Nanomaterial Databases: Data Sources for Promoting Design and Risk Assessment of Nanomaterials
Abstract
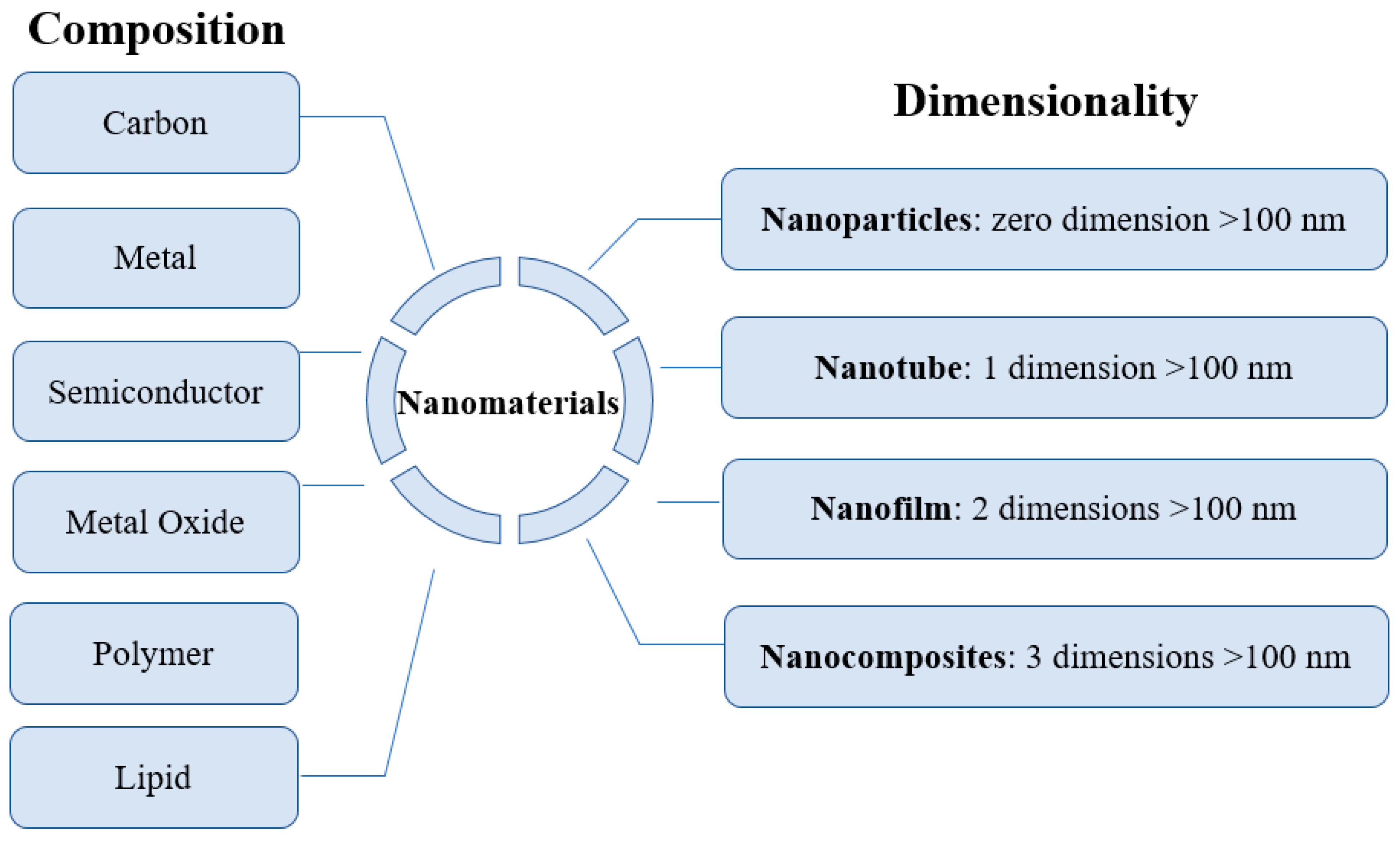
:1. Introduction
2. Brief Description of the Databases
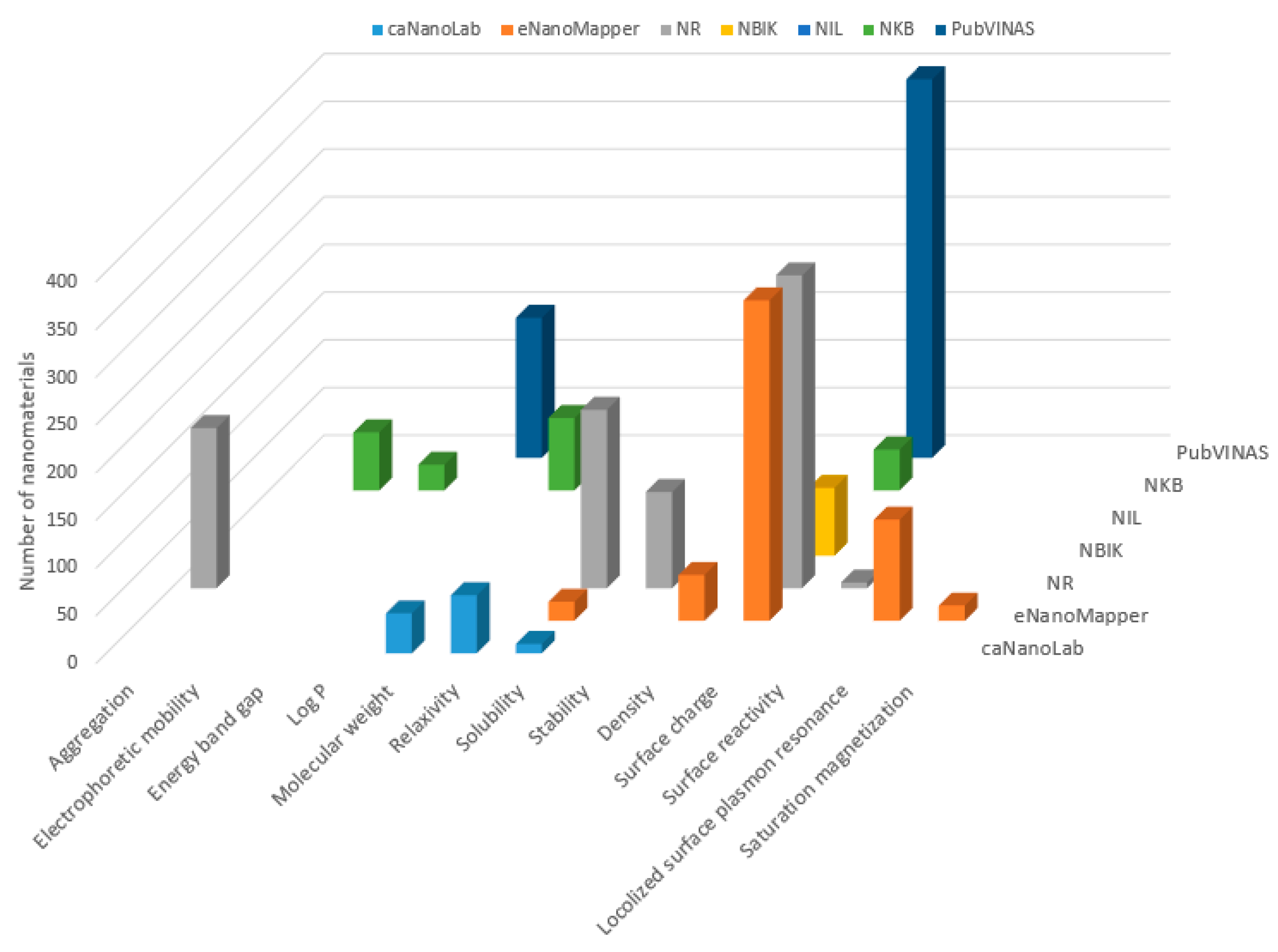
3. Comparative Analysis of the Databases
4. Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Initiative, N.N. National Nanotechnology Initiative Strategic Plan. December 2007. Available online: https://www.nano.gov/2007-Strategic-Plan (accessed on 16 February 2021).
