Validation of a Standard Luminescence Method for the Fast Determination of the Antimicrobial Activity of Nanoparticles in Escherichia coli
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis of Nanoparticles
2.1.1. Mesoporous Silica Nanoparticles Synthesis
2.1.2. Oleic Acid Stabilized SPION Synthesis
2.1.3. Single-Layer Mesoporous Silica Surface Modification
2.1.4. Template Removal
2.1.5. Drugs Loading and Release Assays
2.2. Confirmation Assays
2.2.1. CFU Counting
2.2.2. Luminescence Assay
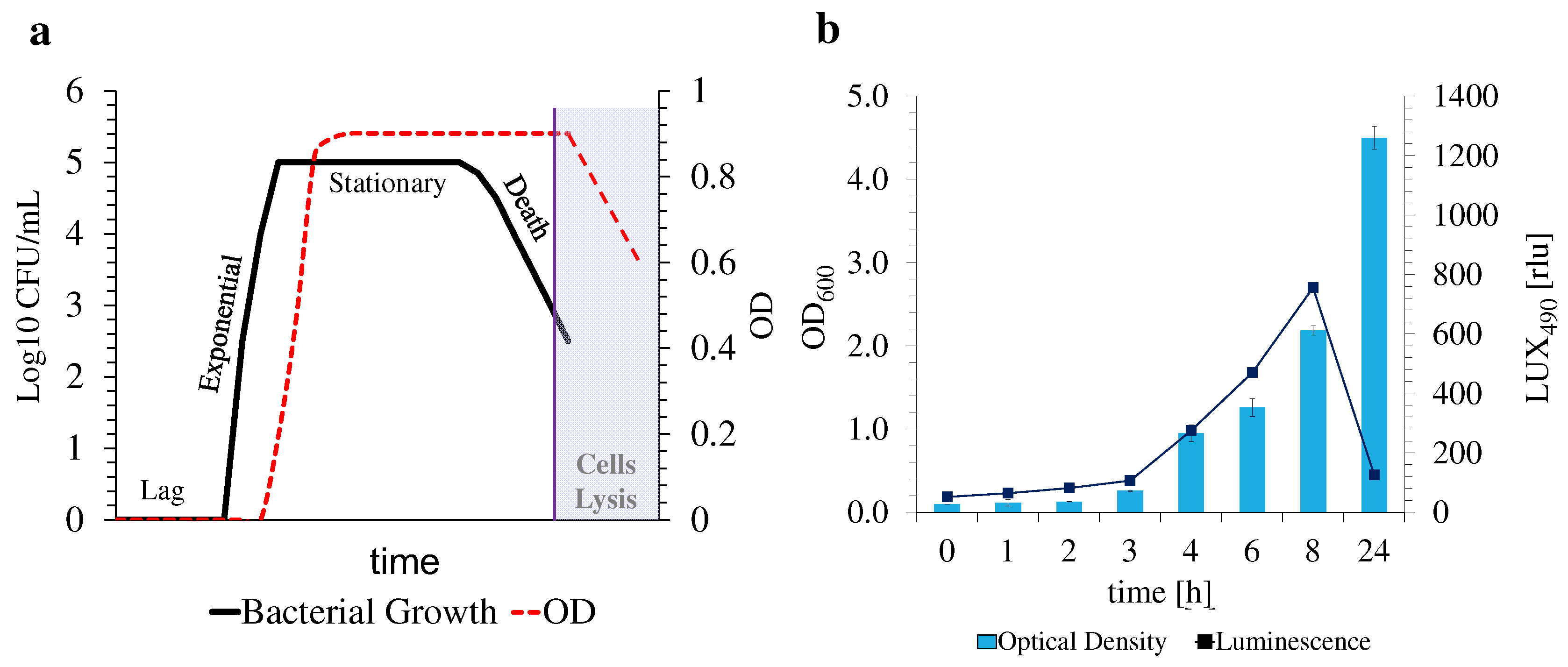
2.2.3. Optimization of Antimicrobial Assay Conditions with E. coli Lux
2.2.4. Antimicrobial Activity Assessment
2.2.5. Statistical Analysis
3. Results and Discussion
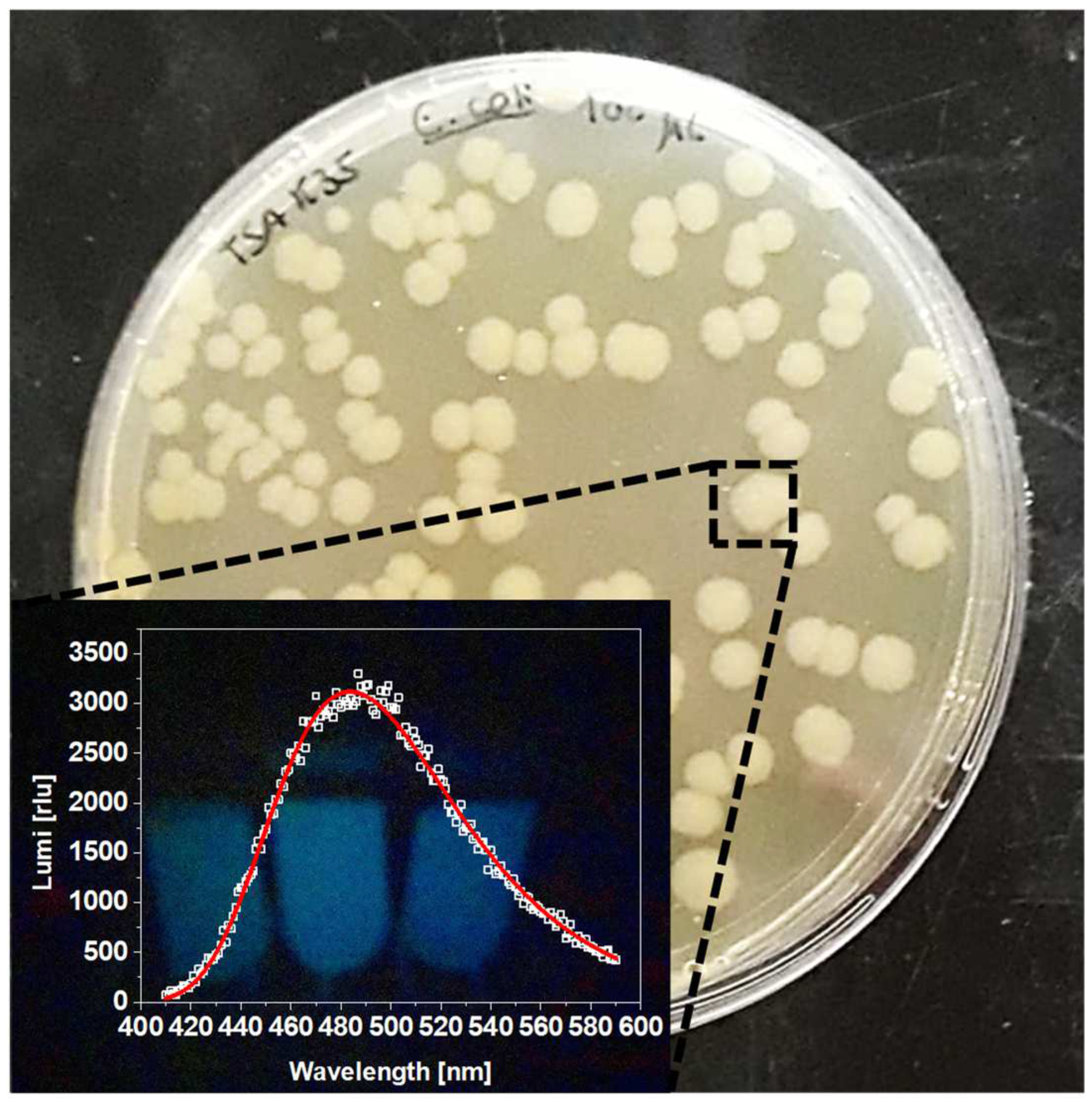
3.1. Bacterial Transformation and Validation
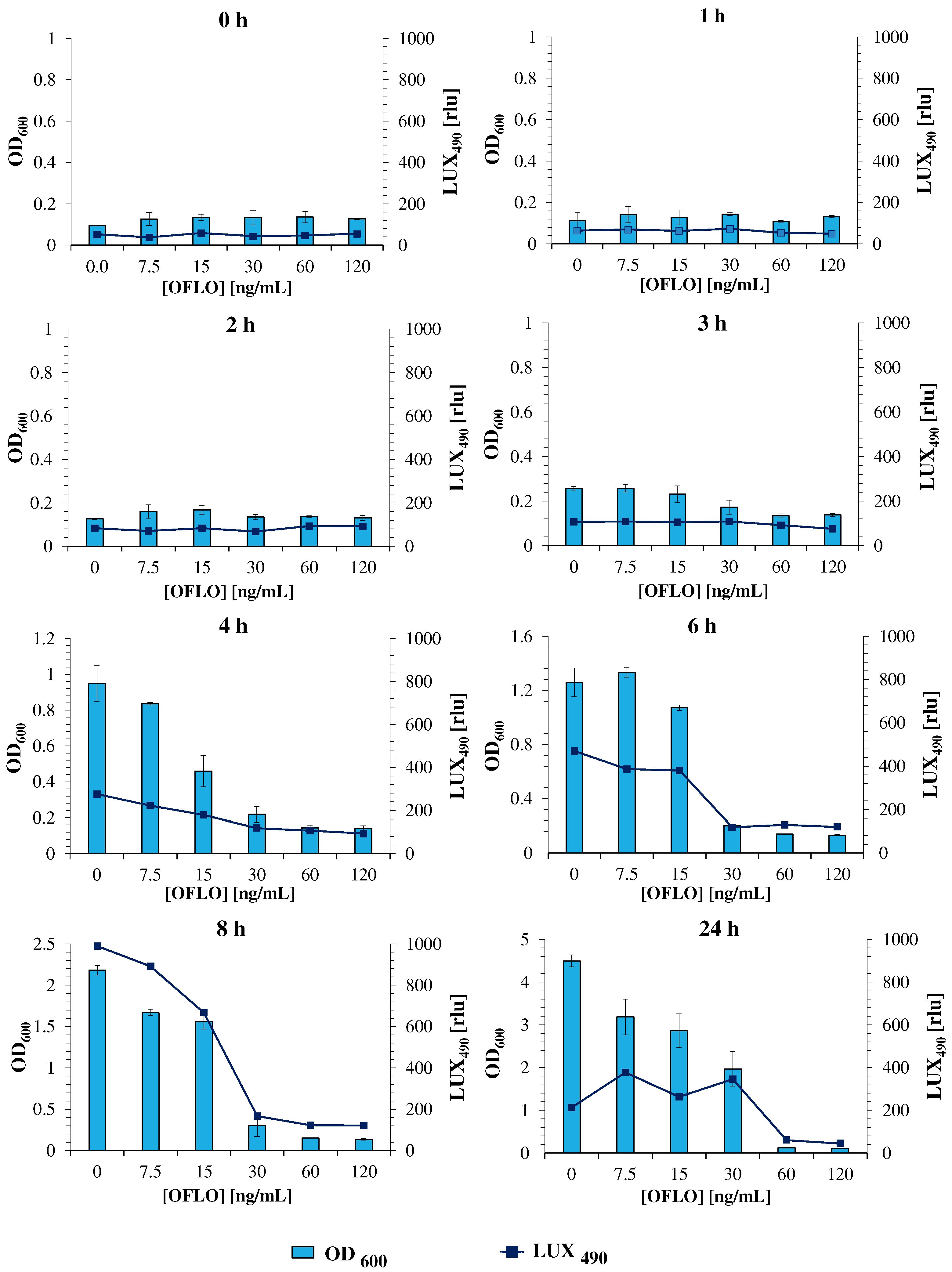
3.2. E. coli Lux Susceptibility to Ofloxacin
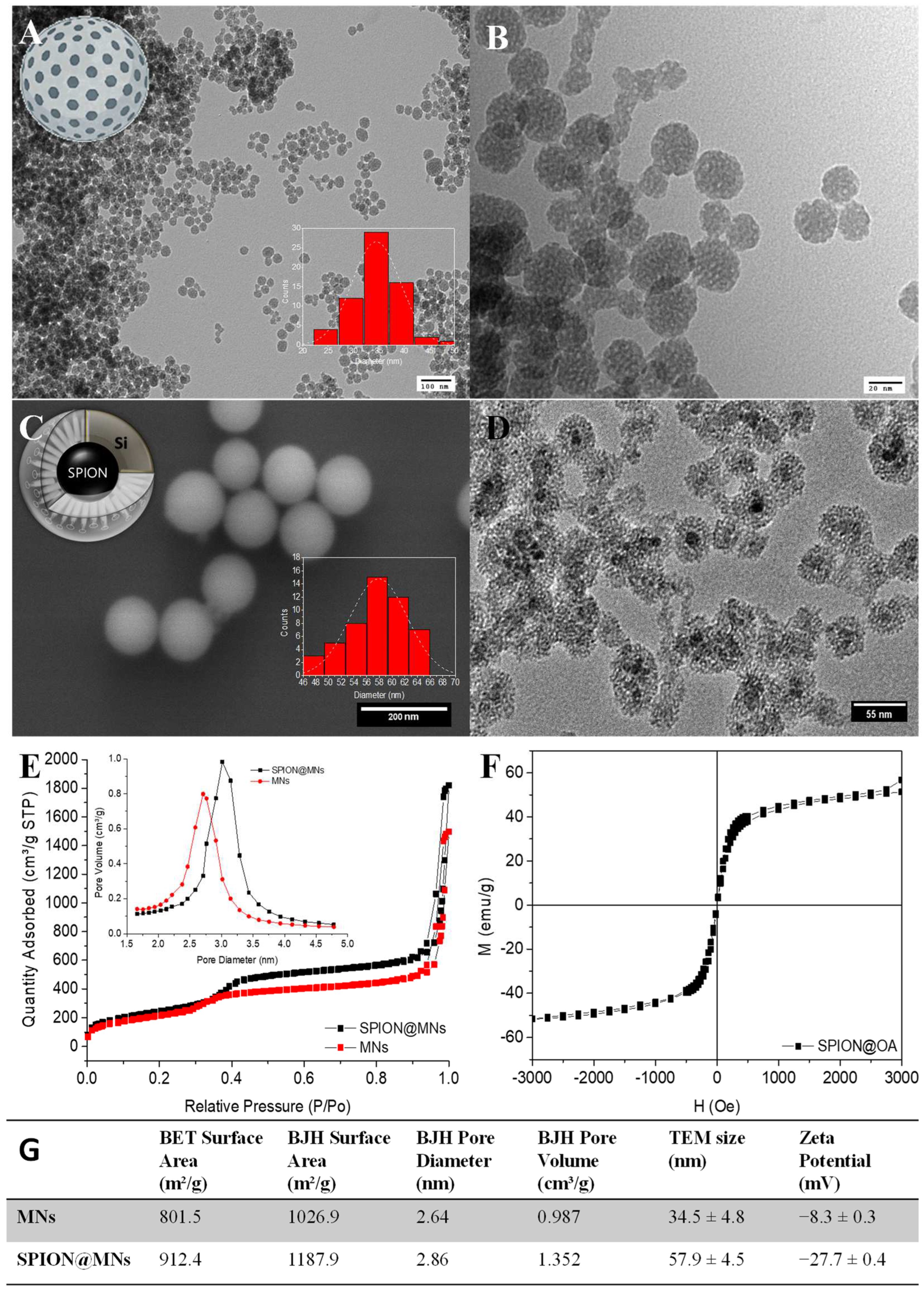
3.3. Nanoformulations Synthesis
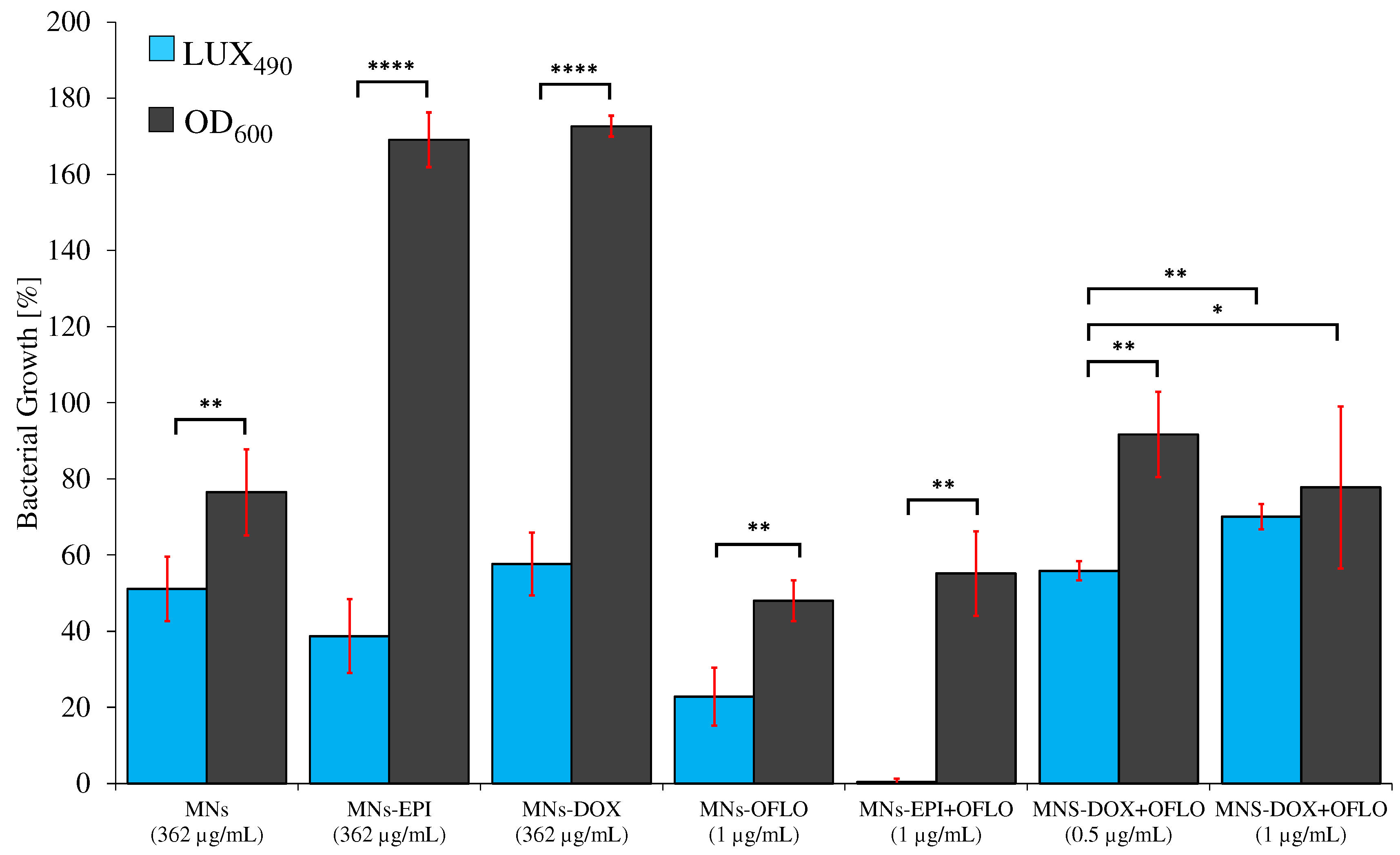
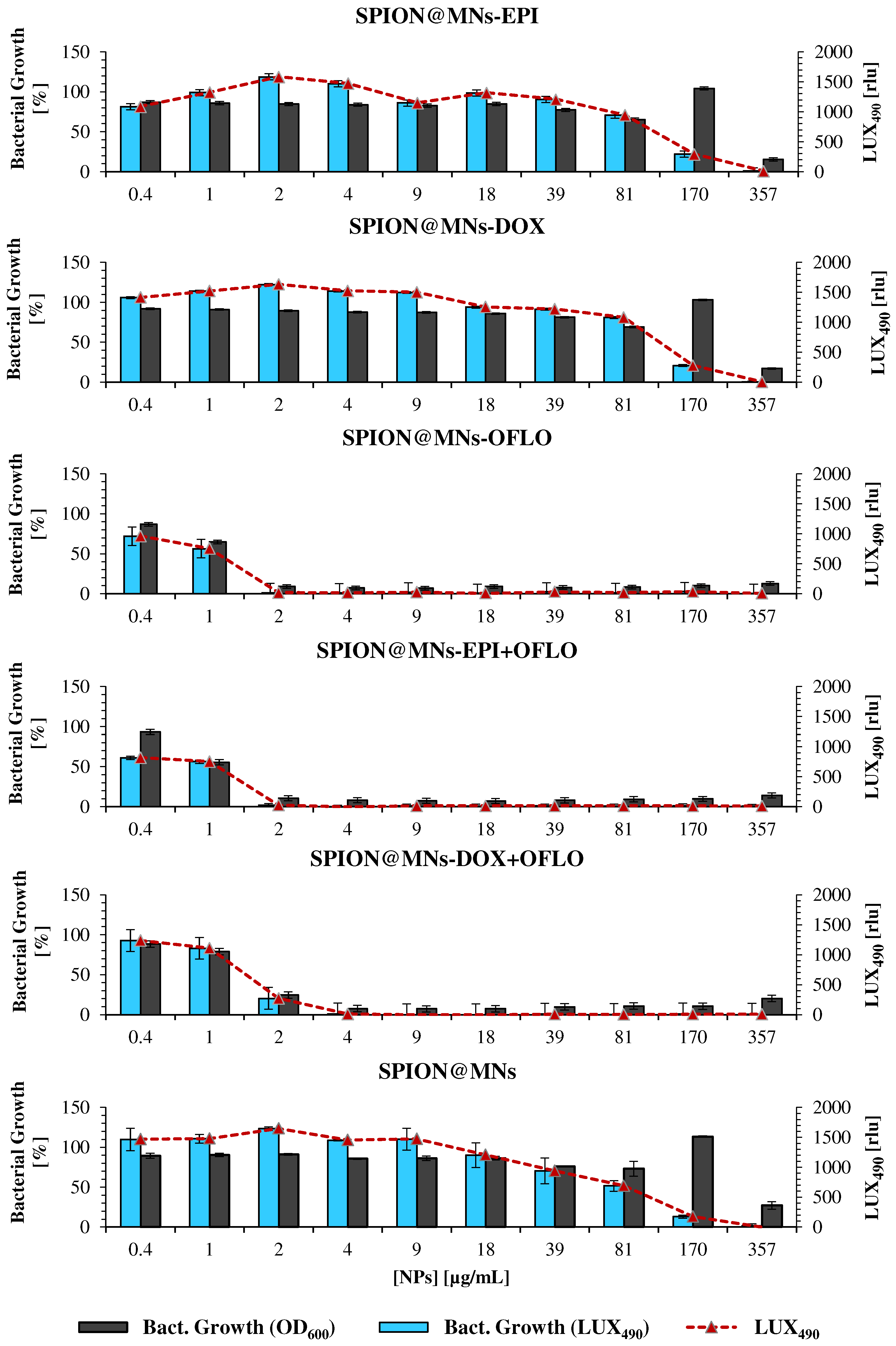
3.4. Nanoparticles’ Antimicrobial Activity against E. coli and E. coli Lux
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hofmann-Amtenbrink, M.; Grainger, D.W.; Hofmann, H. Nanoparticles in medicine: Current challenges facing inorganic nanoparticle toxicity assessments and standardizations. Nanomedicine 2015, 11, 1689–1694. [Google Scholar] [CrossRef]
