Protein Hydrogels: The Swiss Army Knife for Enhanced Mechanical and Bioactive Properties of Biomaterials
Abstract
:1. Hydrogels
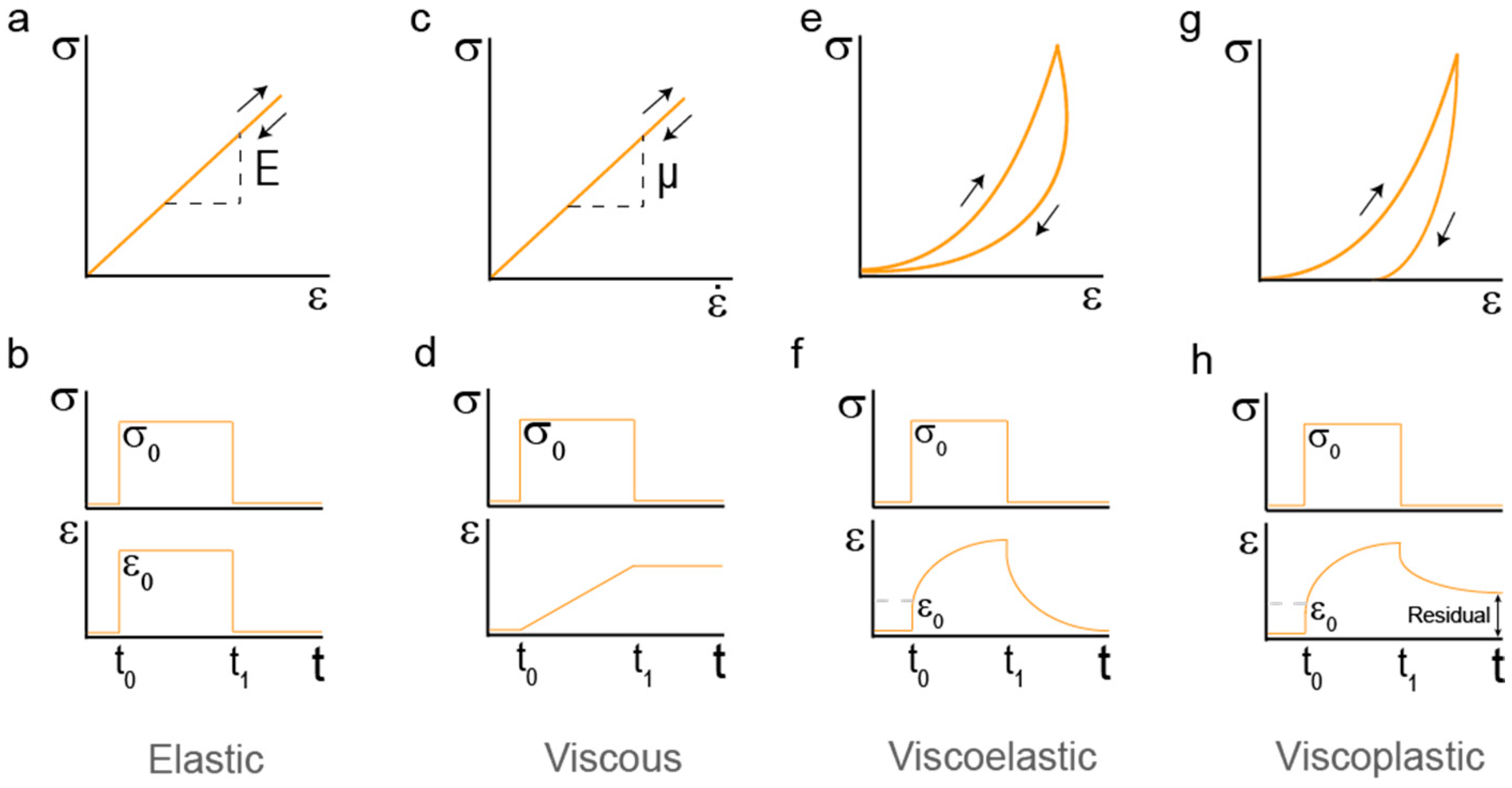
2. Mechanical Properties of Hydrogels
3. Composition and Structural Properties of Hydrogels
4. Protein Hydrogels
4.1. Types of Protein Hydrogels
4.1.1. Natural-Protein-Based Hydrogels
4.1.2. Engineered Protein-Based Hydrogels
5. Bottom-Up Mechanical Design of Protein Hydrogels: Lessons from Titin

6. Smart Protein Hydrogels
7. Applications of Protein Hydrogels
7.1. Drug and Cell Delivery
7.2. Tissue Engineering
7.3. Cell Mechanobiology

8. Limitations of Protein Hydrogels
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wichterle, O.; Lím, D. Hydrophilic Gels for Biological Use. Nature 1960, 185, 117–118. [Google Scholar] [CrossRef]
- Das, N. Preparation methods and properties of hydrogel: A review. Int. J. Pharm. Pharm. Sci. 2013, 5, 112–117. [Google Scholar]
- Buwalda, S.J.; Boere, K.W.; Dijkstra, P.J.; Feijen, J.; Vermonden, T.; Hennink, W.E. Hydrogels in a historical perspective: From simple networks to smart materials. J. Control. Release 2014, 190, 254–273. [Google Scholar] [CrossRef]
- Deligkaris, K.; Tadele, T.S.; Olthuis, W.; Berg, A.V.D. Hydrogel-based devices for biomedical applications. Sens. Actuators B Chem. 2010, 147, 765–774. [Google Scholar] [CrossRef]
- Zhu, J.; Marchant, R.E. Design properties of hydrogel tissue-engineering scaffolds. Expert Rev. Med. Devices 2011, 8, 607–626. [Google Scholar] [CrossRef]
- Gyles, D.A.; Castro, L.D.; Silva, J.O.C.; Ribeiro-Costa, R.M. A review of the designs and prominent biomedical advances of natural and synthetic hydrogel formulations. Eur. Polym. J. 2017, 88, 373–392. [Google Scholar] [CrossRef]
- Ahmed, E.M. Hydrogel: Preparation, characterization, and applications: A review. J. Adv. Res. 2015, 6, 105–121. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Kong, N.; Laver, B.; Liu, J. Hydrogels Constructed from Engineered Proteins. Small 2016, 12, 973–987. [Google Scholar] [CrossRef]
- Nayak, A.K.; Das, B. Introduction to polymeric gels. In Polymeric Gels; Pal, K., Banerjee, I., Eds.; Woodhead Publishing: Sawston, UK, 2018; pp. 3–27. [Google Scholar]
- Haque, M.A.; Kurokawa, T.; Gong, J.P. Super tough double network hydrogels and their application as biomaterials. Polymer 2012, 53, 1805–1822. [Google Scholar] [CrossRef]
- Courtney, T.H. Mechanical Behavior of Materials; McGraw-Hill: New York, NY, USA, 1990. [Google Scholar]
- Roylance, D. Mechanical Properties of Materials; Wiley: Hoboken, NJ, USA, 2008. [Google Scholar]
- Wineman, A.S.; Rajagopal, K.R. Mechanical Response of Polymers: An Introduction; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar]
- Vedadghavami, A.; Minooei, F.; Mohammadi, M.H.; Khetani, S.; Kolahchi, A.R.; Mashayekhan, S.; Sanati-Nezhad, A. Manufacturing of hydrogel biomaterials with controlled mechanical properties for tissue engineering applications. Acta Biomater. 2017, 62, 42–63. [Google Scholar] [CrossRef]
- Anseth, K.S.; Bowman, C.N.; Brannon-Peppas, L. Mechanical properties of hydrogels and their experimental determination. Biomaterials 1996, 17, 1647–1657. [Google Scholar] [CrossRef]
- Gibson, L.J.; Ashby, M.F.; Schajer, G.S.; Robertson, C.I. The mechanics of two-dimensional cellular materials. Proc. R. Soc. Lond. A Math. Phys. Sci. 1982, 382, 25–42. [Google Scholar] [CrossRef]
- Maaß, R.; Derlet, P.M. Micro-plasticity and recent insights from intermittent and small-scale plasticity. Acta Mater. 2018, 143, 338–363. [Google Scholar] [CrossRef] [Green Version]
- Chai, Q.; Jiao, Y.; Yu, X. Hydrogels for Biomedical Applications: Their Characteristics and the Mechanisms behind Them. Gels 2017, 3, 6. [Google Scholar] [CrossRef] [Green Version]
- Peppas, N.A.; Huang, Y.; Torres-Lugo, M.; Ward, J.H.; Zhang, J. Physicochemical Foundations and Structural Design of Hydrogels in Medicine and Biology. Annu. Rev. Biomed. Eng. 2000, 2, 9–29. [Google Scholar] [CrossRef] [Green Version]
- De Leon Rodriguez, L.M.; Hemar, Y.; Cornish, J.; Brimble, M.A. Structure-mechanical property correlations of hydrogel forming beta-sheet peptides. Chem. Soc. Rev. 2016, 45, 4797–4824. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Tang, S.; Olsen, B.D. Physics of engineered protein hydrogels. J. Polym. Sci. Part B Polym. Phys. 2013, 51, 587–601. [Google Scholar] [CrossRef]
- Koh, L.-D.; Cheng, Y.; Teng, C.-P.; Khin, Y.-W.; Loh, X.J.; Tee, S.-Y.; Low, M.; Ye, E.; Yu, H.-D.; Zhang, Y.-W.; et al. Structures, mechanical properties and applications of silk fibroin materials. Prog. Polym. Sci. 2015, 46, 86–110. [Google Scholar] [CrossRef]
- Boothroyd, S.; Saiani, A.; Miller, A.F. Controlling network topology and mechanical properties of co-assembling peptide hydrogels. Biopolymers 2013, 101, 669–680. [Google Scholar] [CrossRef]
- Ramachandran, S.; Trewhella, J.; Tseng, Y.; Yu, Y.B. Coassembling Peptide-Based Biomaterials: Effects of Pairing Equal and Unequal Chain Length Oligopeptides. Chem. Mater. 2006, 18, 6157–6162. [Google Scholar] [CrossRef]
- Kajiwara, K.; Hiragi, Y. Chapter 6 Structure analysis by small-angle X-ray scattering. In Analytical Spectroscopy Library; Saisho, H., Gohshi, Y., Eds.; Elsevier: Amsterdam, The Netherlands, 1996; pp. 353–404. [Google Scholar]
- Walenta, E. Small angle x-ray scattering. Acta Polym. 1985, 36, 296. [Google Scholar] [CrossRef]
- Roessle, M.; Svergun, D.I. Small Angle X-ray Scattering. In Encyclopedia of Biophysics; Roberts, G.C.K., Ed.; Springer Science and Business Media LLC: Berlin, Germany, 2013; pp. 2382–2389. [Google Scholar]
- Guilbaud, J.-B.; Saiani, A. Using small angle scattering (SAS) to structurally characterise peptide and protein self-assembled materials. Chem. Soc. Rev. 2011, 40, 1200–1210. [Google Scholar] [CrossRef]
- Lin, Y.-J.; Chuang, W.-T.; Hsu, S.-H. Gelation Mechanism and Structural Dynamics of Chitosan Self-Healing Hydrogels by In Situ SAXS and Coherent X-ray Scattering. ACS Macro Lett. 2019, 8, 1449–1455. [Google Scholar] [CrossRef]
- Messaoud, G.B.; Le Griel, P.; Hermida-Merino, D.; Baccile, N. Effects of pH, temperature and shear on the structure-property relationship of lamellar hydrogels from microbial glucolipids probed by in situ rheo-SAXS. Soft Matter 2020, 16, 2540–2551. [Google Scholar] [CrossRef]
- Windsor, C.G. An introduction to small-angle neutron scattering. J. Appl. Crystallogr. 1988, 21, 582–588. [Google Scholar] [CrossRef]
- Gerold, V.; Kostorz, G. Small-angle scattering applications to materials science. J. Appl. Crystallogr. 1978, 11, 376–404. [Google Scholar] [CrossRef]
- Ricciardi, R.; Mangiapia, G.; Celso, F.L.; Paduano, L.; Triolo, R.; Auriemma, F.; De Rosa, C.; Lauprêtre, F. Structural Organization of Poly(vinyl alcohol) Hydrogels Obtained by Freezing and Thawing Techniques: A SANS Study. Chem. Mater. 2005, 17, 1183–1189. [Google Scholar] [CrossRef] [Green Version]
- Saffer, E.M.; Lackey, M.A.; Griffin, D.M.; Kishore, S.; Tew, G.N.; Bhatia, S.R. SANS study of highly resilient poly(ethylene glycol) hydrogels. Soft Matter 2014, 10, 1905–1916. [Google Scholar] [CrossRef]
- Wiener, C.G.; Wang, C.; Liu, Y.; Weiss, R.A.; Vogt, B.D. Nanostructure Evolution during Relaxation from a Large Step Strain in a Supramolecular Copolymer-Based Hydrogel: A SANS Investigation. Macromolecules 2017, 50, 1672–1680. [Google Scholar] [CrossRef]
- Gagner, J.E.; Kim, W.; Chaikof, E.L. Designing protein-based biomaterials for medical applications. Acta Biomater. 2014, 10, 1542–1557. [Google Scholar] [CrossRef] [Green Version]