- Authority, E.F.S. The Potential Risks Arising from Nanoscience and Nanotechnologies on Food and Feed Safety. EFSA J. 2009, 7, 958. [Google Scholar]
- The Royal Society; The Royal Academy of Engineering. Nanoscience and Nanotechnologies; Clyvedon Press: Cardiff, UK, 2004; Chapter 2; p. 5. [Google Scholar]
- Kreyling, W.G.; Semmler-Behnke, M.; Chaudhry, Q. A complementary definition of nanomaterial. Nano Today 2010, 5, 165–168. [Google Scholar] [CrossRef]
- SCENIHR (Scientific Committee on Emerging and Newly-Identified Health Risks). The Existing and Proposed Definitions Relating to Products of Nanotechnologies. 2007. Available online: http://ec.europa.eu/health/archive/ph_risk/committees/04_scenihr/docs/scenihr_o_012.pdf (accessed on 16 February 2021).
- Zhang, J.X.J.; Hoshino, K. Nanomaterials for molecular sensing. In Molecular Sensors and Nanodevices, 2nd ed.; Zhang, J.X.J., Hoshino, K., Eds.; Academic Press: Cambridge, MA, USA, 2019; Chapter 7; pp. 413–487. [Google Scholar]
- Podyacheva, O.Y.; Ismagilov, Z. Nitrogen-Doped carbon nanomaterials: To the mechanism of growth, electrical conductivity and application in catalysis. Catal. Today 2015, 249, 12–22. [Google Scholar] [CrossRef]
- Zhou, Q.; Wang, Y.; Xiao, J.; Fan, H.; Chen, C. Preparation and characterization of magnetic nanomaterial and its application for removal of polycyclic aromatic hydrocarbons. J. Hazard. Mater. 2019, 371, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wang, X.; Jia, Y.; Chen, X.; Han, H.; Li, C. Titanium dioxide-based nanomaterials for photocatalytic fuel generations. Chem. Rev. 2014, 114, 9987–10043. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, J.N.; Vij, V.; Kemp, K.C.; Kim, K.S. Engineered carbon-nanomaterial-based electrochemical sensors for biomolecules. ACS Nano 2016, 10, 46–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mlinar, V. Engineered nanomaterials for solar energy conversion. Nanotechnology 2013, 24, 042001. [Google Scholar] [CrossRef]
- Rauf, S.; Hayat Nawaz, M.A.; Badea, M.; Marty, J.L.; Hayat, A. Nano-Engineered biomimetic optical sensors for glucose monitoring in diabetes. Sensors 2016, 16, 1931. [Google Scholar] [CrossRef] [PubMed]
- Fournier, S.; D’errico, J.; Stapleton, P. Engineered nanomaterial applications in perinatal therapeutics. Pharmacol. Res. 2018, 130, 36–43. [Google Scholar] [CrossRef] [PubMed]
- West, J.L.; Halas, N.J. Engineered nanomaterials for biophotonics applications: Improving sensing, imaging, and therapeutics. Annu. Rev. Biomed. Eng. 2003, 5, 285–292. [Google Scholar] [CrossRef] [Green Version]
- Shi, D.; Bedford, N.M.; Cho, H.S. Engineered multifunctional nanocarriers for cancer diagnosis and therapeutics. Small 2011, 7, 2549–2567. [Google Scholar] [CrossRef]
- Yu, H.; Li, L.; Zhang, Y. Silver nanoparticle-based thermal interface materials with ultra-low thermal resistance for power electronics applications. Scr. Mater. 2012, 66, 931–934. [Google Scholar] [CrossRef]
- Yeo, J.; Kim, G.; Hong, S.; Kim, M.S.; Kim, D.; Lee, J.; Lee, H.B.; Kwon, J.; Suh, Y.D.; Kang, H.W.; et al. Flexible supercapacitor fabrication by room temperature rapid laser processing of roll-to-roll printed metal nanoparticle ink for wearable electronics application. J. Power Sources 2014, 246, 562–568. [Google Scholar] [CrossRef]
