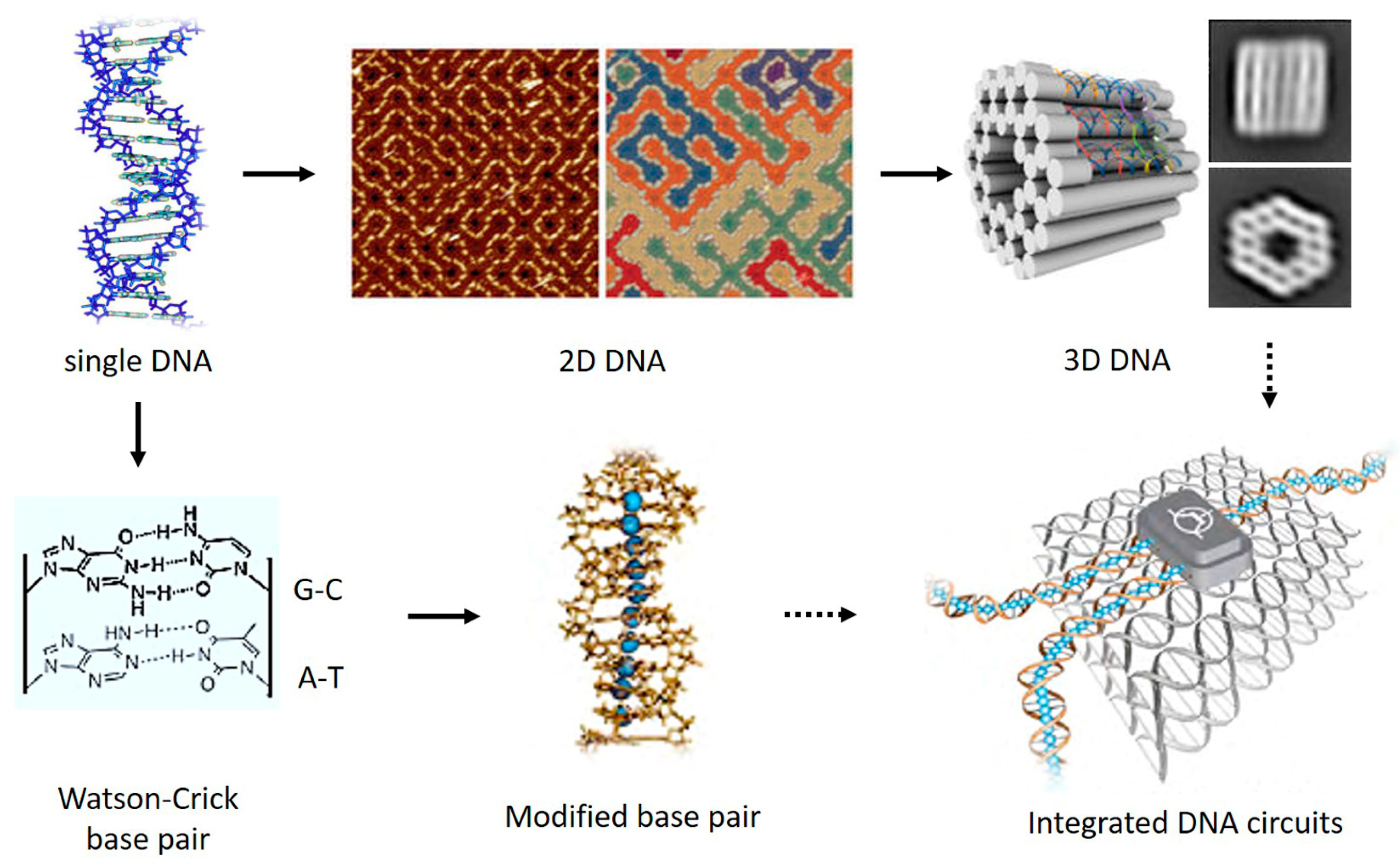
:1. Introduction
2. Experimental Approaches towards DNA Single-Molecule Conductance
3. Charge Transport through Native DNA Molecules
3.1. Environmental Effect: Wet vs. Dry
3.2. Single Strand and Mismatched DNA
3.3. Sequence- and Length-Dependent CT through DNA
3.4. Structure-Dependent Transport in DNA
4. Charge Transport through Modified DNA Molecules
4.1. DNA Methylation
4.2. Metallo-DNA
4.3. DNA-Small Molecule Complex
5. Toward DNA Molecular Diode and Transistor
6. Beyond Simple Charge Transport in DNA
6.1. DNA Spintronics
6.2. DNA Piezoresistivity
6.3. DNA Thermoelectricity
7. Conclusions and Outlook
Acknowledgments
Conflicts of Interest
References
- Aviram, A.; Ratner, M.A. Molecular rectifiers. Chem. Phys. Lett. 1974, 29, 277–283. [Google Scholar] [CrossRef]
- Ratner, M. A brief history of molecular electronics. Nat. Nanotechnol. 2013, 8, 378–381. [Google Scholar] [CrossRef] [PubMed]
- Reed, M.A.; Zhou, C.; Muller, C.J.; Burgin, T.P.; Tour, J.M. Conductance of a molecular junction. Science 1997, 278, 252–254. [Google Scholar] [CrossRef]
- Xu, B.Q.; Tao, N.J. Measurement of single-molecule resistance by repeated formation of molecular junctions. Science 2003, 301, 1221–1223. [Google Scholar] [CrossRef] [PubMed]
- Di Ventra, M.; Pantelides, S.T.; Lang, N.D. First-principles calculation of transport properties of a molecular device. Phys. Rev. Lett. 2000, 84, 979–982. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Datta, S.; Ratner, M.A. First-principles-based matrix green’s function approach to molecular electronic devices: General formalism. Chem. Phys. 2002, 281, 151–170. [Google Scholar] [CrossRef]
- Porath, D.; Bezryadin, A.; de Vries, S.; Dekker, C. Direct measurement of electrical transport through DNA molecules. Nature 2000, 403, 635–638. [Google Scholar] [CrossRef] [PubMed]
- Reichert, J.; Ochs, R.; Beckmann, D.; Weber, H.B.; Mayor, M.; Löhneysen, H.V. Driving current through single organic molecules. Phys. Rev. Lett. 2002, 88, 176804. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Wang, K.; Zerah-Harush, E.; Hamill, J.; Wang, B.; Dubi, Y.; Xu, B. Molecular rectifier composed of DNA with high rectification ratio enabled by intercalation. Nat. Chem. 2016, 8, 484–490. [Google Scholar] [CrossRef] [PubMed]
- Venkataraman, L.; Klare, J.E.; Nuckolls, C.; Hybertsen, M.S.; Steigerwald, M.L. Dependence of single-molecule junction conductance on molecular conformation. Nature 2006, 442, 904–907. [Google Scholar] [CrossRef] [PubMed]
- Jia, C.; Migliore, A.; Xin, N.; Huang, S.; Wang, J.; Yang, Q.; Wang, S.; Chen, H.; Wang, D.; Feng, B.; et al. Covalently bonded single-molecule junctions with stable and reversible photoswitched conductivity. Science 2016, 352, 1443–1445. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.H.; Kim, B.; Frisbie, C.D. Electrical resistance of long conjugated molecular wires. Science 2008, 320, 1482–1486. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Gorodetsky, A.A.; Hone, J.; Barton, J.K.; Nuckolls, C. Conductivity of a single DNA duplex bridging a carbon nanotube gap. Nat. Nanotechnol. 2008, 3, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Quek, S.Y.; Kamenetska, M.; Steigerwald, M.L.; Choi, H.J.; Louie, S.G.; Hybertsen, M.S.; Neaton, J.B.; Venkataraman, L. Mechanically controlled binary conductance switching of a single-molecule junction. Nat. Nanotechnol. 2009, 4, 230–234. [Google Scholar] [CrossRef] [PubMed]
- Ismael, A.K.; Wang, K.; Vezzoli, A.; Al-Khaykanee, M.K.; Gallagher, H.E.; Grace, I.M.; Lambert, C.J.; Xu, B.; Nichols, R.J.; Higgins, S.J. Side-group-mediated mechanical conductance switching in molecular junctions. Angew. Chem. Int. Ed. 2017, 56, 15378–15382. [Google Scholar] [CrossRef] [PubMed]
