Biogenic Selenium Nanoparticles: A Fine Characterization to Unveil Their Thermodynamic Stability
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Bacterial Growth and Loss of Thiol Pool as a Consequence of SeO32− Bioprocessing
2.3. Biogenic Selenium Nanoparticles Preparation
2.4. SeNP Characterization
2.5. Fourier Transform Infrared Spectroscopy in Attenuated Totale Reflectance (ATR-FITR) Mode
2.6. Density Functional Theory (DFT) Calculations
2.7. Multivariate Statistical Analysis of ATR-FTIR Spectra
3. Results and Discussion
3.1. Bacterial Tolerance towards SeO32− Oxyanion
3.2. Characterization of Biogenic SeNPs
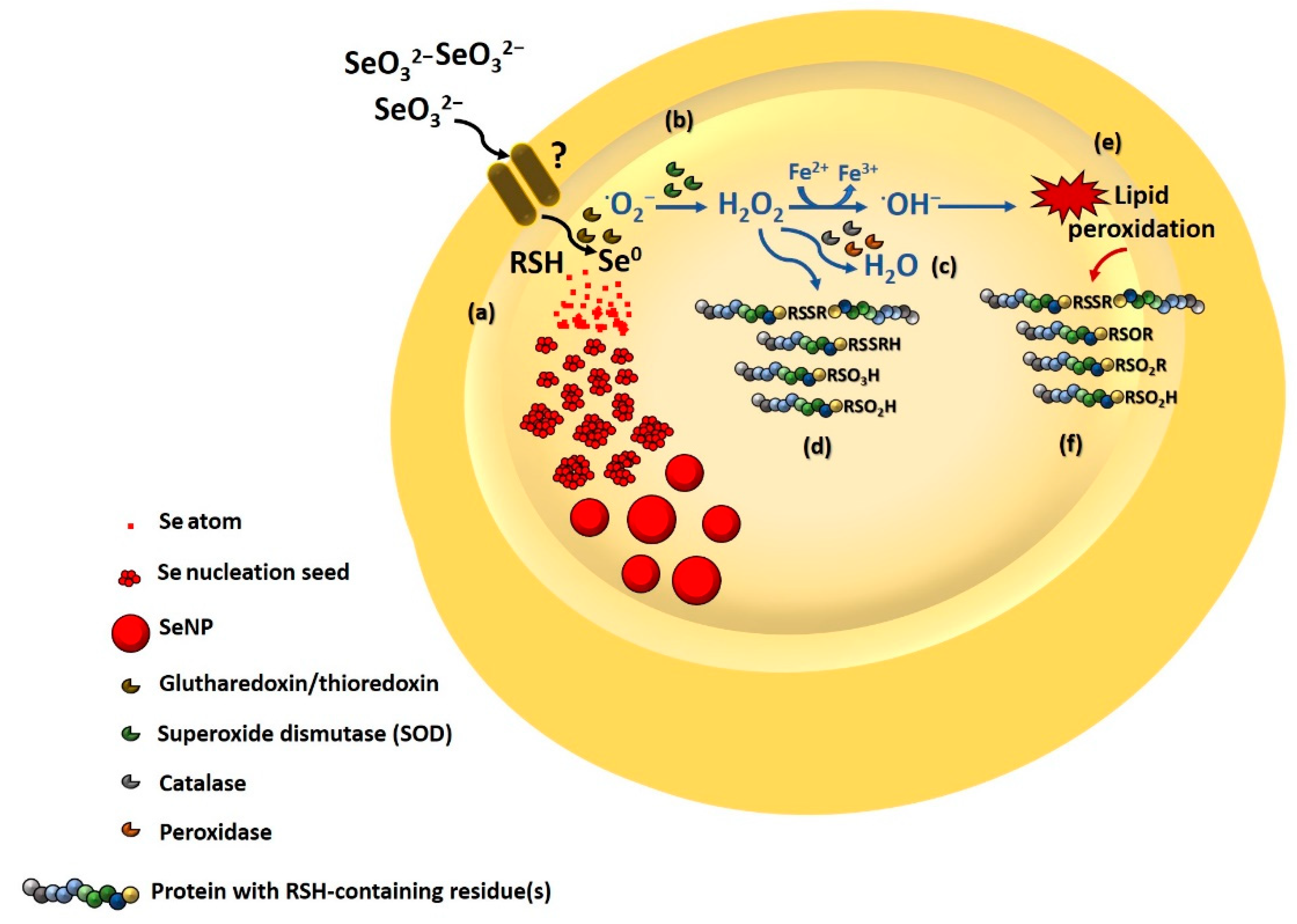
3.3. Biomolecules Involved in SeO32− Bioprocessing and SeNP Stabilization
3.3.1. Lipids
3.3.2. Proteins
3.3.3. Polysaccharides and Nucleic Acids
3.3.4. Involvement of Thiol Chemistry
3.3.5. Biomolecule Contribution to SeNP Stability
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rao, C.N.R.; Muller, A.; Cheetham, A.K. The Chemistry of Nanomaterials: Synthesis, Properties and Applications; WILEY-VCH Verlag GmbH & Co. KgaA: Weinheim, Germany, 2004; pp. 1–11. [Google Scholar] [CrossRef]
- Piacenza, E.; Presentato, A.; Zonaro, E.; Lampis, S.; Vallini, G.; Turner, R.J. Selenium and Tellurium Nanomaterials. Phys. Sci. Rev. 2018, 3, 20170100. [Google Scholar] [CrossRef]
- Ilyas, S.; Kim, M.S.; Lee, J.C.; Jabeen, A.; Bhatti, H.N. Bio-Reclamation of Strategic and Energy Critical Metals from Secondary Resources. Metals 2017, 7, 207. [Google Scholar] [CrossRef]
- Khurana, A.; Tekula, S.; Saifi, M.A.; Venkatesh, P.; Godugu, C. Therapeutic applications of selenium nanoparticles. Biomed. Pharmacother. 2019, 111, 802–812. [Google Scholar] [CrossRef] [PubMed]
- Shoeibi, S.; Mozdziak, P.; Golkar-Narenji, A. Biogenesis of Selenium Nanoparticles Using Green Chemistry. Top. Curr. Chem. Z 2017, 375, 88. [Google Scholar] [CrossRef] [PubMed]
- Wadhwani, S.A.; Shedbalkar, U.U.; Singh, R.; Chopade, B.A. Biogenic selenium nanoparticles: Current status and future pro-spects. Appl. Microbiol. Biotechnol. 2016, 100, 2555–2566. [Google Scholar] [CrossRef] [PubMed]
- Painter, E.P. The chemistry and toxicity of selenium compounds with special reference to the selenium problem. Chem. Rev. 1941, 28, 179–213. [Google Scholar] [CrossRef]
- Ganther, H.E. Reduction of the selenotrisulfide derivative of glutathione to a persulfide analog by glutathione reductase. Biochemistry 1971, 10, 4089–4098. [Google Scholar] [CrossRef]
- Kessi, J.; Hanselmann, K.W. Similarities between the abiotic reduction of selenite with glutathione and the dissimilatory reaction mediated by Rhodospirillum rubrum and Escherichia coli. J. Biol. Chem. 2004, 279, 50662–50669. [Google Scholar] [CrossRef]
- Piacenza, E.; Presentato, A.; Turner, R.J. Stability of biogenic metal(loid) nanomaterials related to the colloidal stabilization theory of chemical nanostructures. Crit. Rev. Biotechnol. 2018, 38, 1137–1156. [Google Scholar] [CrossRef]