- Ning, F.; Shao, M.; Xu, S.; Fu, Y.; Zhang, R.; Wei, M.; Evans, D.G.; Duan, X. TiO2/graphene/NiFe-layered double hydroxide nanorod array photoanodes for efficient photoelectrochemical water splitting. Energy Environ. Sci. 2016, 9, 2633–2643. [Google Scholar] [CrossRef]
- Nie, H.; Li, M.; Li, Q.; Liang, S.; Tan, Y.; Sheng, L.; Shi, W.; Zhang, S.X.-A. Carbon dots with continuously tunable full-color emission and their application in ratiometric pH sensing. Chem. Mater. 2014, 26, 3104–3112. [Google Scholar] [CrossRef]
- Lin, X.; Gao, G.; Zheng, L.; Chi, Y.; Chen, G. Encapsulation of strongly fluorescent carbon quantum dots in metal–organic frameworks for enhancing chemical sensing. Anal. Chem. 2014, 86, 1223–1228. [Google Scholar] [CrossRef]
- Nikalje, A.P. Nanotechnology and its applications in medicine. Med. Chem. 2015, 5, 81–89. [Google Scholar] [CrossRef]
- Hofmann-Amtenbrink, M.; Grainger, D.W.; Hofmann, H. Nanoparticles in medicine: Current challenges facing inorganic nanoparticle toxicity assessments and standardizations. Nanomed. NBM 2015, 11, 1689–1694. [Google Scholar] [CrossRef]
- Grand View Research, I. Nanomaterials Market Size, Share & Trends Analysis Report By Product (Carbon Nanotubes, Titanium Dioxide), By Application (Medical, Electronics, Paints & Coatings), By Region, And Segment Forecasts, 2020–2027. Available online: https://www.giiresearch.com/report/grvi940783-nanomaterials-market-size-share-trends-analysis.html (accessed on 14 January 2021).
- Borm, P.J.A.; Robbins, D.; Haubold, S.; Kuhlbusch, T.; Fissan, H.; Donaldson, K.; Schins, R.; Stone, V.; Kreyling, W.; Lademann, J.; et al. The potential risks of nanomaterials: A review carried out for ECETOC. Part Fibre Toxicol. 2006, 3, 11. [Google Scholar] [CrossRef] [Green Version]
- Vollath, D. Nanomaterials an introduction to synthesis, properties and application. Environ. Eng. Manag. J. 2008, 7, 865–870. [Google Scholar]
- Ray, S.; Saha, A.; Jana, N.R.; Sarkar, R. Fluorescent carbon nanoparticles: Synthesis, characterization, and bioimaging application. J. Phys. Chem. C 2009, 113, 18546–18551. [Google Scholar] [CrossRef]
- Amendola, V.; Meneghetti, M. What controls the composition and the structure of nanomaterials generated by laser ablation in liquid solution? Phys. Chem. Chem. Phys. 2013, 15, 3027–3046. [Google Scholar] [CrossRef]
- Zhou, J.; Booker, C.; Li, R.; Zhou, X.; Sham, T.K.; Sun, X.; Ding, Z. An electrochemical avenue to blue luminescent nanocrystals from multiwalled carbon nanotubes (MWCNTs). J. Am. Chem. Soc. 2007, 129, 744–745. [Google Scholar] [CrossRef] [PubMed]
- Thiruvengadathan, R.; Korampally, V.; Ghosh, A.; Chanda, N.; Gangopadhyay, K.; Gangopadhyay, S. Nanomaterial processing using self-assembly-bottom-up chemical and biological approaches. Rep. Prog. Phys. 2013, 76, 066501. [Google Scholar] [CrossRef] [PubMed]
- Nafees, M.; Ali, S.; Rasheed, K.; Idrees, S. The novel and economical way to synthesize CuS nanomaterial of different morphologies by aqueous medium employing microwaves irradiation. Appl. Nanosci. 2012, 2, 157–162. [Google Scholar] [CrossRef] [Green Version]
