Impact of Fluorinated Ionic Liquids on Human Phenylalanine Hydroxylase—A Potential Drug Delivery System
Abstract
:1. Introduction
2. Materials and Methods
2.1. hPAH Production
2.2. Enzymatic Assays
2.3. Nano Differential Scanning Fluorimetry
2.4. Limited Proteolysis
2.5. Circular Dichroism
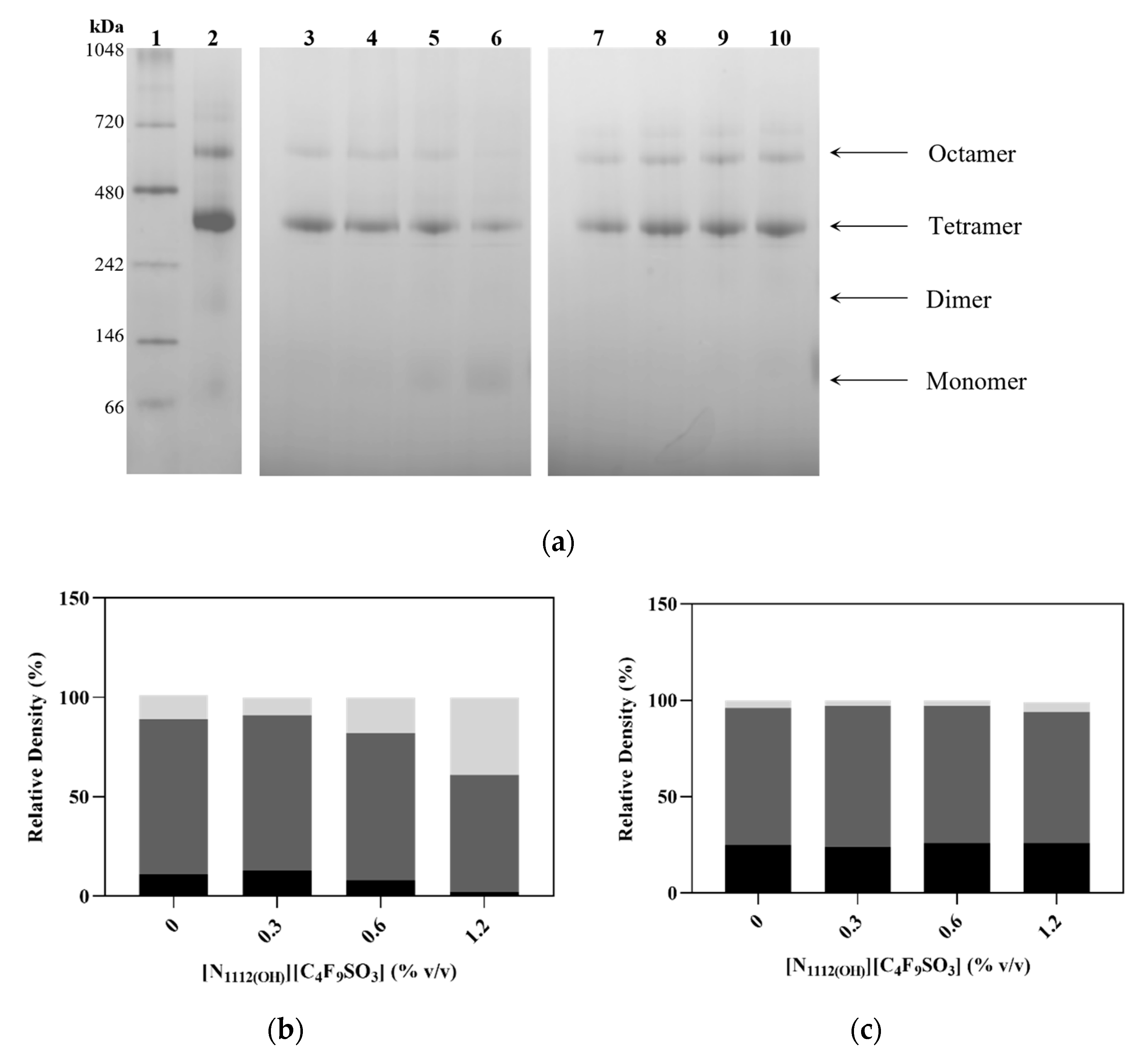
2.6. Blue Native Polyacrylamide Gel Electrophoresis (BN-PAGE)
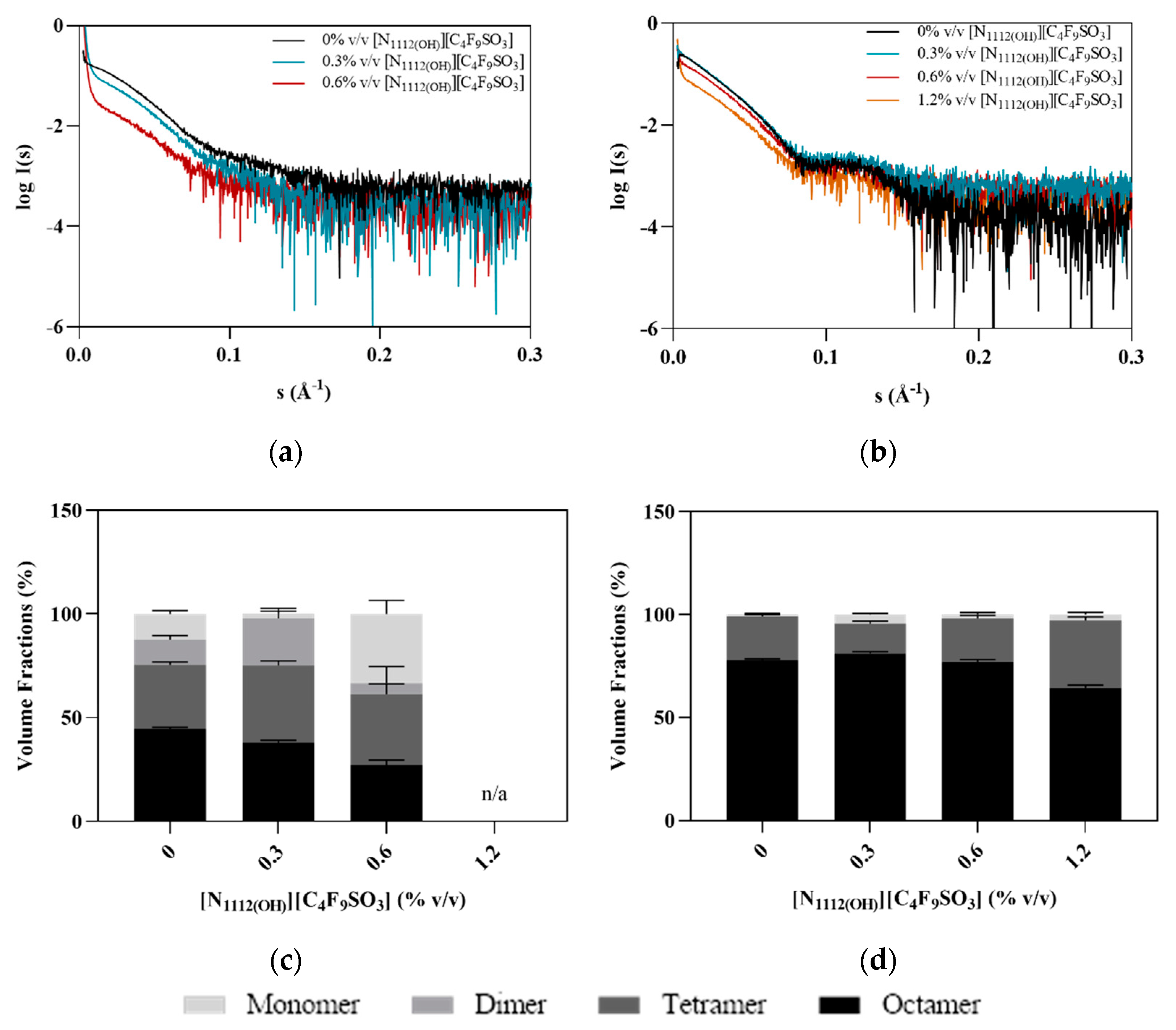
2.7. Small Angle X-ray Scattering
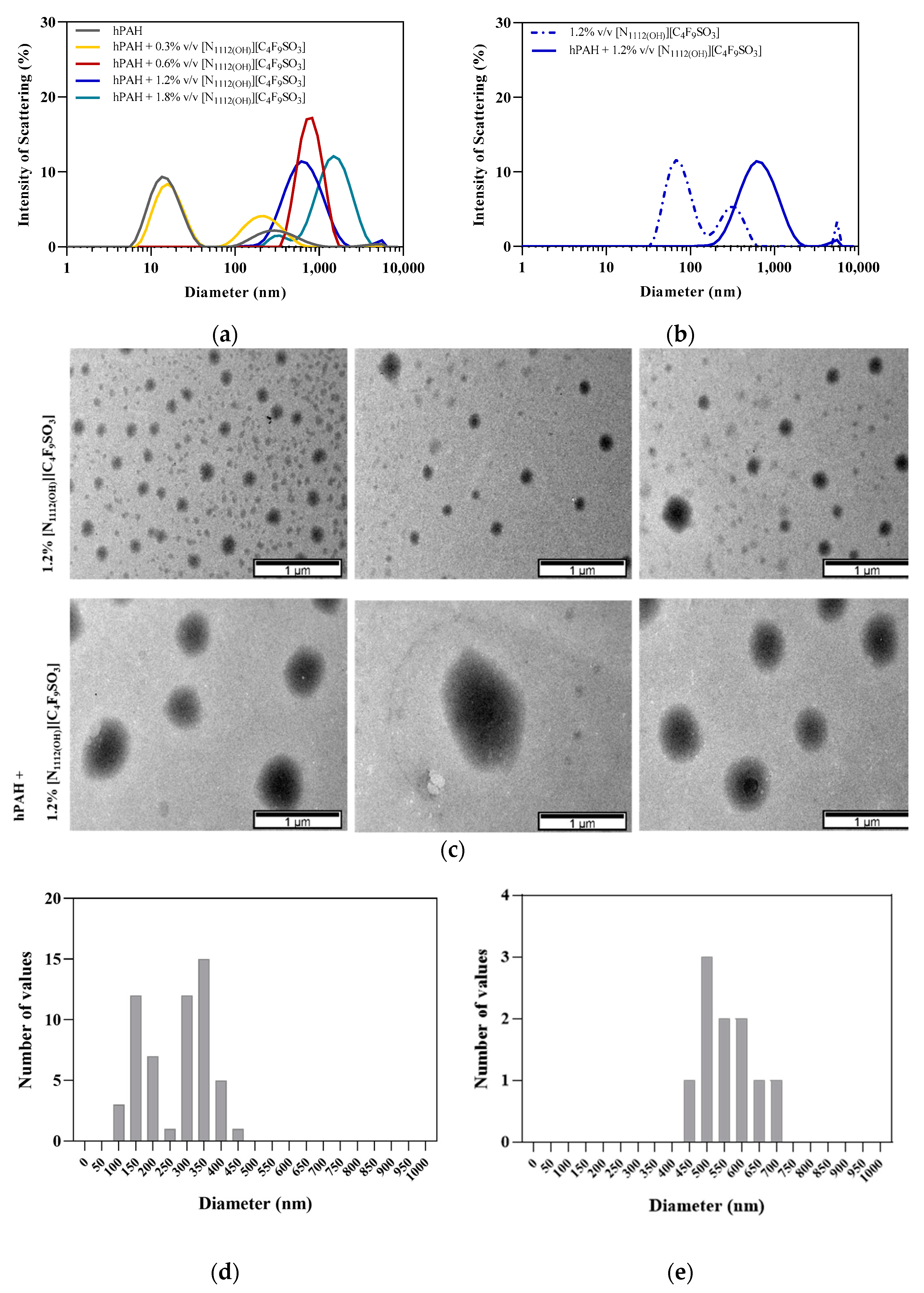
2.8. Dynamic Light Scattering
2.9. Transmission Electron Microscopy
2.10. Statistical Analysis
3. Results and Discussion
3.1. Stability and Function of hPAH
3.2. Effect of FIL on hPAH Structure
3.3. hPAH Encapsulation by FIL
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Adawiyah, N.; Moniruzzaman, M.; Hawatulaila, S.; Goto, M. Ionic liquids as a potential tool for drug delivery systems. MedChemComm 2016, 7, 1881–1897. [Google Scholar] [CrossRef]
- Pedro, S.N.; Freire, C.S.R.; Silvestre, A.J.D.; Freire, M.G. The role of ionic liquids in the pharmaceutical field: An overview of relevant applications. Int. J. Mol. Sci. 2020, 21, 8298. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, T.S.; Júlio, A.; Saraiva, N.; Fernandes, A.S.; Araújo, M.E.M.; Baby, A.R.; Rosado, C.; Mota, J.P. Choline-versus imidazole-based ionic liquids as functional ingredients in topical delivery systems: Cytotoxicity, solubility, and skin permeation studies. Drug Dev. Ind. Pharm. 2017, 43, 1858–1865. [Google Scholar] [CrossRef] [PubMed]