- Piacenza, E.; Presentato, A.; Di Salvo, F.; Alduina, R.; Ferrara, V.; Minore, V.; Giannusa, A.; Sancataldo, G.; Chillura Martino, D.F. A combined physical-chemical and microbiological approach to unveil the fabrication, provenance, and state of conservation of the Kinkarakawa-gami art. Sci. Rep. 2020, 10, 16072. [Google Scholar] [CrossRef] [PubMed]
- Piacenza, E.; Presentato, A.; Ambrosi, E.; Speghini, A.; Turner, R.J.; Vallini, G.; Lampis, S. Physical-chemical properties of biogenic selenium nanostructures produced by Stenotrophomonas maltophilia SeITE02 and Ochrobactrum sp. MPV1 strains. Front. Microbiol. 2018, 9, 3178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, R.J.; Weiner, J.H.; Taylor, D.E. Tellurite-mediate thiol oxidation in Escherichia coli. Microbiology 1999, 145, 2549–2557. [Google Scholar] [CrossRef] [Green Version]
- Piacenza, E.; Presentato, A.; Bardelli, M.; Lampis, S.; Vallini, G.; Turner, R.J. Influence of bacterial physiology on processing of selenite, biogenesis of nanomaterials and their thermodynamic stability. Molecules 2019, 24, 2532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Presentato, A.; Piacenza, E.; Darbandi, A.; Anikovskiy, M.; Cappelletti, M.; Zannoni, D.; Turner, R.J. Assembly, growth and conductive properties of tellurium nanorods produced by Rhodococcus aetherivorans BCP1. Sci. Rep. 2018, 8, 3923. [Google Scholar] [CrossRef] [Green Version]
- Byler, M.; Susi, H. Examination of the secondary structure of proteins by deconvolved FTIR spectra. Biopolymers 1986, 25, 469–487. [Google Scholar] [CrossRef] [PubMed]
- Zannoni, D.; Borsetti, F.; Harrison, J.J.; Turner, R.J. The bacterial response to the chalcogen metalloids Se and Te. Adv. Microb. Physiol. 2008, 53, 1–71. [Google Scholar] [CrossRef]
- Frish, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16, Revision C.01; Gaussian, Inc.: Wallingford, CT, USA, 2019. [Google Scholar]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti Correlation-Energy Formula into a Functional of the Electron Density. Phys. Rev. B 1988, 37, 785. [Google Scholar] [CrossRef] [Green Version]
- Wilson, A.K.; Woon, D.E.; Peterson, K.A.; Dunning, T.H. Gaussian basis sets for use in correlated molecular calculations. IX. The atoms gallium through krypton. J. Chem. Phys. 1999, 110, 7667. [Google Scholar] [CrossRef]
- Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. A consistent and accurate ab initio parameterization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010, 132, 154104. [Google Scholar] [CrossRef] [Green Version]
- Somerville, G.A.; Said-Salim, B.; Wickman, J.; Raffael, S.J.; Kreiswirth, B.N.; Musser, J.M. Correlation of acetate catabolism and growth yield in Staphylococcus aureus implications for host-pathogen interactions. Infect. Immun. 2003, 71, 4724–4732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rice, K.C.; Bayles, K.W. Molecular control of bacterial death and lysis. Microbiol. Mol. Biol. Rev. 2008, 72, 85–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bebien, M.; Chauvin, J.P.; Adriano, J.M.; Grosse, S.; Vermeglio, A. Effect of selenite on growth and protein synthesis in the phototrophic bacterium Rhodobacter sphaeroides. Appl. Environ. Microbiol. 2001, 67, 4440–4447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piacenza, E.; Presentato, A.; Heyne, B.; Turner, R.J. Tunable photoluminescence properties of Selenium nanoparticles: Biogenic vs chemogenic synthesis. Nanophotonics 2020, 9, 3615–3628. [Google Scholar] [CrossRef]