- Silva, N.H.C.S.; Vilela, C.; Marrucho, I.M.; Freire, C.S.R.; Neto, C.P.; Silvestre, A. Protein-based materials: From sources to innovative sustainable materials for biomedical applications. J. Mater. Chem. B 2014, 2, 3715–3740. [Google Scholar] [CrossRef]
- Almine, J.F.; Bax, D.V.; Mithieux, S.; Nivison-Smith, L.; Rnjak, J.; Waterhouse, A.; Wise, S.; Weiss, A.S. Elastin-based materials. Chem. Soc. Rev. 2010, 39, 3371–3379. [Google Scholar] [CrossRef]
- An, B.; Kaplan, D.L.; Brodsky, B. Engineered recombinant bacterial collagen as an alternative collagen-based biomaterial for tissue engineering. Front. Chem. 2014, 2, 40. [Google Scholar] [CrossRef] [Green Version]
- Yan, C.; Pochan, D.J. Rheological properties of peptide-based hydrogels for biomedical and other applications. Chem. Soc. Rev. 2010, 39, 3528–3540. [Google Scholar] [CrossRef] [Green Version]
- Tang, Z.; Chen, Q.; Chen, F.; Zhu, L.; Lu, S.; Ren, B.; Zhang, Y.; Yang, J.; Zheng, J. General Principle for Fabricating Natural Globular Protein-Based Double-Network Hydrogels with Integrated Highly Mechanical Properties and Surface Adhesion on Solid Surfaces. Chem. Mater. 2018, 31, 179–189. [Google Scholar] [CrossRef]
- Fang, J.; Zhang, X.; Cai, Y.; Wei, Y. Small Globular Protein Motif Forms Particulate Hydrogel under Various pH Conditions. Biomacromolecules 2011, 12, 1578–1584. [Google Scholar] [CrossRef]
- Achilli, M.; Mantovani, D. Tailoring Mechanical Properties of Collagen-Based Scaffolds for Vascular Tissue Engineering: The Effects of pH, Temperature and Ionic Strength on Gelation. Polymers 2010, 2, 664–680. [Google Scholar] [CrossRef] [Green Version]
- Antoine, E.E.; Vlachos, P.P.; Rylander, M.N. Review of Collagen I Hydrogels for Bioengineered Tissue Microenvironments: Characterization of Mechanics, Structure, and Transport. Tissue Eng. Part B Rev. 2014, 20, 683–696. [Google Scholar] [CrossRef] [Green Version]
- Bielajew, B.J.; Hu, J.C.; Athanasiou, K.A. Collagen: Quantification, biomechanics and role of minor subtypes in cartilage. Nat. Rev. Mater. 2020, 5, 1–18. [Google Scholar] [CrossRef]
- Catoira, M.C.; Fusaro, L.; Di Francesco, D.; Ramella, M.; Boccafoschi, F. Overview of natural hydrogels for regenerative medicine applications. J. Mater. Sci. Mater. Med. 2019, 30, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Caliari, S.; Burdick, J.A. A practical guide to hydrogels for cell culture. Nat. Methods 2016, 13, 405–414. [Google Scholar] [CrossRef] [Green Version]
- Nair, M.; Johal, R.K.; Hamaia, S.W.; Best, S.M.; Cameron, R.E. Tunable bioactivity and mechanics of collagen-based tissue engineering constructs: A comparison of EDC-NHS, genipin and TG2 crosslinkers. Biomaterials 2020, 254, 120109. [Google Scholar] [CrossRef]
- Gasperini, L.; Mano, J.F.; Reis, R.L. Natural polymers for the microencapsulation of cells. J. R. Soc. Interface 2014, 11, 20140817. [Google Scholar] [CrossRef] [Green Version]
- Young, S.; Wong, M.; Tabata, Y.; Mikos, A.G. Gelatin as a delivery vehicle for the controlled release of bioactive molecules. J. Control. Release 2005, 109, 256–274. [Google Scholar] [CrossRef]
- Kuijpers, A.J.; Engbers, G.H.M.; Feijen, J.; De Smedt, S.C.; Meyvis, T.K.L.; Demeester, J.; Krijgsveld, J.; Zaat, S.A.J.; Dankert, J. Characterization of the Network Structure of Carbodiimide Cross-Linked Gelatin Gels. Macromolecules 1999, 32, 3325–3333. [Google Scholar] [CrossRef]
- Debelle, L.; Tamburro, A.M. Elastin: Molecular description and function. Int. J. Biochem. Cell Biol. 1999, 31, 261–272. [Google Scholar] [CrossRef]
- Bellingham, C.M.; Lillie, M.A.; Gosline, J.M.; Wright, G.M.; Starcher, B.C.; Bailey, A.J.; Woodhouse, K.A.; Keeley, F.W. Recombinant human elastin polypeptides self-assemble into biomaterials with elastin-like properties. Biopolymers 2003, 70, 445–455. [Google Scholar] [CrossRef]
- Annabi, N.; Mithieux, S.; Boughton, E.A.; Ruys, A.J.; Weiss, A.S.; Dehghani, F. Synthesis of highly porous crosslinked elastin hydrogels and their interaction with fibroblasts in vitro. Biomaterials 2009, 30, 4550–4557. [Google Scholar] [CrossRef]
- Leach, J.B.; Wolinsky, J.B.; Stone, P.J.; Wong, J.Y. Crosslinked α-elastin biomaterials: Towards a processable elastin mimetic scaffold. Acta Biomater. 2005, 1, 155–164. [Google Scholar] [CrossRef]
- Vrhovski, B.; Weiss, A.S. Biochemistry of tropoelastin. Eur. J. Biochem. 1998, 258, 1–18. [Google Scholar] [CrossRef]
- Mithieux, S.M.; Tu, Y.; Korkmaz, E.; Braet, F.; Weiss, A.S. In situ polymerization of tropoelastin in the absence of chemical cross-linking. Biomaterials 2009, 30, 431–435. [Google Scholar] [CrossRef]
- Barros, D.; Conde-Sousa, E.; Gonçalves, A.M.; Han, W.M.; García, A.J.; Amaral, I.F.; Pêgo, A.P. Engineering hydrogels with affinity-bound laminin as 3D neural stem cell culture systems. Biomater. Sci. 2019, 7, 5338–5349. [Google Scholar] [CrossRef] [PubMed]
- Hyysalo, A.; Ristola, M.; Mäkinen, M.E.-L.; Häyrynen, S.; Nykter, M.; Narkilahti, S. Laminin α5 substrates promote survival, network formation and functional development of human pluripotent stem cell-derived neurons in vitro. Stem Cell Res. 2017, 24, 118–127. [Google Scholar] [CrossRef]
- Seifu, D.G.; Purnama, A.; Mequanint, K.; Mantovani, D. Small-diameter vascular tissue engineering. Nat. Rev. Cardiol. 2013, 10, 410–421. [Google Scholar] [CrossRef] [PubMed]
- Briganti, E.; Spiller, D.; Mirtelli, C.; Kull, S.; Counoupas, C.; Losi, P.; Senesi, S.; Di Stefano, R.; Soldani, G. A composite fibrin-based scaffold for controlled delivery of bioactive pro-angiogenetic growth factors. J. Control. Release 2010, 142, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, D.; Adams, W.W.; Farmer, B.; Viney, C. Silk: Biology, Structure, Properties, and Genetics. In Silk Polymers; American Chemical Society (ACS): Washington, DC, USA, 1993; pp. 2–16. [Google Scholar]
- Lazaris, A.; Arcidiacono, S.; Huang, Y.; Zhou, J.-F.; Duguay, F.; Chretien, N.; Welsh, E.A.; Soares, J.W.; Karatzas, C.N. Spider silk fibers spun from soluble recombinant silk produced in mammalian cells. Science 2002, 295, 472–476. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.H.; Yeon, Y.K.; Lee, J.M.; Chao, J.R.; Lee, Y.J.; Seo, Y.B.; Sultan, M.T.; Lee, O.J.; Lee, J.S.; Yoon, S.I.; et al. Precisely printable and biocompatible silk fibroin bioink for digital light processing 3D printing. Nat. Commun. 2018, 9, 1620. [Google Scholar] [CrossRef]
- Orkin, R.W.; Gehron, P.; McGoodwin, E.B.; Martin, G.R.; Valentine, T.; Swarm, R. A murine tumor producing a matrix of basement membrane. J. Exp. Med. 1977, 145, 204–220. [Google Scholar] [CrossRef] [Green Version]
- Kleinman, H.K.; Martin, G.R. Matrigel: Basement membrane matrix with biological activity. Semin. Cancer Biol. 2005, 15, 378–386. [Google Scholar] [CrossRef]
- Petka, W.A.; Harden, J.L.; McGrath, K.P.; Wirtz, D.; Tirrell, D.A. Reversible Hydrogels from Self-Assembling Artificial Proteins. Science 1998, 281, 389–392. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Stewart, R.J.; Kopeček, J. Hybrid hydrogels assembled from synthetic polymers and coiled-coil protein domains. Nature 1999, 397, 417–420. [Google Scholar] [CrossRef]
- Li, Y.; Xue, B.; Cao, Y. 100th Anniversary of Macromolecular Science Viewpoint: Synthetic Protein Hydrogels. ACS Macro Lett. 2020, 9, 512–524. [Google Scholar] [CrossRef]
- Gomes, S.; Leonor, I.B.; Mano, J.F.; Reis, R.L.; Kaplan, D.L. Natural and genetically engineered proteins for tissue engineering. Prog. Polym. Sci. 2012, 37, 1–17. [Google Scholar] [CrossRef] [PubMed]
- DiMarco, R.L.; Heilshorn, S.C. Multifunctional Materials through Modular Protein Engineering. Adv. Mater. 2012, 24, 3923–3940. [Google Scholar] [CrossRef] [PubMed]