- Dhawan, A.; Sharma, V. Toxicity assessment of nanomaterials: Methods and challenges. Anal. Bioanal. Chem. 2010, 398, 589–605. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Singh, S.; Shanker, R.; Dhawan, A. Chapter 1. Nanotoxicology: Challenges for Biologists. In Nanotoxicology; Royal Society of Chemistry: London, UK, 2017; pp. 1–16. [Google Scholar]
- Kumar, V.; Sharma, N.; Maitra, S.S. In vitro and in vivo toxicity assessment of nanoparticles. Int. Nano Lett. 2017, 7, 243–256. [Google Scholar] [CrossRef] [Green Version]
- Dusinska, M.; Mariussen, E.; Runden-Pran, E.; Hudecova, A.M.; Elje, E.; Kazimirova, A.; El Yamani, N.; Dommershausen, N.; Tharmann, J.; Fieblinger, D.; et al. In Vitro Approaches for Assessing the Genotoxicity of Nanomaterials. Methods Mol. Biol. 2019, 1894, 83–122. [Google Scholar] [CrossRef] [PubMed]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [Green Version]
- Veiga, A.; Toledo, M.; Rossa, L.S.; Mengarda, M.; Stofella, N.C.F.; Oliveira, L.J.; Goncalves, A.G.; Murakami, F.S. Colorimetric microdilution assay: Validation of a standard method for determination of MIC, IC50%, and IC90% of antimicrobial compounds. J. Microbiol. Methods 2019, 162, 50–61. [Google Scholar] [CrossRef]
- Giannousi, K.; Pantazaki, A.; Dendrinou-Samara, C. Copper-Based Nanoparticles as Antimicrobials. In Nanostructures for Antimicrobial Therapy; Elsevier: Amsterdam, The Netherlands, 2017; pp. 515–529. [Google Scholar]
- Schwarz, S.; Silley, P.; Simjee, S.; Woodford, N.; van Duijkeren, E.; Johnson, A.P.; Gaastra, W. Editorial: Assessing the antimicrobial susceptibility of bacteria obtained from animals. J. Antimicrob. Chemother. 2010, 65, 601–604. [Google Scholar] [CrossRef]