- Patil, S.; Chandrasekaran, R. Biogenic nanoparticles: A comprehensive perspective in synthesis, characterization, application and its challenges. J. Genet. Eng. Biotechnol. 2020, 18, 67. [Google Scholar] [CrossRef] [PubMed]
- Potocnik, J. Commission recommendation of 18 October 2011 on the definition of nanomaterial. Off. J. Eur. Communities Legis. 2011, L275, 38–40. [Google Scholar]
- Vreeland, E.C.; Watt, J.; Schober, G.B.; Hance, B.G.; Austin, M.J.; Price, A.D.; Fellows, B.D.; Monson, T.C.; Hudak, N.S.; Maldonado-Camargo, L.; et al. Enhanced Nanoparticle Size Control by Extending LaMer’s Mechanism. Chem. Mater. 2015, 27, 6059–6066. [Google Scholar] [CrossRef]
- Tugarova, A.V.; Mamchenkova, P.V.; Dylatova, Y.A.; Kamnev, A.A. FTIR and Raman spectroscopic studies of selenium nanoparticles synthesized by the bacterium Azospirillum thiophilum. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2018, 192, 458–463. [Google Scholar] [CrossRef]
- Presentato, A.; Piacenza, E.; Anikovkiy, M.; Cappelletti, M.; Zannoni, D.; Turner, R.J. Biosynthesis of selenium-nanoparticles and -nanorods as a product of selenite bioconversion by the aerobic bacterium Rhodococcus aetherivorans BCP1. N. Biotechnol. 2018, 41, 1–8. [Google Scholar] [CrossRef]
- Song, J.M.; Zhu, J.H.; Yu, S.H. Crystallization and shape evolution of single crystalline selenium nanorods at liquid-liquid interface: From monodisperse amorphous Se nanospheres towards Se nanorods. J. Phys. Chem. B 2006, 110, 23790–23795. [Google Scholar] [CrossRef]
- Hotze, E.M.; Phenrat, T.; Lowry, G.V. Nanoparticle aggregation: Challenges to understanding transport and reactivity in the environment. J. Environ. Qual. 2010, 39, 1909–1924. [Google Scholar] [CrossRef] [Green Version]
- Israelachvili, J. Chapter 14, Electrostatic forces between surfaces in liquids. In Intermolecular and Surfaces Forces, 2nd ed.; Israelachvili, J., Ed.; Academic Press: Oxford, UK, 1994; pp. 291–340. [Google Scholar]
- Chandramohan, S.; Sundar, K.; Muthukumaran, A. Monodispersed spherical shaped selenium nanoparticles (SeNPs) synthesized by Bacillus subtilis and its toxicity evaluation in zebrafish embryos. Mater. Res. Express 2018, 5, 025020. [Google Scholar] [CrossRef]
- Kora, A.J. Bacillus cereus, selenite-reducing bacterium from contaminated lake of an industrial area: A renewable nanofactory for the synthesis of selenium nanoparticles. Bioresour. Bioprocess. 2018, 5, 30. [Google Scholar] [CrossRef] [Green Version]
- Abbas, H.S.; Baker, D.H.A.; Ahmed, E.A. Cytotoxicity and antimicrobial efficiency of selenium nanoparticles biosynthesized by Spirulina platensis. Arch. Microbiol. 2021, 203, 523–532. [Google Scholar] [CrossRef]