- Hudalla, G.A.; Sun, T.; Gasiorowski, J.Z.; Han, H.; Tian, Y.F.; Chong, A.S.; Collier, J.H. Gradated assembly of multiple proteins into supramolecular nanomaterials. Nat. Mater. 2014, 13, 829–836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H. ‘Mechanical Engineering’ of Elastomeric Proteins: Toward Designing New Protein Building Blocks for Biomaterials. Adv. Funct. Mater. 2008, 18, 2643–2657. [Google Scholar] [CrossRef]
- Elvin, C.M.; Carr, A.G.; Huson, M.G.; Maxwell, J.M.; Pearson, R.D.; Vuocolo, T.; Liyou, N.E.; Wong, D.C.C.; Merritt, D.; Dixon, N.E. Synthesis and properties of crosslinked recombinant pro-resilin. Nature 2005, 437, 999–1002. [Google Scholar] [CrossRef]
- Lampe, K.J.; Antaris, A.L.; Heilshorn, S.C. Design of three-dimensional engineered protein hydrogels for tailored control of neurite growth. Acta Biomater. 2013, 9, 5590–5599. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, M.A.; Simon, J.R.; Ghoorchian, A.; Scholl, Z.; Lin, S.; Rubinstein, M.; Marszalek, P.; Chilkoti, A.; López, G.P.; Zhao, X. Strong, Tough, Stretchable, and Self-Adhesive Hydrogels from Intrinsically Unstructured Proteins. Adv. Mater. 2017, 29, 1604743. [Google Scholar] [CrossRef]
- Duan, T.; Li, H. In Situ Phase Transition of Elastin-Like Polypeptide Chains Regulates Thermoresponsive Properties of Elastomeric Protein-Based Hydrogels. Biomacromolecules 2020, 21, 2258–2267. [Google Scholar] [CrossRef]
- Fancy, D.A.; Kodadek, T. Chemistry for the analysis of protein-protein interactions: Rapid and efficient cross-linking triggered by long wavelength light. Proc. Natl. Acad. Sci. USA 1999, 96, 6020–6024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, S.; Bu, T.; Kayser, J.; Bausch, A.; Li, H. Towards constructing extracellular matrix-mimetic hydrogels: An elastic hydrogel constructed from tandem modular proteins containing tenascin FnIII domains. Acta Biomater. 2013, 9, 6481–6491. [Google Scholar] [CrossRef] [PubMed]
- Gronenborn, A.M.; Filpula, D.R.; Essig, N.Z.; Achari, A.; Whitlow, M.; Wingfield, P.T.; Clore, G.M. A novel, highly stable fold of the immunoglobulin binding domain of streptococcal protein G. Science 1991, 253, 657–661. [Google Scholar] [CrossRef]
- Lv, S.; Dudek, D.M.; Cao, Y.; Balamurali, M.M.; Gosline, J.M.; Li, H. Designed biomaterials to mimic the mechanical properties of muscles. Nature 2010, 465, 69–73. [Google Scholar] [CrossRef]
- Fang, J.; Li, H. A Facile Way to Tune Mechanical Properties of Artificial Elastomeric Proteins-Based Hydrogels. Langmuir 2012, 28, 8260–8265. [Google Scholar] [CrossRef]
- Zakeri, B.; Fierer, J.O.; Celik, E.; Chittock, E.C.; Schwarz-Linek, U.; Moy, V.T.; Howarth, M. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. USA 2012, 109, E690–E697. [Google Scholar] [CrossRef] [Green Version]
- Sun, F.; Zhang, W.B.; Mahdavi, A.; Arnold, F.H.; Tirrell, D.A. Synthesis of bioactive protein hydrogels by genetically encoded SpyTag-SpyCatcher chemistry. Proc. Natl. Acad. Sci. USA 2014, 111, 11269–11274. [Google Scholar] [CrossRef] [Green Version]
- Zakeri, B.; Howarth, M. Spontaneous Intermolecular Amide Bond Formation between Side Chains for Irreversible Peptide Targeting. J. Am. Chem. Soc. 2010, 132, 4526–4527. [Google Scholar] [CrossRef] [Green Version]
- Keeble, A.H.; Howarth, M. Power to the protein: Enhancing and combining activities using the Spy toolbox. Chem. Sci. 2020, 11, 7281–7291. [Google Scholar] [CrossRef]
- Gao, X.; Lyu, S.; Li, H. Decorating a Blank Slate Protein Hydrogel: A General and Robust Approach for Functionalizing Protein Hydrogels. Biomacromolecules 2017, 18, 3726–3732. [Google Scholar] [CrossRef]
- Liu, X.; Yang, X.; Yang, Z.; Luo, J.; Tian, X.; Liu, K.; Kou, S.; Sun, F. Versatile Engineered Protein Hydrogels Enabling Decoupled Mechanical and Biochemical Tuning for Cell Adhesion and Neurite Growth. ACS Appl. Nano Mater. 2018, 1, 1579–1585. [Google Scholar] [CrossRef]
- Li, B.; Ren, K.; Wang, Y.; Qi, Y.; Chen, X.; Huang, Y. Protein-Cross-Linked Hydrogels with Tailored Swelling and Bioactivity Performance: A Comparative Study. ACS Appl. Mater. Interfaces 2016, 8, 30788–30796. [Google Scholar] [CrossRef] [PubMed]
- Discher, D.E.; Mooney, D.J.; Zandstra, P.W. Growth Factors, Matrices, and Forces Combine and Control Stem Cells. Science 2009, 324, 1673–1677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engler, A.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Discher, D.E.; Janmey, P.; Wang, Y.-L. Tissue Cells Feel and Respond to the Stiffness of Their Substrate. Science 2005, 310, 1139–1143. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Cao, Y. Protein Mechanics: From Single Molecules to Functional Biomaterials. Accounts Chem. Res. 2010, 43, 1331–1341. [Google Scholar] [CrossRef]
- Chung, J.; Kushner, A.M.; Weisman, A.C.; Guan, Z. Direct correlation of single-molecule properties with bulk mechanical performance for the biomimetic design of polymers. Nat. Mater. 2014, 13, 1055–1062. [Google Scholar] [CrossRef]
- Rief, M.; Gautel, M.; Oesterhelt, F.; Fernandez, J.M.; Gaub, H.E. Reversible Unfolding of Individual Titin Immunoglobulin Domains by AFM. Science 1997, 276, 1109–1112. [Google Scholar] [CrossRef] [Green Version]
- Linke, W.A.; Hamdani, N. Gigantic business: Titin properties and function through thick and thin. Circ. Res. 2014, 114, 1052–1068. [Google Scholar] [CrossRef]
- Bang, M.-L.; Centner, T.; Fornoff, F.; Geach, A.J.; Gotthardt, M.; McNabb, M.; Witt, C.C.; Labeit, D.; Gregorio, C.C.; Granzier, H.; et al. The Complete Gene Sequence of Titin, Expression of an Unusual ≈700-kDa Titin Isoform, and Its Interaction With Obscurin Identify a Novel Z-Line to I-Band Linking System. Circ. Res. 2001, 89, 1065–1072. [Google Scholar] [CrossRef] [Green Version]
- Kellermayer, M.S.Z.; Smith, S.B.; Granzier, H.L.; Bustamante, C. Folding-Unfolding Transitions in Single Titin Molecules Characterized with Laser Tweezers. Science 1997, 276, 1112–1116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tskhovrebova, L.; Trinick, J.; Sleep, J.A.; Simmons, R.M. Elasticity and unfolding of single molecules of the giant muscle protein titin. Nature 1997, 387, 308–312. [Google Scholar] [CrossRef]
- Rassier, D.E. Sarcomere mechanics in striated muscles: From molecules to sarcomeres to cells. Am. J. Physiol. Cell Physiol. 2017, 313, C134–C145. [Google Scholar] [CrossRef]
- Krüger, M.; Linke, W.A. The Giant Protein Titin: A Regulatory Node That Integrates Myocyte Signaling Pathways. J. Biol. Chem. 2011, 286, 9905–9912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linke, W.; Stockmeier, M.; Ivemeyer, M.; Hosser, H.; Mundel, P. Characterizing titin’s I-band Ig domain region as an entropic spring. J. Cell Sci. 1998, 111, 1567–1574. [Google Scholar] [CrossRef] [PubMed]
- Freiburg, A.; Trombitas, K.; Hell, W.; Cazorla, O.; Fougerousse, F.; Centner, T.; Kolmerer, B.; Witt, C.; Beckmann, J.S.; Gregorio, C.C.; et al. Series of Exon-Skipping Events in the Elastic Spring Region of Titin as the Structural Basis for Myofibrillar Elastic Diversity. Circ. Res. 2000, 86, 1114–1121. [Google Scholar] [CrossRef] [Green Version]
- Herrero-Galán, E.; Martínez-Martín, I.; Alegre-Cebollada, J. Redox regulation of protein nanomechanics in health and disease: Lessons from titin. Redox Biol. 2019, 21, 101074. [Google Scholar] [CrossRef]
- Li, H.; Linke, W.A.; Oberhauser, A.F.; Carrion-Vazquez, M.; Kerkvliet, J.G.; Lu, H.; Marszalek, P.E.; Fernandez, J.M. Reverse engineering of the giant muscle protein titin. Nature 2002, 418, 998–1002. [Google Scholar] [CrossRef]