- Amaral, M.; Pereiro, A.B.; Gaspar, M.M.; Reis, C.P. Recent advances in ionic liquids and nanotechnology for drug delivery. Nanomedicine 2020, 16, 63–80. [Google Scholar] [CrossRef]
- Reslan, M.; Kayser, V. Ionic liquids as biocompatible stabilizers of proteins. Biophys. Rev. 2018, 10, 781–793. [Google Scholar] [CrossRef]
- Egorova, K.S.; Gordeev, E.G.; Ananikov, V.P. Biological Activity of Ionic Liquids and Their Application in Pharmaceutics and Medicine. Chem. Rev. 2017, 117, 7132–7189. [Google Scholar] [CrossRef]
- Bui-Le, L.; Clarke, C.J.; Bröhl, A.; Brogan, A.P.S.; Arpino, J.A.J.; Polizzi, K.M.; Hallett, J.P. Revealing the complexity of ionic liquid–protein interactions through a multi-technique investigation. Commun. Chem. 2020, 3, 55. [Google Scholar] [CrossRef]
- Pal, A.; Yadav, A. Binding interactions of anesthetic drug with surface active ionic liquid. J. Mol. Liq. 2016, 222, 471–479. [Google Scholar] [CrossRef]
- Zhou, L.; Tian, T.; Xiao, J.; Wang, T.; Yu, L. Aggregation behavior of pyrrolidinium-based surface active ionic liquids in H2O-EAN binary solvents. J. Mol. Liq. 2017, 225, 50–55. [Google Scholar] [CrossRef]
- Ali, M.K.; Moshikur, R.M.; Wakabayashi, R.; Tahara, Y.; Moniruzzaman, M.; Kamiya, N.; Goto, M. Synthesis and characterization of choline–fatty-acid-based ionic liquids: A new biocompatible surfactant. J. Colloid Interface Sci. 2019, 551, 72–80. [Google Scholar] [CrossRef]