- Hiemenz, P.; Rajagopalan, R. Chapter 11, The electrical double layer and double layer interactions. In Principles of Colloidal and Surface Chemistry, 3rd ed.; Hiemenz, P., Rajagopalan, R., Eds.; Marcel Dekker: New York, NY, USA, 1997; pp. 499–531. [Google Scholar]
- Kamnev, A.A.; Mamchenkova, P.V.; Dylatova, Y.A.; Tugarova, A.V. FTIR spectroscopic studies of selenite reduction by cells of the rhizobacterium Azospirillum brasilense Sp7 and the formation of selenium nanoparticles. J. Mol. Struct. 2017, 1140, 106–112. [Google Scholar] [CrossRef]
- Fischer, S.; Krause, T.; Lederer, F.; Merroun, M.L.; Shevchenko, A.; Hubner, R.; Firkala, T.; Stump, T.; Jordan, N.; Jain, R. Bacillus safensis JG-B5T affects the fate of selenium by extracellular production of colloidally less stable selenium nanoparticles. J. Haz. Mat. 2020, 384, 121146. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.W.; Wang, Y.; Tang, C.; Jia, H.L.; Wu, L. Speeding up selenite bioremediation using the highly selenite-tolerant strain Providencia rettgeri HF16- A novel mechanism of selenite reduction based on proteomic analysis. J. Haz. Mat. 2021, 406, 124690. [Google Scholar] [CrossRef]
- Shakibaie, M.; Shahverdi, A.R.; Faramarzi, M.A.; Hassanzadeh, G.R.; Rahimi, H.R.; Sabzevari, O. Acute and subacute toxicity of novel biogenic selenium nanoparticles in mice. Pharm. Biol. 2013, 51, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Sonkusre, P.; Nanduri, R.; Gupta, P.; Cameotra, S.S. Improved extraction of intracellular biogenic selenium nanoparticles and their specificity for cancer chemoprevention. J. Nanomed. Nanotechnol. 2014, 5, 194. [Google Scholar] [CrossRef] [Green Version]
- Ramya, S.; Shanmugasundaram, T.; Balagurunathan, R. Biomedical potential of actinobacterially synthesized selenium nanoparticles with special reference to anti-biofilm, anti-oxidant, wound healing, cytotoxic and anti-viral activities. J. Trace Elem. Med. Biol. 2015, 32, 30–39. [Google Scholar] [CrossRef]
- Lampis, S.; Zonaro, E.; Bertolini, C.; Cecconi, D.; Monti, F.; Micaroni, M.; Turner, R.J.; Butler, C.S.; Vallini, G. Selenite biotransformation and detoxification by Stenotrophomonas maltophilia SeITE02: Novel clues on the route to bacterial biogenesis of selenium nanoparticles. J. Haz. Mat. 2017, 324, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Zonaro, E.; Piacenza, E.; Presentato, A.; Monti, F.; Dell’Anna, R.; Lampis, S.; Vallini, G. Ochrobactrum sp. MPV1 from a dump of roasted pyrites can be exploited as bacterial catalyst for the biogenesis of selenium and tellurium nanoparticles. Microb. Cell Fact. 2017, 16, 215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, D.; Yang, L.; Wang, Y.; Wang, G.; Rensing, C.; Zheng, S. Proteins enriched in charged amino acids control the formation and stabilization of selenium nanoparticles in Comamonas testosteroni S44. Sci. Rep. 2018, 8, 4766. [Google Scholar] [CrossRef] [PubMed]
- Abdollahnia, M.; Makhdoumi, A.; Mashreghi, M.; Eshghi, H. Exploring the potentials of halophilic prokaryotes from a solar saltern for synthesizing nanoparticles: The case of silver and selenium. PLoS ONE 2020, 15, e0229886. [Google Scholar] [CrossRef] [PubMed]