- Herrero-Galán, E.; Domínguez, F.; Martínez-Martín, I.; Sánchez-González, C.; Vicente, N.; Lalaguna, L.; Bonzón-Kulichenko, E.; Calvo, E.; González-López, E.; Cobo-Marcos, M.; et al. Conserved cysteines in titin sustain the mechanical function of cardiomyocytes. bioRxiv 2020. [Google Scholar] [CrossRef]
- Alegre-Cebollada, J.; Kosuri, P.; Giganti, D.; Eckels, E.; Rivas-Pardo, J.A.; Hamdani, N.; Warren, C.M.; Solaro, R.J.; Linke, W.A.; Fernández, J.M. S-Glutathionylation of Cryptic Cysteines Enhances Titin Elasticity by Blocking Protein Folding. Cell 2014, 156, 1235–1246. [Google Scholar] [CrossRef] [Green Version]
- Giganti, D.; Yan, K.; Badilla, C.L.; Fernandez, J.M.; Alegre-Cebollada, J. Disulfide isomerization reactions in titin immunoglobulin domains enable a mode of protein elasticity. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Alegre-Cebollada, J.; Kosuri, P.; Rivas-Pardo, J.A.; Fernández, J.M. Direct observation of disulfide isomerization in a single protein. Nat. Chem. 2011, 3, 882–887. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Carrión-Vázquez, M.; Oberhauser, A.F.; Marszalek, P.E.; Fernandez, J.M. Point mutations alter the mechanical stability of immunoglobulin modules. Nat. Struct. Biol. 2000, 7, 1117–1120. [Google Scholar] [CrossRef]
- Brockwell, D.J.; Paci, E.; Zinober, R.C.; Beddard, G.S.; Olmsted, P.D.; Smith, D.A.; Perham, R.N.; Radford, S. Pulling geometry defines the mechanical resistance of a β-sheet protein. Nat. Struct. Mol. Biol. 2003, 10, 731–737. [Google Scholar] [CrossRef]
- Jagannathan, B.; Elms, P.J.; Bustamante, C.; Marqusee, S. Direct observation of a force-induced switch in the anisotropic mechanical unfolding pathway of a protein. Proc. Natl. Acad. Sci. USA 2012, 109, 17820–17825. [Google Scholar] [CrossRef] [Green Version]
- Dietz, H.; Berkemeier, F.; Bertz, M.; Rief, M. Anisotropic deformation response of single protein molecules. Proc. Natl. Acad. Sci. USA 2006, 103, 12724–12728. [Google Scholar] [CrossRef] [Green Version]
- Eckels, E.C.; Rivas-Pardo, J.A.; Valle-Orero, J.; Popa, I.; Fernandez, J.M. The Science of Stretching: Mechanical Anisotropy in Titin Ig Domains. Biophys. J. 2016, 110, 393a. [Google Scholar] [CrossRef] [Green Version]
- Carrión-Vázquez, M.; Li, H.; Lu, H.; Marszalek, P.E.; Oberhauser, A.F.; Fernandez, J.M. The mechanical stability of ubiquitin is linkage dependent. Nat. Struct. Mol. Biol. 2003, 10, 738–743. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Oberhauser, A.F.; Redick, S.D.; Carrión-Vázquez, M.; Erickson, H.P.; Fernandez, J.M. Multiple conformations of PEVK proteins detected by single-molecule techniques. Proc. Natl. Acad. Sci. USA 2001, 98, 10682–10686. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Fernandez, J.M. Mechanical design of the first proximal Ig domain of human cardiac titin revealed by single molecule force spectroscopy. J. Mol. Biol. 2003, 334, 75–86. [Google Scholar] [CrossRef]
- Linke, W.A.; Kulke, M.; Li, H.; Fujita-Becker, S.; Neagoe, C.; Manstein, D.J.; Gautel, M.; Fernandez, J.M. PEVK Domain of Titin: An Entropic Spring with Actin-Binding Properties. J. Struct. Biol. 2002, 137, 194–205. [Google Scholar] [CrossRef]
- Fu, L.; Haage, A.; Kong, N.; Tanentzapf, G.; Li, H. Dynamic protein hydrogels with reversibly tunable stiffness regulate human lung fibroblast spreading reversibly. Chem. Commun. 2019, 55, 5235–5238. [Google Scholar] [CrossRef]
- Kong, N.; Peng, Q.; Li, H. Rationally Designed Dynamic Protein Hydrogels with Reversibly Tunable Mechanical Properties. Adv. Funct. Mater. 2014, 24, 7310–7317. [Google Scholar] [CrossRef]
- da Silva, M.A.; Lenton, S.; Hughes, M.; Brockwell, D.J.; Dougan, L. Assessing the Potential of Folded Globular Polyproteins As Hydrogel Building Blocks. Biomacromolecules 2017, 18, 636–646. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Li, P.; Dong, C.; Jiang, H.; Xue, B.; Gao, X.; Qin, M.; Wang, W.; Chen, B.; Cao, Y. Rationally designed synthetic protein hydrogels with predictable mechanical properties. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Lei, H.; Dong, L.; Li, Y.; Zhang, J.; Chen, H.; Wu, J.; Zhang, Y.; Fan, Q.; Xue, B.; Qin, M.; et al. Stretchable hydrogels with low hysteresis and anti-fatigue fracture based on polyprotein cross-linkers. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Shmilovich, K.; Popa, I. Modeling Protein-Based Hydrogels under Force. Phys. Rev. Lett. 2018, 121, 168101. [Google Scholar] [CrossRef] [Green Version]
- Fu, L.; Li, H. Toward Quantitative Prediction of the Mechanical Properties of Tandem Modular Elastomeric Protein-Based Hydrogels. Macromolecules 2020, 53, 4704–4710. [Google Scholar] [CrossRef]
- Chaterji, S.; Kwon, I.K.; Park, K. Smart polymeric gels: Redefining the limits of biomedical devices. Prog. Polym. Sci. 2007, 32, 1083–1122. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, N.; Ferreira, L.; Cardoso, V.M.O.; Boni, F.; Souza, A.; Gremião, M. Recent advances in smart hydrogels for biomedical applications: From self-assembly to functional approaches. Eur. Polym. J. 2018, 99, 117–133. [Google Scholar] [CrossRef] [Green Version]
- Bao, X.; Si, X.; Ding, X.; Duan, L.; Xiao, C. pH-responsive hydrogels based on the self-assembly of short polypeptides for controlled release of peptide and protein drugs. J. Polym. Res. 2019, 26, 278. [Google Scholar] [CrossRef]
- Harden, J.L.; Li, H. Editorial: Special Issue on Designer Protein Biomaterials. ACS Biomater. Sci. Eng. 2017, 3, 658–660. [Google Scholar] [CrossRef] [Green Version]
- Roberts, S.; Harmon, T.S.; Schaal, J.L.; Miao, V.; Li, K.J.; Hunt, A.; Wen, Y.; Oas, T.G.; Collier, J.H.; Pappu, R.V.; et al. Injectable tissue integrating networks from recombinant polypeptides with tunable order. Nat. Mater. 2018, 17, 1154–1163. [Google Scholar] [CrossRef]
- Feng, Y.; Taraban, M.; Yu, Y.B. The effect of ionic strength on the mechanical, structural and transport properties of peptide hydrogels. Soft Matter 2012, 8, 11723–11731. [Google Scholar] [CrossRef]
- Pereira, R.N.; Souza, B.W.S.; Cerqueira, M.A.; Teixeira, J.A.; Vicente, A.A. Effects of Electric Fields on Protein Unfolding and Aggregation: Influence on Edible Films Formation. Biomacromolecules 2010, 11, 2912–2918. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Yang, Z.; Luo, J.; Hsing, I.-M.; Sun, F. B12-dependent photoresponsive protein hydrogels for controlled stem cell/protein release. Proc. Natl. Acad. Sci. USA 2017, 114, 5912–5917. [Google Scholar] [CrossRef] [Green Version]
- Parisi-Amon, A.; Mulyasasmita, W.; Chung, C.; Heilshorn, S.C. Protein-Engineered Injectable Hydrogel to Improve Retention of Transplanted Adipose-Derived Stem Cells. Adv. Heal. Mater. 2012, 2, 428–432. [Google Scholar] [CrossRef] [Green Version]
- Parisi-Amon, A.; Lo, D.D.; Montoro, D.; Dewi, R.E.; Longaker, M.T.; Heilshorn, S.C. Protein–Nanoparticle Hydrogels That Self-assemble in Response to Peptide-Based Molecular Recognition. ACS Biomater. Sci. Eng. 2017, 3, 750–756. [Google Scholar] [CrossRef]
- Kong, N.; Li, H. Protein Fragment Reconstitution as a Driving Force for Self-Assembling Reversible Protein Hydrogels. Adv. Funct. Mater. 2015, 25, 5593–5601. [Google Scholar] [CrossRef]
- Knoff, D.S.; Szczublewski, H.; Altamirano, D.; Cortes, K.A.F.; Kim, M. Cytoskeleton-Inspired Artificial Protein Design to Enhance Polymer Network Elasticity. Macromolecules 2020, 53, 3464–3471. [Google Scholar] [CrossRef]
- Cao, Y.; Li, H. Engineering tandem modular protein based reversible hydrogels. Chem. Commun. 2008, 35, 4144–4146. [Google Scholar] [CrossRef]