- Tunney, M.M.; Ramage, G.; Field, T.R.; Moriarty, T.F.; Storey, D.G. Rapid colorimetric assay for antimicrobial susceptibility testing of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2004, 48, 1879–1881. [Google Scholar] [CrossRef] [Green Version]
- Marcelo, G.A.; Duarte, M.P.; Oliveira, E. Gold@mesoporous silica nanocarriers for the effective delivery of antibiotics and by-passing of β-lactam resistance. SN Appl. Sci. 2020, 2, 1354. [Google Scholar] [CrossRef]
- Dhawan, A.; Pandey, A.; Sharma, V. Toxicity assessment of engineered nanomaterials: Resolving the challenges. J. Biomed. Nanotechnol. 2011, 7, 6–7. [Google Scholar] [CrossRef]
- Karaman, D.Ş.; Manner, S.; Fallarero, A.; Rosenholm, J.M. Current Approaches for Exploration of Nanoparticles as Antibacterial Agents. In Antibacterial Agents; BoD—Books on Demand: Nordstedt, Germany, 2017. [Google Scholar]
- Schumacher, A.; Vranken, T.; Malhotra, A.; Arts, J.J.C.; Habibovic, P. In vitro antimicrobial susceptibility testing methods: Agar dilution to 3D tissue-engineered models. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 187–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marquis, B.J.; Love, S.A.; Braun, K.L.; Haynes, C.L. Analytical methods to assess nanoparticle toxicity. Analyst 2009, 134, 425–439. [Google Scholar] [CrossRef] [PubMed]