- San Keskin, N.O.; Vural, O.A.; Abaci, S. Biosynthesis of noble selenium nanoparticles from Lysinibacillus sp. NOSK for antimicrobial, antibiofilm activity, and biocompatibility. Geomicrobiol. J. 2020, 37, 919–928. [Google Scholar] [CrossRef]
- Ashengroph, M.; Hosseini, S.R. A newly isolated Bacillus amyloliquefaciens SRB04 for the synthesis of selenium nanoparticles with potential antibacterial properties. Int. Microbiol. 2021, 24, 103–114. [Google Scholar] [CrossRef]
- Alhazmi, H.A. FT-IR spectroscopy for the identification of binding sites and measurements of the binding interactions of important metal ions with bovine serum albumin. Sci. Pharm. 2019, 87, 5. [Google Scholar] [CrossRef] [Green Version]
- Tugarova, A.V.; Kamnev, A.A. Proteins in microbial synthesis of selenium nanoparticles. Talanta 2017, 174, 539–547. [Google Scholar] [CrossRef]
- Heacock, R.A.; Marion, L. The Infrared spectra of secondary amines and their salts. Can. J. Chem. 1956, 1782–1795. [Google Scholar] [CrossRef]
- Suzuki, M.; Lee, D.Y.; Inyamah, N.; Stadtman, T.C.; Tjandra, N. Solution NMR structure of selenium-binding protein from Methanococcus vannielii. J. Biol. Chem. 2008, 283, 25936–25943. [Google Scholar] [CrossRef] [Green Version]
- Van Laer, K.; Buts, L.; Foloppe, N.; Vertommen, D.; Van Belle, K.; Wahni, K.; Roos, G.; Nilsson, L.; Mateos, L.M.; Mamta, R.; et al. Mycoredoxin-1 is one of the missing links in the oxidative stress defence mechanism of Mycobacteria. Mol. Microbiol. 2012, 86, 787–804. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, K.; Toda, H.; Ikeguchi, M. Dependence of α-helical and β-sheet amino acid propensities on the overall protein fold type. BMC Struct. Biol. 2021, 12, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deponte, M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim. Biophys. Acta. 2013, 1830, 3217–3266. [Google Scholar] [CrossRef] [Green Version]
- Eberle, R.J.; Kawai, L.A.; de Morales, F.R.; Tasic, L.; Arni, R.K.; Coronado, M.A. Biochemical and biophysical characterization of a mycoredoxin protein glutaredoxin A1 from Corynebacterium pseudotuberculosis. Int. J. Biol. Macromol. 2018, 107, 1999–2007. [Google Scholar] [CrossRef]
- Geng, Z.; Song, X.; Xing, Z.; Geng, J.; Zhang, S.; Zhang, X.; Wang, Z. Effects of selenium on the structure and function of recombinant human S-adenosyl-L-methionine dependent arsenic (+3 oxidation state) methyltransferase in E. coli. J. Biol. Inorg. Chem. 2009, 14, 485–496. [Google Scholar] [CrossRef]