- Lv, S.; Cao, Y.; Li, H. Tandem Modular Protein-Based Hydrogels Constructed Using a Novel Two-Component Approach. Langmuir 2012, 28, 2269–2274. [Google Scholar] [CrossRef]
- Ozbas, B.; Kretsinger, J.; Rajagopal, K.; Schneider, J.P.; Pochan, D.J. Salt-Triggered Peptide Folding and Consequent Self-Assembly into Hydrogels with Tunable Modulus. Macromolecules 2004, 37, 7331–7337. [Google Scholar] [CrossRef]
- Khoury, L.R.; Popa, I. Chemical unfolding of protein domains induces shape change in programmed protein hydrogels. Nat. Commun. 2019, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Khoury, L.R.; Slawinski, M.; Collison, D.R.; Popa, I. Cation-induced shape programming and morphing in protein-based hydrogels. Sci. Adv. 2020, 6, eaba6112. [Google Scholar] [CrossRef]
- Ikeda, M.; Tanida, T.; Yoshii, T.; Kurotani, K.; Onogi, S.; Urayama, K.; Hamachi, I. Installing logic-gate responses to a variety of biological substances in supramolecular hydrogel–enzyme hybrids. Nat. Chem. 2014, 6, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Kong, N.; Fu, L.; Peng, Q.; Li, H. Metal Chelation Dynamically Regulates the Mechanical Properties of Engineered Protein Hydrogels. ACS Biomater. Sci. Eng. 2016, 3, 742–749. [Google Scholar] [CrossRef]
- Li, L.; Scheiger, J.M.; Levkin, P.A. Design and Applications of Photoresponsive Hydrogels. Adv. Mater. 2019, 31, e1807333. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Dong, C.; Huang, W.; Wang, H.; Wang, L.; Ding, D.; Zhou, H.; Long, J.; Wang, T.; Yang, Z. Rational design of a photo-responsive UVR8-derived protein and a self-assembling peptide–protein conjugate for responsive hydrogel formation. Nanoscale 2015, 7, 16666–16670. [Google Scholar] [CrossRef]
- Lyu, S.; Fang, J.; Duan, T.; Fu, L.; Liu, J.; Li, H. Optically controlled reversible protein hydrogels based on photoswitchable fluorescent protein Dronpa. Chem. Commun. 2017, 53, 13375–13378. [Google Scholar] [CrossRef]
- Gao, J.; Zhan, J.; Yang, Z. Enzyme-Instructed Self-Assembly (EISA) and Hydrogelation of Peptides. Adv. Mater. 2020, 32, 1805798. [Google Scholar] [CrossRef]
- Parmar, P.A.; Skaalure, S.C.; Chow, L.W.; St-Pierre, J.-P.; Stoichevska, V.; Peng, Y.Y.; Werkmeister, J.A.; Ramshaw, J.A.; Stevens, M.M. Temporally degradable collagen–mimetic hydrogels tuned to chondrogenesis of human mesenchymal stem cells. Biomaterials 2016, 99, 56–71. [Google Scholar] [CrossRef] [Green Version]
- Fang, J.; Mehlich, A.; Koga, N.; Huang, J.; Koga, R.; Gao, X.; Hu, C.; Jin, C.; Rief, M.; Kast, J.; et al. Forced protein unfolding leads to highly elastic and tough protein hydrogels. Nat. Commun. 2013, 4, 1–10. [Google Scholar] [CrossRef]
- Chilkoti, A.; Dreher, M.R.; Meyer, D.E. Design of thermally responsive, recombinant polypeptide carriers for targeted drug delivery. Adv. Drug Deliv. Rev. 2002, 54, 1093–1111. [Google Scholar] [CrossRef]
- Chilkoti, A.; Christensen, T.; Mackay, J.A. Stimulus responsive elastin biopolymers: Applications in medicine and biotechnology. Curr. Opin. Chem. Biol. 2006, 10, 652–657. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Fu, L.; Liu, J.; Li, H. Decorating protein hydrogels reversibly enables dynamic presentation and release of functional protein ligands on protein hydrogels. Chem. Commun. 2019, 55, 12703–12706. [Google Scholar] [CrossRef]
- Saqlain, F.; Popa, I.; Fernández, J.M.; Alegre-Cebollada, J. A Novel Strategy for Utilizing Voice Coil Servoactuators in Tensile Tests of Low Volume Protein Hydrogels. Macromol. Mater. Eng. 2015, 300, 369–376. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Kopeček, J.; Stewart, R.J. Responsive Hybrid Hydrogels with Volume Transitions Modulated by a Titin Immunoglobulin Module. Bioconjug. Chem. 2000, 11, 734–740. [Google Scholar] [CrossRef]
- Zagórska-Dziok, M.; Sobczak, M. Hydrogel-Based Active Substance Release Systems for Cosmetology and Dermatology Application: A Review. Pharmaceutics 2020, 12, 396. [Google Scholar] [CrossRef] [PubMed]
- Sionkowska, A.; Skrzyński, S.; Śmiechowski, K.; Kołodziejczak, A. The review of versatile application of collagen. Polym. Adv. Technol. 2017, 28, 4–9. [Google Scholar] [CrossRef]
- Van Duong, H.; Chau, T.T.L.; Dang, N.T.T.; Van Nguyen, D.; Le, S.L.; Ho, T.S.; Vu, T.P.; Van Tran, T.T.; Nguyen, T.-D. Self-aggregation of water-dispersible nanocollagen helices. Biomater. Sci. 2018, 6, 651–660. [Google Scholar] [CrossRef]
- Murphy, K.C.; Whitehead, J.; Zhou, D.; Ho, S.S.; Leach, J.K. Engineering fibrin hydrogels to promote the wound healing potential of mesenchymal stem cell spheroids. Acta Biomater. 2017, 64, 176–186. [Google Scholar] [CrossRef]
- Mandal, A.; Clegg, J.R.; Anselmo, A.C.; Mitragotri, S. Hydrogels in the clinic. Bioeng. Transl. Med. 2020, 5, e10158. [Google Scholar] [CrossRef] [Green Version]
- Mitura, S.; Sionkowska, A.; Jaiswal, A. Biopolymers for hydrogels in cosmetics: Review. J. Mater. Sci. Mater. Med. 2020, 31, 1–14. [Google Scholar] [CrossRef]
- Zhu, Z.; Ling, S.; Yeo, J.; Zhao, S.; Tozzi, L.; Buehler, M.J.; Omenetto, F.; Li, C.; Kaplan, D.L. High-Strength, Durable All-Silk Fibroin Hydrogels with Versatile Processability toward Multifunctional Applications. Adv. Funct. Mater. 2018, 28. [Google Scholar] [CrossRef]
- Chattopadhyay, S.; Raines, R.T. Review collagen-based biomaterials for wound healing. Biopolymers 2014, 101, 821–833. [Google Scholar] [CrossRef] [Green Version]
- Brett, D. A Review of Collagen and Collagen-based Wound Dressings. Wounds 2008, 20, 347–356. [Google Scholar] [PubMed]
- Kim, S.Y.; Park, B.J.; Lee, Y.; Park, N.J.; Park, K.M.; Hwang, Y.-S.; Park, K.D. Human hair keratin-based hydrogels as dynamic matrices for facilitating wound healing. J. Ind. Eng. Chem. 2019, 73, 142–151. [Google Scholar] [CrossRef]
- He, S.; Shi, D.; Han, Z.; Dong, Z.; Xie, Y.; Zhang, F.; Zeng, W.; Yi, Q. Heparinized silk fibroin hydrogels loading FGF1 promote the wound healing in rats with full-thickness skin excision. Biomed. Eng. Online 2019, 18, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, X.; Cebe, P.; Weiss, A.S.; Omenetto, F.; Kaplan, D.L. Protein-based composite materials. Mater. Today 2012, 15, 208–215. [Google Scholar] [CrossRef]
- Li, L.; Eyckmans, J.; Chen, C. Designer biomaterials for mechanobiology. Nat. Mater. 2017, 16, 1164–1168. [Google Scholar] [CrossRef]
- Langer, R. Drug delivery and targeting. Nature 1998, 392, 5–10. [Google Scholar]
- Werle, M.; Bernkop-Schnürch, A. Strategies to improve plasma half life time of peptide and protein drugs. Amino Acids 2006, 30, 351–367. [Google Scholar] [CrossRef] [PubMed]
- Mathur, D.; Prakash, S.; Anand, P.; Kaur, H.; Agrawal, P.; Mehta, A.; Kumar, R.; Singh, S.; Raghava, G.P.S. PEPlife: A Repository of the Half-life of Peptides. Sci. Rep. 2016, 6, 36617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Mooney, D.J. Designing hydrogels for controlled drug delivery. Nat. Rev. Mater. 2016, 1, 1–17. [Google Scholar] [CrossRef]
- Guziewicz, N.; Best, A.; Perez-Ramirez, B.; Kaplan, D.L. Lyophilized silk fibroin hydrogels for the sustained local delivery of therapeutic monoclonal antibodies. Biomaterials 2011, 32, 2642–2650. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Wang, X.; Wang, S.; Zhao, J.; Xu, L.; Zhu, C.; Zeng, D.; Chen, J.; Zhang, Z.; Kaplan, D.L.; et al. The use of injectable sonication-induced silk hydrogel for VEGF165 and BMP-2 delivery for elevation of the maxillary sinus floor. Biomaterials 2011, 32, 9415–9424. [Google Scholar] [CrossRef] [Green Version]