- Harada, L.; Pereira, J.; Campos, W.; Silva, E.; Moutinho, C.; Vila, M.; Oliveira, J., Jr.; Teixeira, J.; Balcão, V.; Tubino, M.; et al. Insights into Protein-Ionic Liquid Interactions Aiming at Macromolecule Delivery Systems. J. Braz. Chem. Soc. 2018, 29, 1983–1998. [Google Scholar] [CrossRef]
- Singh, O.; Singla, P.; Aswal, V.K.; Mahajan, R.K. Aggregation and Morphological Aptitude of Drug-Based Ionic Liquids in Aqueous Solution. ACS Omega 2017, 2, 3296–3307. [Google Scholar] [CrossRef] [PubMed]
- Singla, P.; Singh, O.; Chabba, S.; Mahajan, R.K. Pluronic-SAILs (surface active ionic liquids) mixed micelles as efficient hydrophobic quercetin drug carriers. J. Mol. Liq. 2018, 249, 294–303. [Google Scholar] [CrossRef]
- Bharmoria, P.; Kumar, A. Thermodynamic investigations of protein’s behaviour with ionic liquids in aqueous medium studied by isothermal titration calorimetry. Biochim. Biophys. Acta-Gen. Subj. 2016, 1860, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Pereiro, A.B.; Araújo, J.M.M.; Teixeira, F.S.; Marrucho, I.M.; Piñeiro, M.M.; Rebelo, L.P.N. Aggregation behavior and total miscibility of fluorinated ionic liquids in water. Langmuir 2015, 31, 1283–1295. [Google Scholar] [CrossRef] [PubMed]
- Vieira, N.S.M.; Bastos, J.C.; Rebelo, L.P.N.; Matias, A.; Araújo, J.M.M.; Pereiro, A.B. Human cytotoxicity and octanol/water partition coefficients of fluorinated ionic liquids. Chemosphere 2019, 216, 576–586. [Google Scholar] [CrossRef] [PubMed]
- Vieira, N.S.M.; Stolte, S.; Araújo, J.M.M.; Rebelo, L.P.N.; Pereiro, A.B.; Markiewicz, M. Acute Aquatic Toxicity and Biodegradability of Fluorinated Ionic Liquids. ACS Sustain. Chem. Eng. 2019, 7, 3733–3741. [Google Scholar] [CrossRef]