- Savage, D.T.; Hilt, J.Z.; Dziubla, T.D. In Vitro Methods for Assessing Nanoparticle Toxicity. Methods Mol. Biol. 2019, 1894, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, B.S.; ElMasry, S.A.; Fahim, N.; Abd ElSattar, M.A.; Shaker, O.A. Detection of antibiotic susceptibility by colorimetric minimum inhibitory concentration in staphylococcal isolates. J. Appl. Microbiol. 2019, 127, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Andraos, C.; Yu, I.J.; Gulumian, M. Interference: A Much-Neglected Aspect in High-Throughput Screening of Nanoparticles. Int. J. Toxicol. 2020, 39, 397–421. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Fernandez, A.; Aznar, E.; Martinez-Manez, R.; Sancenon, F. New Advances in In Vivo Applications of Gated Mesoporous Silica as Drug Delivery Nanocarriers. Small 2020, 16, e1902242. [Google Scholar] [CrossRef]
- Rosli, N.A.; Teow, Y.H.; Mahmoudi, E. Current approaches for the exploration of antimicrobial activities of nanoparticles. Sci Technol. Adv. Mater. 2021, 22, 885–907. [Google Scholar] [CrossRef]
- Sharifian, S.; Homaei, A.; Hemmati, R.; Khajeh, K. Light emission miracle in the sea and preeminent applications of bioluminescence in recent new biotechnology. J. Photochem. Photobiol. B 2017, 172, 115–128. [Google Scholar] [CrossRef]
- Suominen, E.N.; Putus, T.; Atosuo, J. Investigating the short- and long-term effects of antibacterial agents using a real-time assay based on bioluminescent E. coli-lux. Heliyon 2020, 6, e04232. [Google Scholar] [CrossRef]
- Duncan, S.; Glover, L.A.; Killham, K.; Prosser, J.I. Luminescence-based detection of activity of starved and viable but nonculturable bacteria. Appl. Environ. Microbiol. 1994, 60, 1308–1316. [Google Scholar] [CrossRef] [Green Version]