- Numata, K.; Reagan, M.; Goldstein, R.H.; Rosenblatt, M.; Kaplan, D.L. Spider Silk-Based Gene Carriers for Tumor Cell-Specific Delivery. Bioconjug. Chem. 2011, 22, 1605–1610. [Google Scholar] [CrossRef] [Green Version]
- Megeed, Z.; Haider, M.; Li, D.; O’Malley, B.W.; Cappello, J.; Ghandehari, H. In vitro and in vivo evaluation of recombinant silk-elastinlike hydrogels for cancer gene therapy. J. Control. Release 2004, 94, 433–445. [Google Scholar] [CrossRef] [PubMed]
- Greish, K.; Araki, K.; Li, D.; O’Malley, J.B.W.; Dandu, R.; Frandsen, J.; Cappello, J.; Ghandehari, H. Silk-Elastinlike Protein Polymer Hydrogels for Localized Adenoviral Gene Therapy of Head and Neck Tumors. Biomacromolecules 2009, 10, 2183–2188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandit, A.; Ashar, R.; Feldman, D. The Effect of TGF-beta Delivered Through a Collagen Scaffold on Wound Healing. J. Investig. Surg. 1999, 12, 89–100. [Google Scholar] [CrossRef]
- Pierce, B.F.; Pittermann, E.; Ma, N.; Gebauer, T.; Neffe, A.T.; Hölscher, M.; Jung, F.; Lendlein, A. Viability of Human Mesenchymal Stem Cells Seeded on Crosslinked Entropy-Elastic Gelatin-Based Hydrogels. Macromol. Biosci. 2012, 12, 312–321. [Google Scholar] [CrossRef]
- Bigi, A.; Cojazzi, G.; Panzavolta, S.; Roveri, N.; Rubini, K. Stabilization of gelatin films by crosslinking with genipin. Biomaterials 2002, 23, 4827–4832. [Google Scholar] [CrossRef]
- Bessa, P.C.; Casal, M.; Reis, R.L. Bone morphogenetic proteins in tissue engineering: The road from laboratory to clinic, part II (BMP delivery). J. Tissue Eng. Regen. Med. 2008, 2, 81–96. [Google Scholar] [CrossRef]
- Maltais, A.; Remondetto, G.E.; Subirade, M. Soy protein cold-set hydrogels as controlled delivery devices for nutraceutical compounds. Food Hydrocoll. 2009, 23, 1647–1653. [Google Scholar] [CrossRef]
- Gao, X.; Fang, J.; Xue, B.; Fu, L.; Li, H. Engineering Protein Hydrogels Using SpyCatcher-SpyTag Chemistry. Biomacromolecules 2016, 17, 2812–2819. [Google Scholar] [CrossRef] [PubMed]
- Grove, T.Z.; Osuji, C.O.; Forster, J.D.; Dufresne, E.R.; Regan, L. Stimuli-Responsive Smart Gels Realized via Modular Protein Design. J. Am. Chem. Soc. 2010, 132, 14024–14026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Andarias, J.; Mejías, S.H.; Sakurai, T.; Matsuda, W.; Seki, S.; Feixas, F.; Osuna, S.; Atienza, C.; Martín, N.; Cortajarena, A.L. Toward Bioelectronic Nanomaterials: Photoconductivity in Protein–Porphyrin Hybrids Wrapped around SWCNT. Adv. Funct. Mater. 2018, 28, 1704031. [Google Scholar] [CrossRef]
- Yu, Z.; Xu, Q.; Dong, C.; Lee, S.S.; Gao, L.; Li, Y.; D’Ortenzio, M.; Wu, J. Self-Assembling Peptide Nanofibrous Hydrogel as a Versatile Drug Delivery Platform. Curr. Pharm. Des. 2015, 21, 4342–4354. [Google Scholar] [CrossRef] [PubMed]
- Moyer, T.J.; Finbloom, J.A.; Chen, F.; Toft, D.J.; Cryns, V.L.; Stupp, S.I. pH and Amphiphilic Structure Direct Supramolecular Behavior in Biofunctional Assemblies. J. Am. Chem. Soc. 2014, 136, 14746–14752. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zeng, R.; Li, C.; Qiao, R. Self-assembled polypeptide-block-poly(vinylpyrrolidone) as prospective drug-delivery systems. Colloids Surf. B Biointerfaces 2009, 74, 284–292. [Google Scholar] [CrossRef]
- Huang, J.; Hastings, C.L.; Duffy, G.; Kelly, H.M.; Raeburn, J.; Adams, D.; Heise, A. Supramolecular Hydrogels with Reverse Thermal Gelation Properties from (Oligo)tyrosine Containing Block Copolymers. Biomacromolecules 2013, 14, 200–206. [Google Scholar] [CrossRef]
- Karimi, M.; Ghasemi, A.; Zangabad, P.S.; Rahighi, R.; Basri, S.M.M.; Mirshekari, H.; Amiri, M.; Pishabad, Z.S.; Aslani, A.; Bozorgomid, M.; et al. Smart micro/nanoparticles in stimulus-responsive drug/gene delivery systems. Chem. Soc. Rev. 2016, 45, 1457–1501. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.-Y.; Hu, J.-J.; Xu, Q.; Chen, S.; Jia, H.-Z.; Sun, Y.-X.; Zhuo, R.-X.; Zhang, X.-Z. A redox-responsive drug delivery system based on RGD containing peptide-capped mesoporous silica nanoparticles. J. Mater. Chem. B 2014, 3, 39–44. [Google Scholar] [CrossRef]
- Huo, M.; Yuan, J.; Tao, L.; Wei, Y. Redox-responsive polymers for drug delivery: From molecular design to applications. Polym. Chem. 2014, 5, 1519–1528. [Google Scholar] [CrossRef]
- Waters, R.; Alam, P.; Pacelli, S.; Chakravarti, A.R.; Ahmed, R.P.; Paul, A. Stem cell-inspired secretome-rich injectable hydrogel to repair injured cardiac tissue. Acta Biomater. 2018, 69, 95–106. [Google Scholar] [CrossRef]
- Lyu, Y.; Azevedo, H.S. Supramolecular Hydrogels for Protein Delivery in Tissue Engineering. Molecules 2021, 26, 873. [Google Scholar] [CrossRef] [PubMed]
- Hoare, T.R.; Kohane, D.S. Hydrogels in drug delivery: Progress and challenges. Polymer 2008, 49, 1993–2007. [Google Scholar] [CrossRef] [Green Version]
- Schloss, A.C.; Williams, D.M.; Regan, L.J. Protein-Based Hydrogels for Tissue Engineering. In Protein-Based Engineered Nanostructures; Cortajarena, A.L., Grove, T.Z., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 167–177. [Google Scholar]
- LeMieux, J.; Hava, D.L.; Basset, A.; Camilli, A. RrgA and RrgB Are Components of a Multisubunit Pilus Encoded by the Streptococcus pneumoniae rlrA Pathogenicity Islet. Infect. Immun. 2006, 74, 2453–2456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miranda-Nieves, D.; Chaikof, E.L. Collagen and Elastin Biomaterials for the Fabrication of Engineered Living Tissues. ACS Biomater. Sci. Eng. 2017, 3, 694–711. [Google Scholar] [CrossRef]
- Tulloch, N.L.; Muskheli, V.; Razumova, M.V.; Korte, F.S.; Regnier, M.; Hauch, K.D.; Pabon, L.; Reinecke, H.; Murry, C.E. Growth of Engineered Human Myocardium With Mechanical Loading and Vascular Coculture. Circ. Res. 2011, 109, 47–59. [Google Scholar] [CrossRef]
- Foster, A.A.; Dewi, R.E.; Cai, L.; Hou, L.; Strassberg, Z.; Alcazar, C.A.; Heilshorn, S.C.; Huang, N.F. Protein-engineered hydrogels enhance the survival of induced pluripotent stem cell-derived endothelial cells for treatment of peripheral arterial disease. Biomater. Sci. 2018, 6, 614–622. [Google Scholar] [CrossRef]
- Amini, A.R.; Laurencin, C.T.; Nukavarapu, S.P. Bone Tissue Engineering: Recent Advances and Challenges. Crit. Rev. Biomed. Eng. 2012, 40, 363–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhumiratana, S.; Vunjak-Novakovic, G. Engineering physiologically stiff and stratified human cartilage by fusing condensed mesenchymal stem cells. Methods 2015, 84, 109–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhumiratana, S.; Bernhard, J.C.; Alfi, D.M.; Yeager, K.; Eton, R.E.; Bova, J.; Shah, F.; Gimble, J.M.; Lopez, M.J.; Eisig, S.B.; et al. Tissue-engineered autologous grafts for facial bone reconstruction. Sci. Transl. Med. 2016, 8, 343ra83. [Google Scholar] [CrossRef] [Green Version]
- Kuo, C.K.; Li, W.-J.; Mauck, R.L.; Tuan, R.S. Cartilage tissue engineering: Its potential and uses. Curr. Opin. Rheumatol. 2006, 18, 64–73. [Google Scholar] [CrossRef]
- Liu, M.; Zeng, X.; Ma, C.; Yi, H.; Ali, Z.; Mou, X.; Li, S.; Deng, Y.; He, N. Injectable hydrogels for cartilage and bone tissue engineering. Bone Res. 2017, 5, 17014. [Google Scholar] [CrossRef]
- Tavakol, D.N.; Tratwal, J.; Bonini, F.; Genta, M.; Campos, V.; Burch, P.; Hoehnel, S.; Béduer, A.; Alessandrini, M.; Naveiras, O.; et al. Injectable, scalable 3D tissue-engineered model of marrow hematopoiesis. Biomaterials 2020, 232, 119665. [Google Scholar] [CrossRef] [PubMed]
- Lamme, E.N.; van Leeuwen, R.T.; Jonker, A.; van Marle, J.; Middelkoop, E. Living Skin Substitutes: Survival and Function of Fibroblasts Seeded in a Dermal Substitute in Experimental Wounds. J. Investig. Dermatol. 1998, 111, 989–995. [Google Scholar] [CrossRef]