- Alves, M.; Vieira, N.S.M.; Rebelo, L.P.N.; Araújo, J.M.M.; Pereiro, A.B.; Archer, M. Fluorinated ionic liquids for protein drug delivery systems: Investigating their impact on the structure and function of lysozyme. Int. J. Pharm. 2017, 526, 309–320. [Google Scholar] [CrossRef]
- Alves, M.M.S.; Araújo, J.M.M.; Martins, I.C.; Pereiro, A.B.; Archer, M. Insights into the interaction of Bovine Serum Albumin with Surface-Active Ionic Liquids in aqueous solution. J. Mol. Liq. 2021, 322, 114537. [Google Scholar] [CrossRef]
- Pey, A.L.; Thórólfsson, M.; Teigen, K.; Ugarte, M.; Martínez, A. Thermodynamic characterization of the binding of tetrahydropterins to phenylalanine hydroxylase. J. Am. Chem. Soc. 2004, 126, 13670–13678. [Google Scholar] [CrossRef]
- Cerreto, M.; Cavaliere, P.; Carluccio, C.; Amato, F.; Zagari, A.; Daniele, A.; Salvatore, F. Natural phenylalanine hydroxylase variants that confer a mild phenotype affect the enzyme’s conformational stability and oligomerization equilibrium. Biochim. Biophys. Acta-Mol. Basis Dis. 2011, 1812, 1435–1445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomé, C.S.; Lopes, R.R.; Sousa, P.M.F.; Amaro, M.P.; Leandro, J.; Mertens, H.D.T.; Leandro, P.; Vicente, J.B. Structure of full-length wild-type human phenylalanine hydroxylase by small angle X-ray scattering reveals substrate-induced conformational stability. Sci. Rep. 2019, 9, 13615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Spronsen, F.J.; Blau, N.; Harding, C.; Burlina, A.; Longo, N.; Bosch, A.M. Phenylketonuria. Nat. Rev. Dis. Prim. 2021, 7, 36. [Google Scholar] [CrossRef]