- Lehtinen, J.; Virta, M.; Lilius, E.-M. Fluoro-luminometric real-time measurement of bacterial viability and killing. J. Microbiol. Methods 2003, 55, 173–186. [Google Scholar] [CrossRef]
- Baban, C.K.; Cronin, M.; Akin, A.R.; O’Brien, A.; Gao, X.; Tabirca, S.; Francis, K.P.; Tangney, M. Bioluminescent bacterial imaging in vivo. J. Vis. Exp. 2012, 69, e4318. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; He, X.; Zhu, W.; Li, H.; Wang, W. Bacterial bioluminescence assay for bioanalysis and bioimaging. Anal. Bioanal. Chem. 2022, 414, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Abbas, M.; Adil, M.; Ehtisham-Ul-Haque, S.; Munir, B.; Yameen, M.; Ghaffar, A.; Shar, G.A.; Asif Tahir, M.; Iqbal, M. Vibrio fischeri bioluminescence inhibition assay for ecotoxicity assessment: A review. Sci. Total Environ. 2018, 626, 1295–1309. [Google Scholar] [CrossRef]
- Andreu, N.; Zelmer, A.; Sampson, S.L.; Ikeh, M.; Bancroft, G.J.; Schaible, U.E.; Wiles, S.; Robertson, B.D. Rapid in vivo assessment of drug efficacy against Mycobacterium tuberculosis using an improved firefly luciferase. J. Antimicrob. Chemother. 2013, 68, 2118–2127. [Google Scholar] [CrossRef]
- Andreu, N.; Zelmer, A.; Wiles, S. Noninvasive biophotonic imaging for studies of infectious disease. FEMS Microbiol. Rev. 2011, 35, 360–394. [Google Scholar] [CrossRef] [Green Version]
- Frolov, G.; Lyagin, I.; Senko, O.; Stepanov, N.; Pogorelsky, I.; Efremenko, E. Metal Nanoparticles for Improving Bactericide Functionality of Usual Fibers. Nanomaterials 2020, 10, 1724. [Google Scholar] [CrossRef]