- Polacheck, W.J.; Kutys, M.L.; Tefft, J.B.; Chen, C.S. Microfabricated blood vessels for modeling the vascular transport barrier. Nat. Protoc. 2019, 14, 1425–1454. [Google Scholar] [CrossRef]
- Nguyen, D.-H.T.; Lee, E.; Alimperti, S.; Norgard, R.J.; Wong, A.; Lee, J.J.-K.; Eyckmans, J.; Stanger, B.Z.; Chen, C.S. A biomimetic pancreatic cancer on-chip reveals endothelial ablation via ALK7 signaling. Sci. Adv. 2019, 5, eaav6789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemme, M.; Ulmer, B.M.; Lemoine, M.D.; Zech, A.T.; Flenner, F.; Ravens, U.; Reichenspurner, H.; Rol-Garcia, M.; Smith, G.; Hansen, A.; et al. Atrial-like Engineered Heart Tissue: An In Vitro Model of the Human Atrium. Stem Cell Rep. 2018, 11, 1378–1390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronaldson-Bouchard, K.; Ma, S.P.; Yeager, K.; Chen, T.; Song, L.; Sirabella, D.; Morikawa, K.; Teles, D.; Yazawa, M.; Vunjak-Novakovic, G. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 2018, 556, 239–243. [Google Scholar] [CrossRef]
- Feric, N.T.; Radisic, M. Maturing human pluripotent stem cell-derived cardiomyocytes in human engineered cardiac tissues. Adv. Drug Deliv. Rev. 2016, 96, 110–134. [Google Scholar] [CrossRef] [Green Version]
- Gelain, F.; Luo, Z.; Rioult, M.; Zhang, S. Self-assembling peptide scaffolds in the clinic. NPJ Regen. Med. 2021, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hynes, R.O. The Extracellular Matrix: Not Just Pretty Fibrils. Science 2009, 326, 1216–1219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prince, E.; Kumacheva, E. Design and applications of man-made biomimetic fibrillar hydrogels. Nat. Rev. Mater. 2019, 4, 99–115. [Google Scholar] [CrossRef]
- Jansen, K.A.; Donato, D.M.; Balcioglu, H.E.; Schmidt, T.; Danen, E.H.; Koenderink, G.H. A guide to mechanobiology: Where biology and physics meet. Biochim. Biophys. Acta BBA Mol. Cell Res. 2015, 1853, 3043–3052. [Google Scholar] [CrossRef] [Green Version]
- Rosales, A.M.; Anseth, K.S. The design of reversible hydrogels to capture extracellular matrix dynamics. Nat. Rev. Mater. 2016, 1, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Vining, K.H.; Mooney, D.J. Mechanical forces direct stem cell behaviour in development and regeneration. Nat. Rev. Mol. Cell Biol. 2017, 18, 728–742. [Google Scholar] [CrossRef] [PubMed]
- Mammoto, T.; Mammoto, A.; Ingber, D.E. Mechanobiology and Developmental Control. Annu. Rev. Cell Dev. Biol. 2013, 29, 27–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tajik, A.; Zhang, Y.; Wei, F.; Sun, J.; Jia, Q.; Zhou, W.; Singh, R.; Khanna, N.; Belmont, A.S.; Wang, N. Transcription upregulation via force-induced direct stretching of chromatin. Nat. Mater. 2016, 15, 1287–1296. [Google Scholar] [CrossRef] [Green Version]
- Totaro, A.; Panciera, T.; Piccolo, S. YAP/TAZ upstream signals and downstream responses. Nat. Cell Biol. 2018, 20, 888–899. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.; Trappmann, B.; Wang, W.Y.; Sakar, M.S.; Kim, I.L.; Shenoy, V.B.; Burdick, J.A.; Chen, C. Cell-mediated fibre recruitment drives extracellular matrix mechanosensing in engineered fibrillar microenvironments. Nat. Mater. 2015, 14, 1262–1268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphries, J.; Chastney, M.; Askari, J.A.; Humphries, M.J. Signal transduction via integrin adhesion complexes. Curr. Opin. Cell Biol. 2019, 56, 14–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aragona, M.; Panciera, T.; Manfrin, A.; Giulitti, S.; Michielin, F.; Elvassore, N.; Dupont, S.; Piccolo, S. A Mechanical Checkpoint Controls Multicellular Growth through YAP/TAZ Regulation by Actin-Processing Factors. Cell 2013, 154, 1047–1059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lo, C.-M.; Wang, H.-B.; Dembo, M.; Wang, Y.-L. Cell Movement Is Guided by the Rigidity of the Substrate. Biophys. J. 2000, 79, 144–152. [Google Scholar] [CrossRef] [Green Version]
- Murphy, W.L.; McDevitt, T.C.; Engler, A. Materials as stem cell regulators. Nat. Mater. 2014, 13, 547–557. [Google Scholar] [CrossRef]
- Paszek, M.J.; Zahir, N.; Johnson, K.R.; Lakins, J.N.; Rozenberg, G.I.; Gefen, A.; Reinhart-King, C.A.; Margulies, S.S.; Dembo, M.; Boettiger, D.; et al. Tensional homeostasis and the malignant phenotype. Cancer Cell 2005, 8, 241–254. [Google Scholar] [CrossRef] [Green Version]
- Engler, A.; Bacakova, L.; Newman, C.; Hategan, A.; Griffin, M.; Discher, D. Substrate Compliance versus Ligand Density in Cell on Gel Responses. Biophys. J. 2004, 86, 617–628. [Google Scholar] [CrossRef] [Green Version]
- Engler, A.J.; Griffin, M.A.; Sen, S.; Bonnemann, C.G.; Sweeney, H.L.; Discher, D.E. Myotubes differentiate optimally on substrates with tissue-like stiffness: Pathological implications for soft or stiff microenvironments. J. Cell Biol. 2004, 166, 877–887. [Google Scholar] [CrossRef] [Green Version]
- Butcher, D.T.; Alliston, T.; Weaver, V.M. A tense situation: Forcing tumour progression. Nat. Rev. Cancer 2009, 9, 108–122. [Google Scholar] [CrossRef]
- Herron, T.J.; Da Rocha, A.M.; Campbell, K.F.; Ponce-Balbuena, D.; Willis, B.C.; Guerrero-Serna, G.; Liu, Q.; Klos, M.; Musa, H.; Zarzoso, M.; et al. Extracellular Matrix–Mediated Maturation of Human Pluripotent Stem Cell–Derived Cardiac Monolayer Structure and Electrophysiological Function. Circ. Arrhythmia Electrophysiol. 2016, 9, e003638. [Google Scholar] [CrossRef] [Green Version]
- Wen, Q.; Janmey, P.A. Effects of non-linearity on cell–ECM interactions. Exp. Cell Res. 2013, 319, 2481–2489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janmey, P.A.; McCulloch, C.A. Cell Mechanics: Integrating Cell Responses to Mechanical Stimuli. Annu. Rev. Biomed. Eng. 2007, 9, 1–34. [Google Scholar] [CrossRef] [Green Version]
- Seo, B.R.; Chen, X.; Ling, L.; Song, Y.H.; Shimpi, A.A.; Choi, S.; Gonzalez, J.; Sapudom, J.; Wang, K.; Eguiluz, R.C.A.; et al. Collagen microarchitecture mechanically controls myofibroblast differentiation. Proc. Natl. Acad. Sci. USA 2020, 117, 11387–11398. [Google Scholar] [CrossRef] [PubMed]
- Denisin, A.K.; Pruitt, B.L. Tuning the Range of Polyacrylamide Gel Stiffness for Mechanobiology Applications. ACS Appl. Mater. Interfaces 2016, 8, 21893–21902. [Google Scholar] [CrossRef]
- Haugh, M.G.; Heilshorn, S.C. Integrating concepts of material mechanics, ligand chemistry, dimensionality and degradation to control differentiation of mesenchymal stem cells. Curr. Opin. Solid State Mater. Sci. 2016, 20, 171–179. [Google Scholar] [CrossRef] [Green Version]
- Haugh, M.G.; Vaughan, T.J.; Madl, C.M.; Raftery, R.; McNamara, L.M.; O’Brien, F.J.; Heilshorn, S.C. Investigating the interplay between substrate stiffness and ligand chemistry in directing mesenchymal stem cell differentiation within 3D macro-porous substrates. Biomaterials 2018, 171, 23–33. [Google Scholar] [CrossRef]
- Harley, B.A.; Kim, H.-D.; Zaman, M.H.; Yannas, I.V.; Lauffenburger, D.A.; Gibson, L.J. Microarchitecture of Three-Dimensional Scaffolds Influences Cell Migration Behavior via Junction Interactions. Biophys. J. 2008, 95, 4013–4024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarz, U.S.; Bischofs, I.B. Physical determinants of cell organization in soft media. Med Eng. Phys. 2005, 27, 763–772. [Google Scholar] [CrossRef]
- McCoy, R.J.; Jungreuthmayer, C.; O’Brien, F.J. Influence of flow rate and scaffold pore size on cell behavior during mechanical stimulation in a flow perfusion bioreactor. Biotechnol. Bioeng. 2012, 109, 1583–1594. [Google Scholar] [CrossRef]