- Devaraju, M.K.; Honma, I. Hydrothermal and solvothermal process towards development of LiMPO4 (M = Fe, Mn) nanomaterials for lithium-ion batteries. Adv. Energy Mater. 2012, 2, 284–297. [Google Scholar] [CrossRef]
- Malhotra, B.D.; Ali, M.A. Nanomaterials in Biosensors: Fundamentals and Applications. In Nanomaterials for Biosensors; Malhotra, B.D., Ali, M.A., Eds.; William Andrew Publishing: Norwich, NY, USA, 2018; Chapter 1; pp. 1–74. [Google Scholar]
- Rao, C.N.R.; Müller, A.; Cheetham, A.K. The Chemistry of Nanomaterials: Synthesis, Properties and Applications; John Wiley & Sons: Hoboken, NJ, USA, 2006. [Google Scholar]
- Edelstein, A.S.; Cammaratra, R. Nanomaterials: Synthesis, Properties and Applications; CRC Press: Boca Raton, FL, USA, 1998. [Google Scholar]
- Cao, G. Nanostructures & Nanomaterials: Synthesis, Properties & Applications; Imperial College Press: London, UK, 2004. [Google Scholar]
- Rodríguez, J.A.; Fernández-García, M. Synthesis, Properties, and Applications of Oxide Nanomaterials; John Wiley & Sons: Hoboken, NJ, USA, 2007. [Google Scholar]
- Aillon, K.L.; Xie, Y.; El-Gendy, N.; Berkland, C.J.; Forrest, M.L. Effects of nanomaterial physicochemical properties on in vivo toxicity. Adv. Drug Deliv. Rev. 2009, 61, 457–466. [Google Scholar] [CrossRef] [Green Version]
- Jung, S.-K.; Qu, X.; Aleman-Meza, B.; Wang, T.; Riepe, C.; Liu, Z.; Li, Q.; Zhong, W. Multi-Endpoint, High-Throughput Study of Nanomaterial Toxicity in Caenorhabditis elegans. Environ. Sci. Technol. 2015, 49, 2477–2485. [Google Scholar] [CrossRef] [Green Version]
- Colvin, V.L. The potential environmental impact of engineered nanomaterials. Nat. Biotechnol. 2003, 21, 1166–1170. [Google Scholar] [CrossRef] [PubMed]
- Fojtů, M.; Teo, W.Z.; Pumera, M. Environmental impact and potential health risks of 2D nanomaterials. Environ. Sci. Nano 2017, 4, 1617–1633. [Google Scholar] [CrossRef]
- Nowack, B.; Ranville, J.F.; Diamond, S.; Gallego-Urrea, J.A.; Metcalfe, C.; Rose, J.; Horne, N.; Koelmans, A.A.; Klaine, S.J. Potential scenarios for nanomaterial release and subsequent alteration in the environment. Environ. Toxicol. Chem. 2012, 31, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Foltête, A.-S.; Masfaraud, J.-F.; Bigorgne, E.; Nahmani, J.; Chaurand, P.; Botta, C.; Labille, J.; Rose, J.; Férard, J.-F.; Cotelle, S. Environmental impact of sunscreen nanomaterials: Ecotoxicity and genotoxicity of altered TiO2 nanocomposites on Vicia faba. Environ. Pollut. 2011, 159, 2515–2522. [Google Scholar] [CrossRef]
- Zhu, M.; Nie, G.; Meng, H.; Xia, T.; Nel, A.; Zhao, Y. Physicochemical Properties Determine Nanomaterial Cellular Uptake, Transport, and Fate. Acc. Chem. Res. 2013, 46, 622–631. [Google Scholar] [CrossRef] [Green Version]
- Magrez, A.; Kasas, S.; Salicio, V.; Pasquier, N.; Seo, J.W.; Celio, M.; Catsicas, S.; Schwaller, B.; Forró, L. Cellular Toxicity of Carbon-Based Nanomaterials. Nano Lett. 2006, 6, 1121–1125. [Google Scholar] [CrossRef] [PubMed]
- Fairbairn, E.A.; Keller, A.A.; Mädler, L.; Zhou, D.; Pokhrel, S.; Cherr, G.N. Metal oxide nanomaterials in seawater: Linking physicochemical characteristics with biological response in sea urchin development. J. Hazard. Mater. 2011, 192, 1565–1571. [Google Scholar] [CrossRef] [PubMed]