- Borges, A.C.; Broersen, K.; Leandro, P.; Fernandes, T.G. Engineering Organoids for in vitro Modeling of Phenylketonuria. Front. Mol. Neurosci. 2022, 14, 787242. [Google Scholar] [CrossRef] [PubMed]
- Flydal, M.I.; Martinez, A. Critical Review Phenylalanine Hydroxylase: Function, Structure, and Regulation. IUBMB Life 2013, 65, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Leandro, P.; Rivera, I.; Lechner, M.C.; De Almeida, I.T.; Konecki, D. The V388M Mutation Results in a Kinetic Variant Form of Phenylalanine Hydroxylase. Mol. Genet. Metab. 2000, 69, 204–212. [Google Scholar] [CrossRef] [Green Version]
- Montalbano, F.; Leandro, J.; Farias, G.D.V.F.; Lino, P.R.; Guedes, R.C.; Vicente, J.B.; Leandro, P.; Gois, P.M.P. Phenylalanine iminoboronates as new phenylalanine hydroxylase modulators. RSC Adv. 2014, 4, 61022–61027. [Google Scholar] [CrossRef]
- Lopes, R.R.; Tomé, C.S.; Russo, R.; Paterna, R.; Leandro, J.; Candeias, N.R.; Gonçalves, L.M.D.; Teixeira, M.; Sousa, P.M.F.; Guedes, R.C.; et al. Modulation of human phenylalanine hydroxylase by 3-hydroxyquinolin-2(1h)-one derivatives. Biomolecules 2021, 11, 462. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Whitmore, L.; Wallace, B.A. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers 2008, 89, 392–400. [Google Scholar] [CrossRef]