- Su, T.A.; Li, H.; Steigerwald, M.L.; Venkataraman, L.; Nuckolls, C. Stereoelectronic switching in single-molecule junctions. Nat. Chem. 2015, 7, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Capozzi, B.; Xia, J.; Adak, O.; Dell, E.J.; Liu, Z.-F.; Taylor, J.C.; Neaton, J.B.; Campos, L.M.; Venkataraman, L. Single-molecule diodes with high rectification ratios through environmental control. Nat. Nanotechnol. 2015, 10, 522–527. [Google Scholar] [CrossRef] [PubMed]
- Diez-Perez, I.; Hihath, J.; Lee, Y.; Yu, L.; Adamska, L.; Kozhushner, M.A.; Oleynik, I.I.; Tao, N. Rectification and stability of a single molecular diode with controlled orientation. Nat. Chem. 2009, 1, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Atesci, H.; Kaliginedi, V.; Celis Gil, J.A.; Ozawa, H.; Thijssen, J.M.; Broekmann, P.; Haga, M.-A.; van der Molen, S.J. Humidity-controlled rectification switching in ruthenium-complex molecular junctions. Nat. Nanotechnol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Reed, M.A.; Rawlett, A.M.; Tour, J.M. Large on-off ratios and negative differential resistance in a molecular electronic device. Science 1999, 286, 1550–1552. [Google Scholar] [CrossRef] [PubMed]
- Perrin, M.L.; Frisenda, R.; Koole, M.; Seldenthuis, J.S.; GilJose, A.C.; Valkenier, H.; Hummelen, J.C.; Renaud, N.; Grozema, F.C.; Thijssen, J.M.; et al. Large negative differential conductance in single-molecule break junctions. Nat. Nanotechnol. 2014, 9, 830–834. [Google Scholar] [CrossRef] [PubMed]
- Guédon, C.M.; Valkenier, H.; Markussen, T.; Thygesen, K.S.; Hummelen, J.C.; van der Molen, S.J. Observation of quantum interference in molecular charge transport. Nat. Nanotechnol. 2012, 7, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Sangtarash, S.; Reber, D.; Zhang, D.; Sadeghi, H.; Shi, J.; Xiao, Z.-Y.; Hong, W.; Lambert, C.J.; Liu, S.-X. Gating of quantum interference in molecular junctions by heteroatom substitution. Angew. Chem. Int. Ed. 2017, 56, 173–176. [Google Scholar] [CrossRef] [PubMed]
- Manrique, D.Z.; Huang, C.; Baghernejad, M.; Zhao, X.; Al-Owaedi, O.A.; Sadeghi, H.; Kaliginedi, V.; Hong, W.; Gulcur, M.; Wandlowski, T.; et al. A quantum circuit rule for interference effects in single-molecule electrical junctions. Nat. Commun. 2015, 6, 6389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.; Jeong, W.; Kim, K.; Lee, W.; Reddy, P. Electrostatic control of thermoelectricity in molecular junctions. Nat. Nanotechnol. 2014, 9, 881–885. [Google Scholar] [CrossRef] [PubMed]
- Reddy, P.; Jang, S.-Y.; Segalman, R.A.; Majumdar, A. Thermoelectricity in molecular junctions. Science 2007, 315, 1568–1571. [Google Scholar] [CrossRef] [PubMed]
- Battacharyya, S.; Kibel, A.; Kodis, G.; Liddell, P.A.; Gervaldo, M.; Gust, D.; Lindsay, S. Optical modulation of molecular conductance. Nano Lett. 2011, 11, 2709–2714. [Google Scholar] [CrossRef] [PubMed]
- Galperin, M.; Nitzan, A. Molecular optoelectronics: The interaction of molecular conduction junctions with light. Phys. Chem. Chem. Phys. 2012, 14, 9421–9438. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, K.; Xu, B.; Dubi, Y. Photo-conductance from exciton binding in molecular junctions. J. Am. Chem. Soc. 2018, 140, 70–73. [Google Scholar] [CrossRef] [PubMed]
- Aragonès, A.C.; Aravena, D.; Cerdá, J.I.; Acís-Castillo, Z.; Li, H.; Real, J.A.; Sanz, F.; Hihath, J.; Ruiz, E.D. Large conductance switching in a single-molecule device through room temperature spin-dependent transport. Nano Lett. 2016, 16, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Schwöbel, J.; Fu, Y.; Brede, J.; Dilullo, A.; Hoffmann, G.; Klyatskaya, S.; Ruben, M.; Wiesendanger, R. Real-space observation of spin-split molecular orbitals of adsorbed single-molecule magnets. Nat. Commun. 2012, 3, 953. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Miao, R.; Wang, K.; Thompson, D.; Zotti, L.A.; Cuevas, J.C.; Meyhofer, E.; Reddy, P. Peltier cooling in molecular junctions. Nat. Nanotechnol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Meyer, T.A.; Pan, V.; Dutta, P.K.K. The beauty and utility of DNA origami. Chem 2017, 2, 359–382. [Google Scholar] [CrossRef]