- Morris, S.A.; Gaheen, S.; Lijowski, M.; Heiskanen, M.; Klemm, J. CaNanoLab: A nanomaterial data repository for biomedical research. In Proceedings of the 2014 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Belfast, UK, 2–5 November 2014; pp. 29–33. [Google Scholar]
- Morris, S.A.; Gaheen, S.; Lijowski, M.; Heiskanen, M.; Klemm, J. Experiences in supporting the structured collection of cancer nanotechnology data using caNanoLab. Beilstein J. Nanotechnol. 2015, 6, 1580–1593. [Google Scholar] [CrossRef] [Green Version]
- Jeliazkova, N.; Chomenidis, C.; Doganis, P.; Fadeel, B.; Grafström, R.; Hardy, B.; Hastings, J.; Hegi, M.; Jeliazkov, V.; Kochev, N.; et al. The eNanoMapper database for nanomaterial safety information. Beilstein J. Nanotechnol. 2015, 6, 1609–1634. [Google Scholar] [CrossRef] [Green Version]
- Hastings, J.; Jeliazkova, N.; Owen, G.; Tsiliki, G.; Munteanu, C.R.; Steinbeck, C.; Willighagen, E. ENanoMapper: Harnessing ontologies to enable data integration for nanomaterial risk assessment. J. Biomed. Semant. 2015, 6, 10. [Google Scholar] [CrossRef] [Green Version]
- Helma, C.; Rautenberg, M.; Gebele, D. Nano-Lazar: Read across Predictions for Nanoparticle Toxicities with Calculated and Measured Properties. Front. Pharmacol. 2017, 8, 377. [Google Scholar] [CrossRef] [Green Version]
- Comandella, D.; Gottardo, S.; Rio-Echevarria, I.M.; Rauscher, H. Quality of physicochemical data on nanomaterials: An assessment of data completeness and variability. Nanoscale 2020, 12, 4695–4708. [Google Scholar] [CrossRef] [Green Version]
- Ostraat, M.L.; Mills, K.C.; Guzan, K.A.; Murry, D. The Nanomaterial Registry: Facilitating the sharing and analysis of data in the diverse nanomaterial community. Int. J. Nanomed. 2013, 8 (Suppl. 1), 7–13. [Google Scholar]
- Miller, A.L.; Hoover, M.D.; Mitchell, D.M.; Stapleton, B.P. The Nanoparticle Information Library (NIL): A Prototype for Linking and Sharing Emerging Data. J. Occup. Environ. Hyg. 2007, 4, D131–D134. [Google Scholar] [CrossRef]
- Yan, X.; Sedykh, A.; Wang, W.; Yan, B.; Zhu, H. Construction of a web-based nanomaterial database by big data curation and modeling friendly nanostructure annotations. Nat. Commun. 2020, 11, 2519. [Google Scholar] [CrossRef]
- Labouta, H.I.; Asgarian, N.; Rinker, K.; Cramb, D.T. Meta-Analysis of Nanoparticle Cytotoxicity via Data-Mining the Literature. ACS Nano 2019, 13, 1583–1594. [Google Scholar] [CrossRef] [PubMed]
- Sayes, C.; Ivanov, I. Comparative study of predictive computational models for nanoparticle-induced cytotoxicity. Risk analysis. Off. Publ. Soc. Risk Anal. 2010, 30, 1723–1734. [Google Scholar] [CrossRef] [PubMed]
- Puzyn, T.; Rasulev, B.; Gajewicz, A.; Hu, X.; Dasari, T.P.; Michalkova, A.; Hwang, H.-M.; Toropov, A.; Leszczynska, D.; Leszczynski, J. Using nano-QSAR to predict the cytotoxicity of metal oxide nanoparticles. Nat. Nanotechnol. 2011, 6, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Rallo, R.; George, S.; Ji, Z.; Nair, S.; Nel, A.E.; Cohen, Y. Classification NanoSAR development for cytotoxicity of metal oxide nanoparticles. Small 2011, 7, 1118–1126. [Google Scholar] [CrossRef] [PubMed]