- Andrade, M.A.A.; Chacón, P.; Merelo, J.J.J.; Morán, F. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. Des. Sel. 1993, 6, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Whitmore, L.; Wallace, B.A. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004, 32, W668–W673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanchet, C.E.; Spilotros, A.; Schwemmer, F.; Graewert, M.A.; Kikhney, A.; Jeffries, C.M.; Franke, D.; Mark, D.; Zengerle, R.; Cipriani, F.; et al. Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III, DESY). J. Appl. Crystallogr. 2015, 48, 431–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franke, D.; Petoukhov, M.V.; Konarev, P.V.; Panjkovich, A.; Tuukkanen, A.; Mertens, H.D.T.; Kikhney, A.G.; Hajizadeh, N.R.; Franklin, J.M.; Jeffries, C.M.; et al. ATSAS 2.8: A comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 2017, 50, 1212–1225. [Google Scholar] [CrossRef] [Green Version]
- Konarev, P.V.; Volkov, V.V.; Sokolova, A.V.; Koch, M.H.J.; Svergun, D.I. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003, 36, 1277–1282. [Google Scholar] [CrossRef]
- Arturo, E.C.; Gupta, K.; Hansen, M.R.; Borne, E.; Jaffe, E.K. Biophysical characterization of full-length human phenylalanine hydroxylase provides a deeper understanding of its quaternary structure equilibrium. J. Biol. Chem. 2019, 294, 10131–10145. [Google Scholar] [CrossRef]
- Jaffe, E.K.; Stith, L.; Lawrence, S.H.; Andrake, M.; Dunbrack, R.L. A new model for allosteric regulation of phenylalanine hydroxylase: Implications for disease and therapeutics. Arch. Biochem. Biophys. 2013, 530, 73–82. [Google Scholar] [CrossRef] [Green Version]