- Wadhwani, S.A.; Gorain, M.; Banerjee, P.; Shedbalkar, U.U.; Singh, R.; Kundu, G.C.; Chopade, B.A. Green synthesis of selenium nanoparticles using Acinetobacter sp. SW30: Optimization, characterization and its anticancer activity in breast cancer cells. Int. J. Nanomed. 2017, 12, 6841–6855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Qiao, L.; Guo, Y.; Ma, L.; Cheng, Y. Preparation, characteristics and antioxidant activity of polysaccharides and proteins-capped selenium nanoparticles synthesized by Lactobacillus casei ATCC 393. Carbohydr. Polym. 2018, 195, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Kopec, K.; Pedziwiatr, M.; Gront, D.; Sztatelman, O.; Slawski, J.; Lazicka, M.; Worch, R.; Zawada, K.; Makarova, K.; Nyk, M.; et al. Comparison of α-helix and β-sheet structure adaptation to a quantum dot geometry: Toward the identification of an optimal motif for a protein nanoparticle cover. ACS Omega 2019, 4, 13086–13099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chothia, C.; Janin, J. Orthogonal packing of β -pleated sheets in proteins. Biochemistry 1982, 21, 3955–3965. [Google Scholar] [CrossRef]
- Yang, H.; Yang, S.; Kong, J.; Dong, A.; Yu, S. Obtaining information about protein secondary structures in aqueous solution using Fourier transform IR spectroscopy. Nat. Protoc. 2015, 10, 3. [Google Scholar] [CrossRef]
- Kamnev, A.A. FTIR spectroscopic studies of bacterial cellular responses to environmental factors, plant-bacterial interactions and signalling. Spectroscopy 2008, 22, 83–85. [Google Scholar] [CrossRef]
- Nikonenko, N.A.; Buslov, D.K.; Sushko, N.J.; Zhbankov, R.G. Investigation of stretching vibrations of glycosidic linkages in disaccharides and polysaccharides with use of IR spectra deconvolution. Biopolymers 2000, 57, 257–262. [Google Scholar] [CrossRef]
- Holst, O.; Muller-Loennies, S. 1.04 Microbial polysaccharide structures. In Comprehensive Glycoscience. From Chemistry to System Biology; Kamerling, J.P., Boons, G.J., Lee, Y.C., Suzuki, A., Taniguchi, N., Voragen, A.G.J., Eds.; Elsevier: Oxford, UK, 2007; Volume 1, pp. 123–179. [Google Scholar]
- Lasch, P.; Naumann, D. Infrared spectroscopy in microbiology. In Encyclopedia of Analytical Chemistry; Meyers, R.A., Ed.; Wiley Online Library: Hoboken, NJ, USA, 2015. [Google Scholar] [CrossRef]
- Kim, S.J.; Chang, J.; Singh, M. Peptidoglycan architecture of Gram-positive bacteria by solid-state NMR. Biochim. Biophys. Acta 2015, 1848, 350–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohamed, M.E.; Mohammed, A.M.A. Experimental and computation vibration study of amino acids. Inter. Lett. Chem. Phys. Astr. 2013, 10, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Imber, M.; Pietrzyk-Brzezinska, A.J.; Antelmann, H. Redox regulation by reversible protein S-thiolation in Gram-positive bacteria. Red. Biol. 2019, 20, 130–145. [Google Scholar] [CrossRef] [PubMed]
- Ayala, A.; Munoz, M.F.; Arguelles, S. Lipid peroxidation: Production, metabolism, and signaling mechanisms of malondialdehyde and 4-Hydroxy-2-Nonenal. Oxid. Med. Cell. Long. 2014, 2014, 360438. [Google Scholar] [CrossRef]
- Gardner, H.W. Lipid hydroperoxide reactivity with proteins and amino acids: A review. J. Agric. Food Chem. 1979, 27, 220–229. [Google Scholar] [CrossRef]
- Kiwi, J.; Nadtochenko, V. Evidence for the mechanism of photocatalytic degradation of the bacterial wall membrane at the TiO2 interface by ATR-FTIR and laser kinetic spectroscopy. Langmuir 2005, 21, 4631–4641. [Google Scholar] [CrossRef]