- Horev-Azaria, L.; Baldi, G.; Beno, D.; Bonacchi, D.; Golla-Schindler, U.; Kirkpatrick, J.C.; Kolle, S.; Landsiedel, R.; Maimon, O.; Marche, P.N.; et al. Predictive Toxicology of cobalt ferrite nanoparticles: Comparative in-vitro study of different cellular models using methods of knowledge discovery from data. Part. Fibre Toxicol. 2013, 10, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fourches, D.; Pu, D.; Tassa, C.; Weissleder, R.; Shaw, S.Y.; Mumper, R.J.; Tropsha, A. Quantitative Nanostructure−Activity Relationship Modeling. ACS Nano 2010, 4, 5703–5712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2015, 44, D1202–D1213. [Google Scholar] [CrossRef]
- Rose, P.W.; Prlić, A.; Altunkaya, A.; Bi, C.; Bradley, A.R.; Christie, C.H.; Costanzo, L.D.; Duarte, J.M.; Dutta, S.; Feng, Z.; et al. The RCSB protein data bank: Integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 2017, 45, D271–D281. [Google Scholar]
- Shen, J.; Xu, L.; Fang, H.; Richard, A.M.; Bray, J.D.; Judson, R.S.; Zhou, G.; Colatsky, T.J.; Aungst, J.L.; Teng, C.; et al. EADB: An estrogenic activity database for assessing potential endocrine activity. Toxicol. Sci. 2013, 135, 277–291. [Google Scholar] [CrossRef] [Green Version]
- Hong, H.; Su, Z.; Ge, W.; Shi, L.; Perkins, R.; Fang, H.; Xu, J.; Chen, J.J.; Han, T.; Kaput, J.; et al. Assessing batch effects of genotype calling algorithm BRLMM for the Affymetrix GeneChip Human Mapping 500 K array set using 270 HapMap samples. BMC Bioinform. 2008, 9, S17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Soika, V.; Meehan, J.; Su, Z.; Ge, W.; Ng, H.W.; Perkins, R.; Simonyan, V.; Tong, W.; Hong, H. Quality control metrics improve repeatability and reproducibility of single-nucleotide variants derived from whole-genome sequencing. Pharm. J. 2015, 15, 298–309. [Google Scholar] [CrossRef]
- Hong, H.; Hong, Q.; Liu, J.; Tong, W.; Shi, L. Estimating relative noise to signal in DNA microarray data. Int. J. Bioinform. Res. Appl. 2013, 9, 433–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, Z.; Hong, H.; Fang, H.; Shi, L.; Perkins, R.; Tong, W. Very Important Pool (VIP) genes—An application for microarray-based molecular signatures. BMC Bioinform. 2008, 9 (Suppl. 9), S9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, W.; Wu, L.; Yavas, G.; Simonyan, V.; Ning, B.; Hong, H. Challenges, solutions, and quality metrics of personal genome assembly in advancing precision medicine. Pharmaceutics 2016, 8, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, W.; Archer, J.; Moore, M.; Bruce, J.; McLain, M.; Shojaee, S.; Zou, W.; Benjamin, L.A.; Adeuya, A.; Fairchild, R.; et al. QUICK: Quality and Usability Investigation and Control Kit for Mass Spectrometric Data from Detection of Persistent Organic Pollutants. Int. J. Environ. Res. Public Health 2019, 16, 4203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeliazkova, N.; Apostolova, M.D.; Andreoli, C.; Barone, F.; Barrick, A.; Battistelli, C.; Bossa, C.; Botea-Petcu, A.; Châtel, A.; de Angelis, I.; et al. Towards FAIR nanosafety data. Nat. Nanotechnol. 2021, 16, 644–654. [Google Scholar] [CrossRef]
- Hu, X.; Li, D.; Gao, Y.; Mu, L.; Zhou, Q. Knowledge gaps between nanotoxicological research and nanomaterial safety. Environ. Int. 2016, 94, 8–23. [Google Scholar] [CrossRef]