- Thouand, G.; Durand, M.J. Bacteria in Ecotoxicology: Recombinant Luminescent Bacteria. In Encyclopedia of Aquatic Ecotoxicology; Springer: Dordrecht, The Netherlands, 2013; pp. 137–150. [Google Scholar]
- Lu, W.; Xinhua, Q.; Lei, G.; Chang, C.; Yi, W. A quantitative method to assess bacterial adhesion using recombinant bioluminescent Pseudomonas aeruginosa. Biophys. Rep. 2021, 7, 55–70. [Google Scholar] [CrossRef]
- Valtonen, S.J.; Kurittu, J.S.; Karp, M.T. A luminescent Escherichia coli biosensor for the high throughput detection of beta-lactams. J. Biomol. Screen. 2002, 7, 127–134. [Google Scholar] [CrossRef] [Green Version]
- Ivask, A.; Rolova, T.; Kahru, A. A suite of recombinant luminescent bacterial strains for the quantification of bioavailable heavy metals and toxicity testing. BMC Biotechnol. 2009, 9, 41. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.H.; Cheng, C.Y.; Tsai, T.H.; Chiang, P.K.; Chung, Y.C. Highly Sensitive Luminescent Bioassay Using Recombinant Escherichia coli Biosensor for Rapid Detection of Low Cr(VI) Concentration in Environmental Water. Biosensors 2021, 11, 357. [Google Scholar] [CrossRef] [PubMed]
- Kaku, T.; Sugiura, K.; Entani, T.; Osabe, K.; Nagai, T. Enhanced brightness of bacterial luciferase by bioluminescence resonance energy transfer. Sci. Rep. 2021, 11, 14994. [Google Scholar] [CrossRef] [PubMed]