- Nadtochenko, V.A.; Rincon, A.G.; Stanca, S.E.; Kiwi, J. Dynamics of E. coli membrane cell peroxidation during TiO2 photocatalysis studied by ATR-FTIR spectroscopy and AFM microscopy. J. Photochem. Photobiol. A Chem. 2005, 169, 131–137. [Google Scholar] [CrossRef]
- Xu, L.; Liang, H.W.; Yang, Y.; Yu, S.H. Stability and Reactivity: Positive and Negative Aspects for Nanoparticle Processing. Chem. Rev. 2018, 118, 3209–3250. [Google Scholar] [CrossRef] [PubMed]




| Samples | Description |
|---|---|
| M9_24 h | Aliquot of 24 h-unchallenged Micrococcus sp. culture |
| M9_72 h | Aliquot of 72 h-unchallenged Micrococcus sp. culture |
| M9_120 h | Aliquot of 120 h-unchallenged Micrococcus sp. culture |
| M9_0.5 mM SeO32−_24 h | Aliquot of Micrococcus sp. culture exposed for 24 h to SeO32− |
| M9_0.5 mM SeO32−_72 h | Aliquot of Micrococcus sp. culture exposed for 72 h to SeO32− |
| M9_0.5 mM SeO32−_120 h | Aliquot of Micrococcus sp. culture exposed for 120 h to SeO32− |
| Bio SeNP extract | Biogenic SeNP extract recovered from Micrococcus sp. cells incubated for 120 h with SeO32− |
| Bio SeNP extract_w | Biogenic SeNP extract washed in 1 mL of ddH2O |
| OM | Organic material recovered from biogenic SeNP extract |
| Sample | ζ Potential Value (mV) |
|---|---|
| Bio SeNP extract | −27.2 ± 0.7 |
| Bio SeNP extract_w | −21.1 ± 0.4 |
| OM | −26.4 ± 0.5 |
| Samples | Aamide I | β-Antiparallel (%) | β-Turn (%) | α-Helix (%) | β-Sheet (%) |
|---|---|---|---|---|---|
| M9_24 h | 40.86 | - | - | 100 | - |
| M9_72 h | 38.99 | - | 1 | 99 | - |
| M9_120 h | 39.80 | - | 1 | 99 | - |
| M9_0.5 mM SeO32−_24 h | 55.63 | - | 25 | 19 | 56 |
| M9_0.5 mM SeO32−_72 h | 42.11 | 2 | - | 98 | - |
| M9_0.5 mM SeO32−_120 h | 50.14 | - | 35 | 24 | 41 |
| Bio SeNP extract | 49.79 | - | 41 | 19 | 40 |
| OM | 62.68 | 8 | - | 28 | 64 |
| Bio SeNP extract_w | 46.36 | - | - | 79 | 21 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piacenza, E.; Presentato, A.; Ferrante, F.; Cavallaro, G.; Alduina, R.; Chillura Martino, D.F. Biogenic Selenium Nanoparticles: A Fine Characterization to Unveil Their Thermodynamic Stability. Nanomaterials 2021, 11, 1195. https://doi.org/10.3390/nano11051195
Piacenza E, Presentato A, Ferrante F, Cavallaro G, Alduina R, Chillura Martino DF. Biogenic Selenium Nanoparticles: A Fine Characterization to Unveil Their Thermodynamic Stability. Nanomaterials. 2021; 11(5):1195. https://doi.org/10.3390/nano11051195
Chicago/Turabian StylePiacenza, Elena, Alessandro Presentato, Francesco Ferrante, Giuseppe Cavallaro, Rosa Alduina, and Delia F. Chillura Martino. 2021. "Biogenic Selenium Nanoparticles: A Fine Characterization to Unveil Their Thermodynamic Stability" Nanomaterials 11, no. 5: 1195. https://doi.org/10.3390/nano11051195
APA StylePiacenza, E., Presentato, A., Ferrante, F., Cavallaro, G., Alduina, R., & Chillura Martino, D. F. (2021). Biogenic Selenium Nanoparticles: A Fine Characterization to Unveil Their Thermodynamic Stability. Nanomaterials, 11(5), 1195. https://doi.org/10.3390/nano11051195