- Tikhomirov, G.; Petersen, P.; Qian, L. Programmable disorder in random DNA tilings. Nat. Nanotechnol. 2016, 12, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Hall, D.B.; Holmlin, R.E.; Barton, J.K. Oxidative DNA damage through long-range electron transfer. Nature 1996, 382, 731–735. [Google Scholar] [CrossRef] [PubMed]
- Saito, I.; Nakamura, T.; Nakatani, K.; Yoshioka, Y.; Yamaguchi, K.; Sugiyama, H. Mapping of the hot spots for DNA damage by one-electron oxidation: Efficacy of gg doublets and ggg triplets as a trap in long-range hole migration. J. Am. Chem. Soc. 1998, 120, 12686–12687. [Google Scholar] [CrossRef]
- Eley, D.D.; Spivey, D.I. Semiconductivity of organic substances. Part 9.—Nucleic acid in the dry state. Trans. Faraday Soc. 1962, 58, 411–415. [Google Scholar] [CrossRef]
- Holmlin, R.E.; Dandliker, P.J.; Barton, J.K. Charge transfer through the DNA base stack. Angew. Chem. Int. Ed. 1997, 36, 2714–2730. [Google Scholar] [CrossRef]
- Taniguchi, M.; Kawai, T. DNA electronics. Phys. E 2006, 33, 1–12. [Google Scholar] [CrossRef]
- Genereux, J.C.; Barton, J.K. Molecular electronics: DNA charges ahead. Nat. Chem. 2009, 1, 106–107. [Google Scholar] [CrossRef] [PubMed]
- Genereux, J.C.; Barton, J.K. Mechanisms for DNA charge transport. Chem. Rev. 2010, 110, 1642–1662. [Google Scholar] [CrossRef] [PubMed]
- Kasumov, A.Y.; Kociak, M.; Guéron, S.; Reulet, B.; Volkov, V.T.; Klinov, D.V.; Bouchiat, H. Proximity-induced superconductivity in DNA. Science 2001, 291, 280–282. [Google Scholar] [CrossRef] [PubMed]
- Braun, E.; Eichen, Y.; Sivan, U.; Ben-Yoseph, G. DNA-templated assembly and electrode attachment of a conducting silver wire. Nature 1998, 391, 775–778. [Google Scholar] [CrossRef] [PubMed]
- Tran, P.; Alavi, B.; Gruner, G. Charge transport along the λ-DNA double helix. Phys. Rev. Lett. 2000, 85, 1564. [Google Scholar] [CrossRef] [PubMed]
- Fink, H.W.; Schonenberger, C. Electrical conduction through DNA molecules. Nature 1999, 398, 407–410. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Zhang, P.; Li, X.; Tao, N. Direct conductance measurement of single DNA molecules in aqueous solution. Nano Lett. 2004, 4, 1105–1108. [Google Scholar] [CrossRef]
- Aradhya, S.V.; Venkataraman, L. Single-molecule junctions beyond electronic transport. Nat. Nanotechnol. 2013, 8, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Xu, B. Modulation and control of charge transport through single-molecule junctions. Top. Curr. Chem. 2017, 375, 17. [Google Scholar] [CrossRef] [PubMed]
- Xiang, D.; Wang, X.; Jia, C.; Lee, T.; Guo, X. Molecular-scale electronics: From concept to function. Chem. Rev. 2016, 116, 4318–4440. [Google Scholar] [CrossRef] [PubMed]
- Cohen, H.; Nogues, C.; Naaman, R.; Porath, D. Direct measurement of electrical transport through single DNA molecules of complex sequence. Proc. Natl. Acad. Sci. USA 2005, 102, 11589–11593. [Google Scholar] [CrossRef] [PubMed]
- Aradhya, S.V.; Frei, M.; Hybertsen, M.S.; Venkataraman, L. Van der waals interactions at metal/organic interfaces at the single-molecule level. Nat. Mater. 2012, 11, 872–876. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Hamill, J.M.; Zhou, J.; Xu, B. Mapping the details of contact effect of modulated au-octanedithiol-au break junction by force–conductance cross-correlation. J. Am. Chem. Soc. 2014, 136, 17406–17409. [Google Scholar] [CrossRef] [PubMed]