- Deryabin, D.G.; Aleshina, E.S.; Vasilchenko, A.S.; Deryabina, T.D.; Efremova, L.V.; Karimov, I.F.; Korolevskaya, L.B. Investigation of copper nanoparticles antibacterial mechanisms tested by luminescent Escherichia coli strains. Nanotechnol. Russ. 2013, 8, 402–408. [Google Scholar] [CrossRef]
- Marcelo, G.; Ariana-Machado, J.; Enea, M.; Carmo, H.; Rodriguez-Gonzalez, B.; Luis Capelo, J.; Lodeiro, C.; Oliveira, E. Toxicological Evaluation of Luminescent Silica Nanoparticles as New Drug Nanocarriers in Different Cancer Cell Lines. Materials 2018, 11, 1310. [Google Scholar] [CrossRef] [Green Version]
- Galhano, J.; Marcelo, G.A.; Duarte, M.P.; Oliveira, E. Ofloxacin@Doxorubicin-Epirubicin functionalized MCM-41 mesoporous silica-based nanocarriers as synergistic drug delivery tools for cancer related bacterial infections. Bioorganic Chem. 2022, 118, 105470. [Google Scholar] [CrossRef]
- Zhang, J.; Li, X.; Rosenholm, J.M.; Gu, H.C. Synthesis and characterization of pore size-tunable magnetic mesoporous silica nanoparticles. J. Colloid Interface Sci. 2011, 361, 16–24. [Google Scholar] [CrossRef]
- Andreu, N.; Zelmer, A.; Fletcher, T.; Elkington, P.T.; Ward, T.H.; Ripoll, J.; Parish, T.; Bancroft, G.J.; Schaible, U.; Robertson, B.D.; et al. Optimisation of bioluminescent reporters for use with mycobacteria. PLoS ONE 2010, 5, e10777. [Google Scholar] [CrossRef] [Green Version]
- The Metabolomics Innovation Centre (TMIC). T. M. I. C. Human Metabolome Database: Showing metabocard for FMNH2 (HMDB0001142). Available online: https://hmdb.ca/metabolites/HMDB0001142 (accessed on 20 May 2022).