- Staggers, N.; McCasky, T.; Brazelton, N.; Kennedy, R. Nanotechnology: The coming revolution and its implications for consumers, clinicians, and informatics. Nurs. Outlook 2008, 56, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Nijhara, R.; Balakrishnan, K. Bringing nanomedicines to market: Regulatory challenges, opportunities, and uncertainties. Nanomed. NBM 2006, 2, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Leso, V.; Fontana, L.; Chiara Mauriello, M.; Iavicoli, I. Occupational risk assessment of engineered nanomaterials: Limits, challenges and opportunities. Curr. Nanosci. 2017, 13, 55–78. [Google Scholar] [CrossRef] [Green Version]



| Database | Website | Records | Remark |
|---|---|---|---|
| caNanoLab | https://cananolab.nci.nih.gov/ | 1383 | Nanotechnology in biomedicine |
| eNanoMapper | https://data.enanomapper.net/ | 2380 | Safety assessment of nanomaterials |
| NR | https://nanomaterialregistry.net/ | 2031 | Physicochemical properties |
| Nanowerk | https://www.nanowerk.com/ | 3785 | Commercially available nanomaterials |
| NBIK | http://nbi.oregonstate.edu/ | 147 | Exposure effect in embryo zebrafish |
| NIL | http://nanoparticlelibrary.net/ | 88 | Physicochemical characteristics |
| NKB | https://ssl.biomax.de/nanocommons/ | 598 | Nano-safety knowledge infrastructure |
| PubVINAS | http://www.pubvinas.com/ | 725 | An online nano-modeling tool |
| Carbon | Lipid | Metal | Metal Oxide | Polymer | Semiconductor | Other | |
|---|---|---|---|---|---|---|---|
| caNanoLab | 78 | 97 | 143 | 272 | 528 | 73 | 192 |
| eNanoMapper | 120 | 42 | 723 | 150 | 513 | 226 | 606 |
| NR | 210 | 2 | 551 | 612 | 190 | 235 | 231 |
| NBIK | 4 | 0 | 47 | 22 | 33 | 34 | 7 |
| NIL | 17 | 0 | 15 | 13 | 0 | 25 | 18 |
| NKB | 31 | 0 | 164 | 96 | 0 | 50 | 257 |
| PubVINAS | 147 | 0 | 456 | 32 | 56 | 34 | 0 |
| Function | caNanoLab | eNanoMapper | NR | NBIK | NIL | NKB | PubVINAS |
|---|---|---|---|---|---|---|---|
| Browse | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Search | Yes | Yes | Yes | Yes | Yes | Yes | |
| Filter | Yes | Yes | Yes | Yes | Yes | Yes | |
| Export | Yes | Yes | Yes | Yes | Yes | Yes | |
| Upload | Yes | Yes | Yes | Yes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, Z.; Guo, W.; Sakkiah, S.; Liu, J.; Patterson, T.A.; Hong, H. Nanomaterial Databases: Data Sources for Promoting Design and Risk Assessment of Nanomaterials. Nanomaterials 2021, 11, 1599. https://doi.org/10.3390/nano11061599
Ji Z, Guo W, Sakkiah S, Liu J, Patterson TA, Hong H. Nanomaterial Databases: Data Sources for Promoting Design and Risk Assessment of Nanomaterials. Nanomaterials. 2021; 11(6):1599. https://doi.org/10.3390/nano11061599
Chicago/Turabian StyleJi, Zuowei, Wenjing Guo, Sugunadevi Sakkiah, Jie Liu, Tucker A. Patterson, and Huixiao Hong. 2021. "Nanomaterial Databases: Data Sources for Promoting Design and Risk Assessment of Nanomaterials" Nanomaterials 11, no. 6: 1599. https://doi.org/10.3390/nano11061599
APA StyleJi, Z., Guo, W., Sakkiah, S., Liu, J., Patterson, T. A., & Hong, H. (2021). Nanomaterial Databases: Data Sources for Promoting Design and Risk Assessment of Nanomaterials. Nanomaterials, 11(6), 1599. https://doi.org/10.3390/nano11061599