- Engelkes, V.B.; Beebe, J.M.; Frisbie, C.D. Length-dependent transport in molecular junctions based on sams of alkanethiols and alkanedithiols: Effect of metal work function and applied bias on tunneling efficiency and contact resistance. J. Am. Chem. Soc. 2004, 126, 14287–14296. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Choi, S.H.; Zhu, X.Y.; Frisbie, C.D. Molecular tunnel junctions based on π-conjugated oligoacene thiols and dithiols between ag, au, and pt contacts: Effect of surface linking group and metal work function. J. Am. Chem. Soc. 2011, 133, 19864–19877. [Google Scholar] [CrossRef] [PubMed]
- Xiang, D.; Jeong, H.; Lee, T.; Mayer, D. Mechanically controllable break junctions for molecular electronics. Adv. Mater. 2013, 25, 4845–4867. [Google Scholar] [CrossRef] [PubMed]
- Xiang, D.; Jeong, H.; Kim, D.; Lee, T.; Cheng, Y.; Wang, Q.; Mayer, D. Three-terminal single-molecule junctions formed by mechanically controllable break junctions with side gating. Nano Lett. 2013, 13, 2809–2813. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Small, J.P.; Klare, J.E.; Wang, Y.; Purewal, M.S.; Tam, I.W.; Hong, B.H.; Caldwell, R.; Huang, L.; Brien, S.; et al. Covalently bridging gaps in single-walled carbon nanotubes with conducting molecules. Science 2006, 311, 356–359. [Google Scholar] [CrossRef] [PubMed]
- Mol, J.A.; Lau, C.S.; Lewis, W.J.M.; Sadeghi, H.; Roche, C.; Cnossen, A.; Warner, J.H.; Lambert, C.J.; Anderson, H.L.; Briggs, G.A.D. Graphene-porphyrin single-molecule transistors. Nanoscale 2015, 7, 13181–13185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadeghi, H.; Mol, J.A.; Lau, C.S.; Briggs, G.A.D.; Warner, J.; Lambert, C.J. Conductance enlargement in picoscale electroburnt graphene nanojunctions. Proc. Natl. Acad. Sci. USA 2015, 112, 2658–2663. [Google Scholar] [CrossRef] [PubMed]
- Tao, N.J. Electron transport in molecular junctions. Nat. Nanotechnol. 2006, 1, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Nichols, R.J.; Haiss, W.; Higgins, S.J.; Leary, E.; Martin, S.; Bethell, D. The experimental determination of the conductance of single molecules. Phys. Chem. Chem. Phys. 2010, 12, 2801–2815. [Google Scholar] [CrossRef] [PubMed]
- Endres, R.G.; Cox, D.L.; Singh, R.R.P. Colloquium: The quest for high-conductance DNA. Rev. Mod. Phys. 2004, 76, 195–214. [Google Scholar] [CrossRef]
- Porath, D.; Cuniberti, G.; Di Felice, R. Charge transport in DNA-based devices. Top. Curr. Chem. 2004, 237, 183–228. [Google Scholar]
- Mantz, Y.A.; Gervasio, F.L.; Laino, T.; Parrinello, M. Solvent effects on charge spatial extent in DNA and implications for transfer. Phys. Rev. Lett. 2007, 99, 058104. [Google Scholar] [CrossRef] [PubMed]
- Van Zalinge, H.; Schiffrin, D.J.; Bates, A.D.; Haiss, W.; Ulstrup, J.; Nichols, R.J. Single-molecule conductance measurements of single- and double-stranded DNA oligonucleotides. ChemPhysChem 2006, 7, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, P.; Yamane, K. DNA mismatch repair: Molecular mechanism, cancer, and ageing. Mech. Ageing Dev. 2008, 129, 391–407. [Google Scholar] [CrossRef] [PubMed]
- Iyer, R.R.; Pluciennik, A.; Burdett, V.; Modrich, P.L. DNA mismatch repair: Functions and mechanisms. Chem. Rev. 2006, 106, 302–323. [Google Scholar] [CrossRef] [PubMed]
- Hihath, J.; Xu, B.; Zhang, P.; Tao, N. Study of single-nucleotide polymorphisms by means of electrical conductance measurements. Proc. Natl. Acad. Sci. USA 2005, 102, 16979–16983. [Google Scholar] [CrossRef] [PubMed]
- Lambert, C.J. Basic concepts of quantum interference and electron transport in single-molecule electronics. Chem. Soc. Rev. 2015, 44, 875–888. [Google Scholar] [CrossRef] [PubMed]