- Francisco, W.A.; Abu-Soud, H.M.; Baldwin, T.O.; Raushel, F.M. Interaction of bacterial luciferase with aldehyde substrates and inhibitors. J. Biol. Chem. 1993, 268, 24734–24741. [Google Scholar] [CrossRef]
- Gehl, J. Electroporation: Theory and methods, perspectives for drug delivery, gene therapy and research. Acta Physiol. Scand. 2003, 177, 437–447. [Google Scholar] [CrossRef]
- Thermo Fisher Scientific. T042-TECHNICAL BULLETIN NanoDrop Spectrophotometers, 260/280 and 260/230 Ratios; Thermo Fisher Scientific: Waltham, MA, USA, 2009. [Google Scholar]
- Wu, G.; Yang, Q.; Long, M.; Guo, L.; Li, B.; Meng, Y.; Zhang, A.; Wang, H.; Liu, S.; Zou, L. Evaluation of agar dilution and broth microdilution methods to determine the disinfectant susceptibility. J. Antibiot. 2015, 68, 661–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tahmasbi, L.; Sedaghat, T.; Motamedi, H.; Kooti, M. Mesoporous silica nanoparticles supported copper(II) and nickel(II) Schiff base complexes: Synthesis, characterization, antibacterial activity and enzyme immobilization. J. Solid State Chem. 2018, 258, 517–525. [Google Scholar] [CrossRef]
- Scholar, E. Ofloxacin. In xPharm: The Comprehensive Pharmacology Reference; Enna, S.J., Bylund, D.B., Eds.; Elsevier: New York, NY, USA, 2007; pp. 1–8. [Google Scholar]
- Sato, K.; Inoue, Y.; Fujii, T.; Aoyama, H.; Mitsuhashi, S. Antibacterial activity of ofloxacin and its mode of action. Infection 1986, 14 (Suppl. S4), S226–S230. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, R.L. Lag Phase Is a Dynamic, Organized, Adaptive, and Evolvable Period That Prepares Bacteria for Cell Division. J. Bacteriol. 2019, 201, e00697-18. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.G.; Acar, H.; Nandipati, A.; Barlow, M. Growth rates made easy. Mol. Biol. Evol. 2014, 31, 232–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bio-Rad Laboratories. MicroPulserTM Electroporation Apparatus Operating Instructions and Applications Guide; Bio-Rad: Hercules, CA, USA, 2016. [Google Scholar]
- Addgene. Sequence Analyzer: pMV306G13+Lux. Available online: https://www.addgene.org/browse/sequence/12288/ (accessed on 21 December 2021).



| Nanomaterial | Drug | Loading % | EE (mg/g) | |
|---|---|---|---|---|
| MNs | EPI | 87.1 ± 9.6 | 21.0 ± 3.9 | |
| DOX | 72.4 ± 4.7 | 19.1 ± 0.1 | ||
| OFLO | 38.8 ± 12.9 | 9.8 ± 3.0 | ||
| EPI + OFLO | Epirubicin | 90.9 ± 6.7 | 22.6 ± 0.9 | |
| Ofloxacin | 58.1 ± 8.3 | 14.4 ± 1.6 | ||
| DOX + OFLO | Doxorubicin | 83.7 ± 14.1 | 21.3 ± 0.6 | |
| Ofloxacin | 57.8 ± 21.2 | 14.5 ± 3.3 | ||
| SPION@MNs | EPI | 100.0 ± 0.0 | 25.0 ± 0.0 | |
| DOX | 100.0 ± 0.0 | 25.0 ± 0.0 | ||
| OFLO | 99.7 ± 0.2 | 24.9 ± 0.1 | ||
| EPI + OFLO | Epirubicin | 88.3 ± 9.1 | 22.1 ± 2.3 | |
| Ofloxacin | 47.3 ± 8.4 | 11.8 ± 2.1 | ||
| DOX + OFLO | Doxorubicin | 80.5 ± 18.7 | 20.1 ± 4.7 | |
| Ofloxacin | 47.2 ± 10.0 | 11.8 ± 2.5 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marcelo, G.A.; Galhano, J.; Duarte, M.P.; Capelo-Martínez, J.L.; Lodeiro, C.; Oliveira, E. Validation of a Standard Luminescence Method for the Fast Determination of the Antimicrobial Activity of Nanoparticles in Escherichia coli. Nanomaterials 2022, 12, 2164. https://doi.org/10.3390/nano12132164
Marcelo GA, Galhano J, Duarte MP, Capelo-Martínez JL, Lodeiro C, Oliveira E. Validation of a Standard Luminescence Method for the Fast Determination of the Antimicrobial Activity of Nanoparticles in Escherichia coli. Nanomaterials. 2022; 12(13):2164. https://doi.org/10.3390/nano12132164
Chicago/Turabian StyleMarcelo, Gonçalo A., Joana Galhano, Maria Paula Duarte, José Luis Capelo-Martínez, Carlos Lodeiro, and Elisabete Oliveira. 2022. "Validation of a Standard Luminescence Method for the Fast Determination of the Antimicrobial Activity of Nanoparticles in Escherichia coli" Nanomaterials 12, no. 13: 2164. https://doi.org/10.3390/nano12132164
APA StyleMarcelo, G. A., Galhano, J., Duarte, M. P., Capelo-Martínez, J. L., Lodeiro, C., & Oliveira, E. (2022). Validation of a Standard Luminescence Method for the Fast Determination of the Antimicrobial Activity of Nanoparticles in Escherichia coli. Nanomaterials, 12(13), 2164. https://doi.org/10.3390/nano12132164