- Chaudhuri, O.; Cooper-White, J.; Janmey, P.A.; Mooney, D.J.; Shenoy, V.B. Effects of extracellular matrix viscoelasticity on cellular behaviour. Nature 2020, 584, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, O.; Gu, L.; Klumpers, D.; Darnell, M.; Bencherif, S.A.; Weaver, J.C.; Huebsch, N.; Lee, H.-P.; Lippens, E.; Duda, G.N.; et al. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat. Mater. 2016, 15, 326–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaudhuri, O.; Gu, L.; Darnell, M.; Klumpers, D.D.; Bencherif, S.A.; Weaver, J.C.; Huebsch, N.; Mooney, D.J. Substrate stress relaxation regulates cell spreading. Nat. Commun. 2015, 6, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nam, S.; Lee, J.; Brownfield, D.; Chaudhuri, O. Viscoplasticity Enables Mechanical Remodeling of Matrix by Cells. Biophys. J. 2016, 111, 2296–2308. [Google Scholar] [CrossRef] [Green Version]
- Vining, K.H.; Stafford, A.; Mooney, D.J. Sequential modes of crosslinking tune viscoelasticity of cell-instructive hydrogels. Biomaterials 2019, 188, 187–197. [Google Scholar] [CrossRef]
- Cameron, A.; Frith, J.; Gomez, G.; Yap, A.S.; Cooper-White, J.J. The effect of time-dependent deformation of viscoelastic hydrogels on myogenic induction and Rac1 activity in mesenchymal stem cells. Biomaterials 2014, 35, 1857–1868. [Google Scholar] [CrossRef]
- Cameron, A.; Frith, J.; Cooper-White, J.J. The influence of substrate creep on mesenchymal stem cell behaviour and phenotype. Biomaterials 2011, 32, 5979–5993. [Google Scholar] [CrossRef]
- Charrier, E.E.; Pogoda, K.; Wells, R.G.; Janmey, P.A. Control of cell morphology and differentiation by substrates with independently tunable elasticity and viscous dissipation. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Marozas, I.A.; Anseth, K.S.; Cooper-White, J.J. Adaptable boronate ester hydrogels with tunable viscoelastic spectra to probe timescale dependent mechanotransduction. Biomaterials 2019, 223, 119430. [Google Scholar] [CrossRef]
- Tang, S.; Ma, H.; Tu, H.; Wang, H.; Lin, P.; Anseth, K.S. Adaptable Fast Relaxing Boronate-Based Hydrogels for Probing Cell–Matrix Interactions. Adv. Sci. 2018, 5, 1800638. [Google Scholar] [CrossRef] [Green Version]
- McKinnon, D.D.; Domaille, D.W.; Cha, J.N.; Anseth, K.S. Biophysically Defined and Cytocompatible Covalently Adaptable Networks as Viscoelastic 3D Cell Culture Systems. Adv. Mater. 2014, 26, 865–872. [Google Scholar] [CrossRef] [Green Version]
- Muller, P.M.; Rademacher, J.; Bagshaw, R.D.; Wortmann, C.; Barth, C.; van Unen, J.; Alp, K.M.; Giudice, G.; Eccles, R.L.; Heinrich, L.E.; et al. Systems analysis of RhoGEF and RhoGAP regulatory proteins reveals spatially organized RAC1 signalling from integrin adhesions. Nat. Cell Biol. 2020, 22, 498–511. [Google Scholar] [CrossRef] [PubMed]
- Batlle, E.; Massague, J. Transforming Growth Factor-beta Signaling in Immunity and Cancer. Immunity 2019, 50, 924–940. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Li, L.; Wang, L.; Wang, C.-Y.; Yu, J.; Guan, K.-L. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012, 26, 54–68. [Google Scholar] [CrossRef] [Green Version]
- Rausch, V.; Hansen, C.G. The Hippo Pathway, YAP/TAZ, and the Plasma Membrane. Trends Cell Biol. 2020, 30, 32–48. [Google Scholar] [CrossRef]
- Meng, Z.; Qiu, Y.; Lin, K.C.; Kumar, A.; Placone, J.K.; Fang, C.; Wang, K.-C.; Lu, S.; Pan, M.; Hong, A.W.; et al. RAP2 mediates mechanoresponses of the Hippo pathway. Nature 2018, 560, 655–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brusatin, G.; Panciera, T.; Gandin, A.; Citron, A.; Piccolo, S. Biomaterials and engineered microenvironments to control YAP/TAZ-dependent cell behaviour. Nat. Mater. 2018, 17, 1063–1075. [Google Scholar] [CrossRef]
- Dupont, S.; Morsut, L.; Aragona, M.; Enzo, E.; Giulitti, S.; Cordenonsi, M.; Zanconato, F.; Le Digabel, J.; Forcato, M.; Bicciato, S.; et al. Role of YAP/TAZ in mechanotransduction. Nature 2011, 474, 179–183. [Google Scholar] [CrossRef]
- Moreno-Vicente, R.; Pavón, D.M.; Martín-Padura, I.; Català-Montoro, M.; Díez-Sánchez, A.; Quílez-Álvarez, A.; Lopez, J.A.; Sanchez-Alvarez, M.; Vázquez, J.; Strippoli, R.; et al. Caveolin-1 Modulates Mechanotransduction Responses to Substrate Stiffness through Actin-Dependent Control of YAP. Cell Rep. 2018, 25, 1622–1635.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Udan, R.S.; Kango-Singh, M.; Nolo, R.; Tao, C.; Halder, G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 2003, 5, 914–920. [Google Scholar] [CrossRef]
- Dooling, L.J.; Buck, M.E.; Zhang, W.-B.; Tirrell, D.A. Programming Molecular Association and Viscoelastic Behavior in Protein Networks. Adv. Mater. 2016, 28, 4651–4657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Huebsch, N.; Mooney, D.; Suo, Z. Stress-relaxation behavior in gels with ionic and covalent crosslinks. J. Appl. Phys. 2010, 107, 063509. [Google Scholar] [CrossRef] [Green Version]
- Billiet, T.; Vandenhaute, M.; Schelfhout, J.; Van Vlierberghe, S.; Dubruel, P. A review of trends and limitations in hydrogel-rapid prototyping for tissue engineering. Biomaterials 2012, 33, 6020–6041. [Google Scholar] [CrossRef]
- Pacak, C.A.; Powers, J.M.; Cowan, D.B. Ultrarapid Purification of Collagen Type I for Tissue Engineering Applications. Tissue Eng. Part C Methods 2011, 17, 879–885. [Google Scholar] [CrossRef] [Green Version]
- Mattanovich, D.; Branduardi, P.; Dato, L.; Gasser, B.; Sauer, M.; Porro, D. Recombinant Protein Production in Yeasts. In Recombinant Gene Expression; Lorence, A., Ed.; Humana Press: Totowa, NJ, USA, 2012; pp. 329–358. [Google Scholar]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5, 172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Y.; Jeong, J.; DeVolder, R.J.; Cha, C.; Wang, F.; Tong, Y.W.; Kong, H. A cell-instructive hydrogel to regulate malignancy of 3D tumor spheroids with matrix rigidity. Biomaterials 2011, 32, 9308–9315. [Google Scholar] [CrossRef]
- Narayanaswamy, R.; Torchilin, V.P. Hydrogels and Their Applications in Targeted Drug Delivery. Molecules 2019, 24, 603. [Google Scholar] [CrossRef] [Green Version]
- Smith, C. Striving for purity: Advances in protein purification. Nat. Methods 2005, 2, 71–77. [Google Scholar] [CrossRef]
- Khoury, L.R.; Nowitzke, J.; Shmilovich, K.; Popa, I. Study of Biomechanical Properties of Protein-Based Hydrogels Using Force-Clamp Rheometry. Macromolecules 2018, 51, 1441–1452. [Google Scholar] [CrossRef]
- Sun, W.; Xue, B.; Fan, Q.; Tao, R.; Wang, C.; Wang, X.; Li, Y.; Qin, M.; Wang, W.; Chen, B.; et al. Molecular engineering of metal coordination interactions for strong, tough, and fast-recovery hydrogels. Sci. Adv. 2020, 6, eaaz9531. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huerta-López, C.; Alegre-Cebollada, J. Protein Hydrogels: The Swiss Army Knife for Enhanced Mechanical and Bioactive Properties of Biomaterials. Nanomaterials 2021, 11, 1656. https://doi.org/10.3390/nano11071656
Huerta-López C, Alegre-Cebollada J. Protein Hydrogels: The Swiss Army Knife for Enhanced Mechanical and Bioactive Properties of Biomaterials. Nanomaterials. 2021; 11(7):1656. https://doi.org/10.3390/nano11071656
Chicago/Turabian StyleHuerta-López, Carla, and Jorge Alegre-Cebollada. 2021. "Protein Hydrogels: The Swiss Army Knife for Enhanced Mechanical and Bioactive Properties of Biomaterials" Nanomaterials 11, no. 7: 1656. https://doi.org/10.3390/nano11071656
APA StyleHuerta-López, C., & Alegre-Cebollada, J. (2021). Protein Hydrogels: The Swiss Army Knife for Enhanced Mechanical and Bioactive Properties of Biomaterials. Nanomaterials, 11(7), 1656. https://doi.org/10.3390/nano11071656