- Lewis, F.D.; Liu, X.; Liu, J.; Miller, S.E.; Hayes, R.T.; Wasielewski, M.R. Direct measurement of hole transport dynamics in DNA. Nature 2000, 406, 51–53. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Palma, J.L.; Bruot, C.; Mujica, V.; Ratner, M.A.; Tao, N. Intermediate tunnelling–hopping regime in DNA charge transport. Nat. Chem. 2015, 7, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Schuster, G.B. Long-range charge transfer in DNA: Transient structural distortions control the distance dependence. Acc. Chem. Res. 2000, 33, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Giese, B. Long-distance charge transport in DNA: The hopping mechanism. Acc. Chem. Res. 2000, 33, 631–636. [Google Scholar] [CrossRef] [PubMed]
- Berlin, Y.A.; Burin, A.L.; Ratner, M.A. Charge hopping in DNA. J. Am. Chem. Soc. 2001, 123, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Giese, B.; Amaudrut, J.; Köhler, A.-K.; Spormann, M.; Wessely, S. Direct observation of hole transfer through DNA by hopping between adenine bases and by tunnelling. Nature 2001, 412, 318–320. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiang, L.; Palma, J.L.; Asai, Y.; Tao, N. Thermoelectric effect and its dependence on molecular length and sequence in single DNA molecules. Nat. Commun. 2016, 7, 11294. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Xiang, L.; Zhang, Y.; Zhang, P.; Beratan, D.N.; Li, Y.; Tao, N. Engineering nanometre-scale coherence in soft matter. Nat. Chem. 2016, 8, 941–945. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Hamill, J.M.; Wang, B.; Guo, C.; Jiang, S.; Huang, Z.; Xu, B. Structure determined charge transport in single DNA molecule break junctions. Chem. Sci. 2014, 5, 3425–3431. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, F.H.C. Molecular structure of nucleic acids. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, V.I.; Minchenkova, L.E.; Minyat, E.E.; Frank-Kamenetskii, M.D.; Schyolkina, A.K. The b to a transition of DNA in solution. J. Mol. Biol. 1974, 87, 817–833. [Google Scholar] [CrossRef]
- Zheng, J.; Li, Z.; Wu, A.; Zhou, H. Afm studies of DNA structures on mica in the presence of alkaline earth metal ions. Biophys. Chem. 2003, 104, 37–43. [Google Scholar] [CrossRef]
- Pohl, F.M.; Jovin, T.M. Salt-induced co-operative conformational change of a synthetic DNA: Equilibrium and kinetic studies with poly(dg-dc). J. Mol. Biol. 1972, 67, 375–396. [Google Scholar] [CrossRef]
- Van de Sande, J.H.; Jovin, T.M. Z* DNA, the left-handed helical form of poly[d(g-c)] in mgcl2-ethanol, is biologically active. EMBO J. 1982, 1, 115–120. [Google Scholar] [PubMed]
- Van de Sande, J.H.; McIntosh, L.P.; Jovin, T.M. Mn2+ and other transition metals at low concentration induce the right-to-left helical transformation of poly[d(G-C)]. EMBO J. 1982, 1, 777–782. [Google Scholar] [PubMed]
- Vasudevaraju, P.; Bharathi; Garruto, R.M.; Sambamurti, K.; Rao, K.S.J. Role of DNA dynamics in alzheimer’s disease. Brain Res. Rev. 2008, 58, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Hamada, H.; Petrino, M.G.; Kakunaga, T. A novel repeated element with z-DNA-forming potential is widely found in evolutionarily diverse eukaryotic genomes. Proc. Natl. Acad. Sci. USA 1982, 79, 6465–6469. [Google Scholar] [CrossRef] [PubMed]
- Rich, A.; Nordheim, A.; Wang, A.H.J. The chemistry and biology of left-handed z-DNA. Annu. Rev. Biochem. 1984, 53, 791–846. [Google Scholar] [CrossRef] [PubMed]
- Artés, J.M.; Li, Y.; Qi, J.; Anantram, M.P.; Hihath, J. Conformational gating of DNA conductance. Nat. Commun. 2015, 6, 8870. [Google Scholar] [CrossRef] [PubMed]
- Cluzel, P.; Lebrun, A.; Heller, C.; Lavery, R.; Viovy, J.-L.; Chatenay, D.; Caron, F. DNA: An extensible molecule. Science 1996, 271, 792–794. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.B.; Cui, Y.; Bustamante, C. Overstretching b-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science 1996, 271, 795–799. [Google Scholar] [CrossRef] [PubMed]
- Lebrun, A.; Lavery, R. Modelling extreme stretching of DNA. Nucleic Acids Res. 1996, 24, 2260–2267. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, H.; Le, S.; Rouzina, I.; Doyle, P.S.; Yan, J. Revealing the competition between peeled ssdna, melting bubbles, and s-DNA during DNA overstretching by single-molecule calorimetry. Proc. Natl. Acad. Sci. USA 2013, 110, 3865–3870. [Google Scholar] [CrossRef] [PubMed]
- Rouzina, I.; Bloomfield, V.A. Force-induced melting of the DNA double helix 1. Thermodynamic analysis. Biophys. J. 2001, 80, 882–893. [Google Scholar] [CrossRef]
- Roe, D.R.; Chaka, A.M. Structural basis of pathway-dependent force profiles in stretched DNA. J. Phys. Chem. B 2009, 113, 15364–15371. [Google Scholar] [CrossRef] [PubMed]
- Nath, S.; Modi, T.; Mishra, R.K.; Giri, D.; Mandal, B.P.; Kumar, S. Statistical mechanics of DNA rupture: Theory and simulations. J. Chem. Phys. 2013, 139, 165101. [Google Scholar] [CrossRef] [PubMed]
- Wolter, M.; Elstner, M.; Kubař, T. On the structure and stretching of microhydrated DNA. J. Phys. Chem. A 2011, 115, 11238–11247. [Google Scholar] [CrossRef] [PubMed]
- Bruot, C.; Xiang, L.; Palma, J.L.; Tao, N. Effect of mechanical stretching on DNA conductance. ACS Nano 2015, 9, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Chaurasiya, K.R.; Paramanathan, T.; McCauley, M.J.; Williams, M.C. Biophysical characterization of DNA binding from single molecule force measurements. Phys. Life Rev. 2010, 7, 299–341. [Google Scholar] [CrossRef] [PubMed]
- De Gennes, P.-G. Maximum pull out force on DNA hybrids. Comptes Rendus Acad. Sci. 2001, 2, 1505–1508. [Google Scholar] [CrossRef] [Green Version]
- Bruot, C.; Xiang, L.; Palma, J.L.; Li, Y.; Tao, N. Tuning the electromechanical properties of single DNA molecular junctions. J. Am. Chem. Soc. 2015, 137, 13933–13937. [Google Scholar] [CrossRef] [PubMed]
- Livshits, G.I.; Stern, A.; Rotem, D.; Borovok, N.; Eidelshtein, G.; Migliore, A.; Penzo, E.; Wind, S.J.; Di Felice, R.; Skourtis, S.S.; et al. Long-range charge transport in single g-quadruplex DNA molecules. Nat. Nanotechnol. 2014, 9, 1040–1046. [Google Scholar] [CrossRef] [PubMed]
- Shapir, E.; Sagiv, L.; Molotsky, T.; Kotlyar, A.B.; Felice, R.D.; Porath, D. Electronic structure of g4-DNA by scanning tunneling spectroscopy. J. Phys. Chem. C 2010, 114, 22079–22084. [Google Scholar] [CrossRef]
- Liu, S.-P.; Weisbrod, S.H.; Tang, Z.; Marx, A.; Scheer, E.; Erbe, A. Direct measurement of electrical transport through g-quadruplex DNA with mechanically controllable break junction electrodes. Angew. Chem. Int. Ed. 2010, 49, 3313–3316. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Vedala, H.; Roy, A.D.; Kim, D.-H.; Doud, M.; Mathee, K.; Shin, H.-K.; Shimamoto, N.; Prasad, V.; Choi, W. Direct electrical measurements on single-molecule genomic DNA using single-walled carbon nanotubes. Nano Lett. 2008, 8, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Kubař, T.; Woiczikowski, P.B.; Cuniberti, G.; Elstner, M. Efficient calculation of charge-transfer matrix elements for hole transfer in DNA. J. Phys. Chem. B 2008, 112, 7937–7947. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.M.; Yang, Z.; Zhu, H.J.; Xiong, S.J. Influence of backbone on the charge transport properties of g4-DNA molecules: A model-based calculation. J. Phys. Condens. Matter 2010, 22, 065102. [Google Scholar] [CrossRef] [PubMed]
- Di Felice, R.; Calzolari, A.; Garbesi, A.; Alexandre, S.S.; Soler, J.M. Strain-dependence of the electronic properties in periodic quadruple helical g4-wires. J. Phys. Chem. B 2005, 109, 22301–22307. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhang, X.; Luo, W.; Wang, Z.; Guo, X.; Steigerwald, M.L.; Fang, X. Single-molecule detection of proteins using aptamer-functionalized molecular electronic devices. Angew. Chem. Int. Ed. 2011, 50, 2496–2502. [Google Scholar] [CrossRef] [PubMed]
- Tsutsui, M.; Matsubara, K.; Ohshiro, T.; Furuhashi, M.; Taniguchi, M.; Kawai, T. Electrical detection of single methylcytosines in a DNA oligomer. J. Am. Chem. Soc. 2011, 133, 9124–9128. [Google Scholar] [CrossRef] [PubMed]
- Hihath, J.; Guo, S.Y.; Zhang, P.M.; Tao, N.J. Effects of cytosine methylation on DNA charge transport. J. Phys. Condens. Matter 2012, 24, 164204. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Clever, G.H.; Takezawa, Y.; Kaneko, M.; Tanaka, K.; Guo, X.; Shionoya, M. Direct conductance measurement of individual metallo-DNA duplexes within single-molecule break junctions. Angew. Chem. Int. Ed. 2011, 50, 8886–8890. [Google Scholar] [CrossRef] [PubMed]
- Harashima, T.; Kojima, C.; Fujii, S.; Kiguchi, M.; Nishino, T. Single-molecule conductance of DNA gated and ungated by DNA-binding molecules. Chem. Commun. 2017, 53, 10378–10381. [Google Scholar] [CrossRef] [PubMed]
- Meissner, A.; Mikkelsen, T.S.; Gu, H.; Wernig, M.; Hanna, J.; Sivachenko, A.; Zhang, X.; Bernstein, B.E.; Nusbaum, C.; Jaffe, D.B.; et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008, 454, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.-M.; et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009, 462, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Clever, G.H.; Shionoya, M. Metal–base pairing in DNA. Coord. Chem. Rev. 2010, 254, 2391–2402. [Google Scholar] [CrossRef]
- Clever, G.H.; Kaul, C.; Carell, T. DNA–metal base pairs. Angew. Chem. Int. Ed. 2007, 46, 6226–6236. [Google Scholar] [CrossRef] [PubMed]
- Nokhrin, S.; Baru, M.; Lee, J.S. A field-effect transistor from m-DNA. Nanotechnology 2007, 18, 095205. [Google Scholar] [CrossRef]
- Joseph, J.; Schuster, G.B. Long-distance radical cation hopping in DNA: The effect of thymine–hg(II)–thymine base pairs. Org. Lett. 2007, 9, 1843–1846. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S.; Tada, T.; Komoto, Y.; Osuga, T.; Murase, T.; Fujita, M.; Kiguchi, M. Rectifying electron-transport properties through stacks of aromatic molecules inserted into a self-assembled cage. J. Am. Chem. Soc. 2015, 137, 5939–5947. [Google Scholar] [CrossRef] [PubMed]
- Ihmels, H.; Otto, D. Intercalation of organic dye molecules into double-stranded DNA. Top. Curr. Chem. 2005, 258, 161–204. [Google Scholar]
- Carrondo, M.A.; Coll, M.; Aymami, J.; Wang, A.H.; van der Marel, G.A.; van Boom, J.H.; Rich, A. Binding of a hoechst dye to d(cgcgatatcgcg) and its influence on the conformation of the DNA fragment. Biochemistry 1989, 28, 7849–7859. [Google Scholar] [CrossRef] [PubMed]
- Ashwell, G.J.; Tyrrell, W.D.; Whittam, A.J. Molecular rectification: Self-assembled monolayers in which donor-(pi-bridge)-acceptor moieties are centrally located and symmetrically coupled to both gold electrodes. J. Am. Chem. Soc. 2004, 126, 7102–7110. [Google Scholar] [CrossRef] [PubMed]
- Nijhuis, C.A.; Reus, W.F.; Whitesides, G.M. Mechanism of rectification in tunneling junctions based on molecules with asymmetric potential drops. J. Am. Chem. Soc. 2010, 132, 18386–18401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yee, S.K.; Sun, J.; Darancet, P.; Tilley, T.D.; Majumdar, A.; Neaton, J.B.; Segalman, R.A. Inverse rectification in donor-acceptor molecular heterojunctions. ACS Nano 2011, 5, 9256–9263. [Google Scholar] [CrossRef] [PubMed]
- Hihath, J.; Bruot, C.; Nakamura, H.; Asai, Y.; Diez-Perez, I.; Lee, Y.; Yu, L.; Tao, N. Inelastic transport and low-bias rectification in a single-molecule diode. ACS Nano 2011, 5, 8331–8339. [Google Scholar] [CrossRef] [PubMed]
- Metzger, R.M. Unimolecular electronics. Chem. Rev. 2015, 115, 5056–5115. [Google Scholar] [CrossRef] [PubMed]
- Jia, C.C.; Guo, X.F. Molecule-electrode interfaces in molecular electronic devices. Chem. Soc. Rev. 2013, 42, 5642–5660. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhou, J.; Hamill, J.M.; Xu, B. Measurement and understanding of single-molecule break junction rectification caused by asymmetric contacts. J. Chem. Phys. 2014, 141, 054712. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Palma, J.L.; Li, Y.; Mujica, V.; Ratner, M.A.; Tao, N. Gate-controlled conductance switching in DNA. Nat. Commun. 2017, 8, 14471. [Google Scholar] [CrossRef] [PubMed]
- Beratan, D.N.; Naaman, R.; Waldeck, D.H. Charge and spin transport through nucleic acids. Curr. Opin. Electrochem. 2017, 4, 175–181. [Google Scholar] [CrossRef]
- Göhler, B.; Hamelbeck, V.; Markus, T.Z.; Kettner, M.; Hanne, G.F.; Vager, Z.; Naaman, R.; Zacharias, H. Spin selectivity in electron transmission through self-assembled monolayers of double-stranded DNA. Science 2011, 331, 894–897. [Google Scholar] [CrossRef] [PubMed]
- Zwang, T.J.; Hürlimann, S.; Hill, M.G.; Barton, J.K. Helix-dependent spin filtering through the DNA duplex. J. Am. Chem. Soc. 2016, 138, 15551–15554. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Markus, T.Z.; Cohen, S.R.; Vager, Z.; Gutierrez, R.; Naaman, R. Spin specific electron conduction through DNA oligomers. Nano Lett. 2011, 11, 4652–4655. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.Q.; Xiao, X.Y.; Tao, N.J. Measurements of single-molecule electromechanical properties. J. Am. Chem. Soc. 2003, 125, 16164–16165. [Google Scholar] [CrossRef] [PubMed]
- Diez-Perez, I.; Hihath, J.; Hines, T.; Wang, Z.-S.; Zhou, G.; Mullen, K.; Tao, N. Controlling single-molecule conductance through lateral coupling of [pi] orbitals. Nat. Nanotechnol. 2011, 6, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Rascón-Ramos, H.; Artés, J.M.; Li, Y.; Hihath, J. Binding configurations and intramolecular strain in single-molecule devices. Nat. Mater. 2015, 14, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Bruot, C.; Palma, J.L.; Xiang, L.; Mujica, V.; Ratner, M.A.; Tao, N. Piezoresistivity in single DNA molecules. Nat. Commun. 2015, 6, 8032. [Google Scholar] [CrossRef] [PubMed]
- Mahan, G.D.; Sofo, J.O. The best thermoelectric. Proc. Natl. Acad. Sci. USA 1996, 93, 7436–7439. [Google Scholar] [CrossRef] [PubMed]
- Rincon-Garcia, L.; Evangeli, C.; Rubio-Bollinger, G.; Agrait, N. Thermopower measurements in molecular junctions. Chem. Soc. Rev. 2016, 45, 4285–4306. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Miao, R.; Jiang, C.; Meyhofer, E.; Reddy, P. Perspective: Thermal and thermoelectric transport in molecular junctions. J. Chem. Phys. 2017, 146, 092201. [Google Scholar] [CrossRef]
- Tikhomirov, G.; Petersen, P.; Qian, L. Fractal assembly of micrometre-scale DNA origami arrays with arbitrary patterns. Nature 2017, 552, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Praetorius, F.; Kick, B.; Behler, K.L.; Honemann, M.N.; Weuster-Botz, D.; Dietz, H. Biotechnological mass production of DNA origami. Nature 2017, 552, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Wagenbauer, K.F.; Sigl, C.; Dietz, H. Gigadalton-scale shape-programmable DNA assemblies. Nature 2017, 552, 78–83. [Google Scholar] [CrossRef] [PubMed]





















© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, K. DNA-Based Single-Molecule Electronics: From Concept to Function. J. Funct. Biomater. 2018, 9, 8. https://doi.org/10.3390/jfb9010008
Wang K. DNA-Based Single-Molecule Electronics: From Concept to Function. Journal of Functional Biomaterials. 2018; 9(1):8. https://doi.org/10.3390/jfb9010008
Chicago/Turabian StyleWang, Kun. 2018. "DNA-Based Single-Molecule Electronics: From Concept to Function" Journal of Functional Biomaterials 9, no. 1: 8. https://doi.org/10.3390/jfb9010008
APA StyleWang, K. (2018). DNA-Based Single-Molecule Electronics: From Concept to Function. Journal of Functional Biomaterials, 9(1), 8. https://doi.org/10.3390/jfb9010008