- Miranda, F.F.; Teigen, K.; Thórólfsson, M.; Svebak, R.M.; Knappskog, P.M.; Flatmark, T.; Martínez, A. Phosphorylation and mutations of Ser16 in human phenylalanine hydroxylase: Kinetic and structural effects. J. Biol. Chem. 2002, 277, 40937–40943. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Yao, S.; Song, H. Application of ionic liquids in liquid chromatography and electrodriven separation. J. Chromatogr. Sci. 2013, 51, 739–752. [Google Scholar] [CrossRef] [Green Version]
- Flydal, M.I.; Alcorlo-Pagés, M.; Johannessen, F.G.; Martínez-Caballero, S.; Skjærven, L.; Fernandez-Leiro, R.; Martinez, A.; Hermoso, J.A. Structure of full-length human phenylalanine hydroxylase in complex with tetrahydrobiopterin. Proc. Natl. Acad. Sci. USA 2019, 166, 11229–11234. [Google Scholar] [CrossRef] [Green Version]
- Gersting, S.W.; Lagler, F.B.; Eichinger, A.; Kemter, K.F.; Danecka, M.K.; Messing, D.D.; Staudigl, M.; Domdey, K.A.; Zsifkovits, C.; Fingerhut, R.; et al. Pahenu1 is a mouse model for tetrahydrobiopterin-responsive phenylalanine hydroxylase deficiency and promotes analysis of the pharmacological chaperone mechanism in vivo. Hum. Mol. Genet. 2010, 19, 2039–2049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
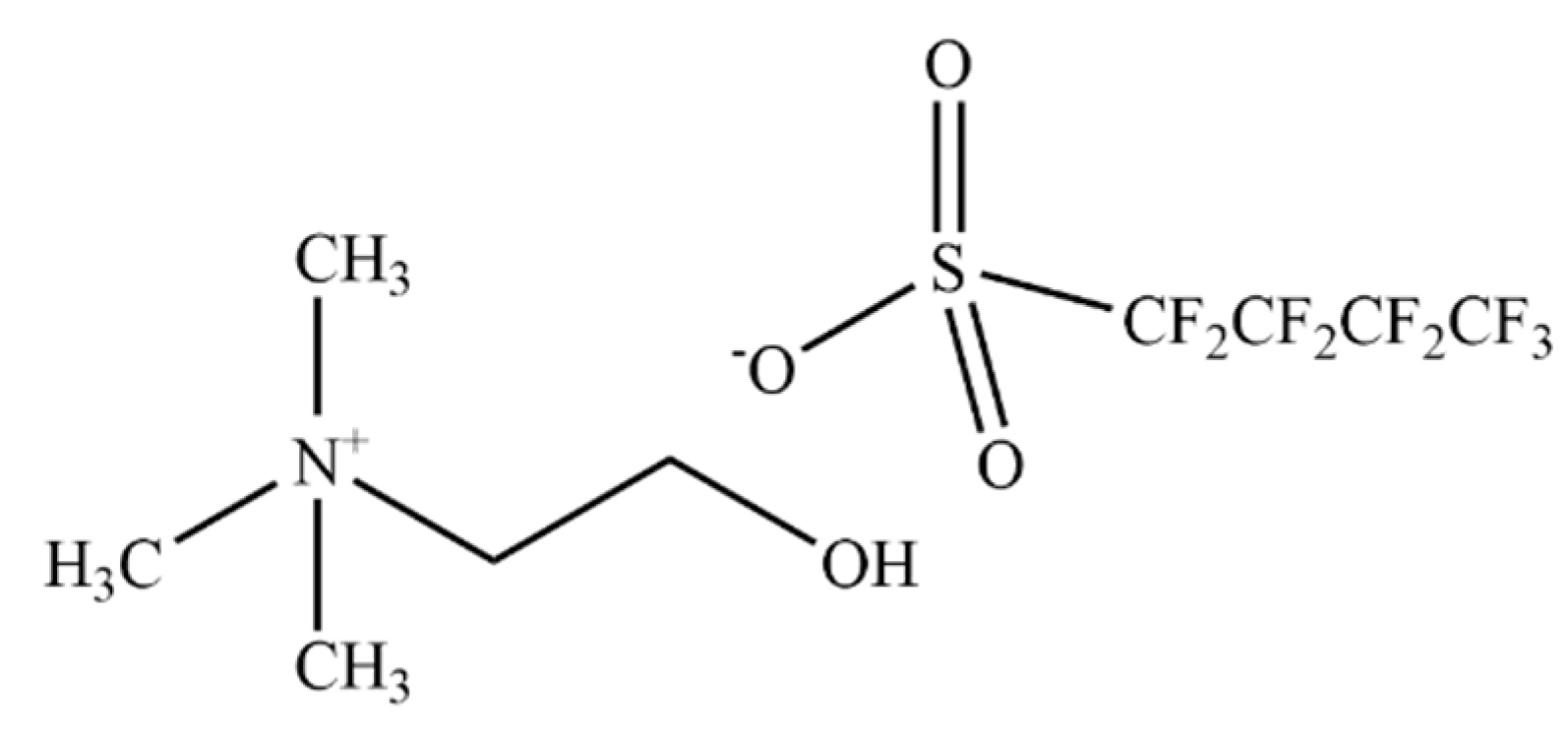





| [N1112(OH)][C4F9SO3] (% v/v) | hPAH Activity (nmol Tyr·min−1·mg−1) | L-Phe Activation Ratio | |
|---|---|---|---|
| L-Phe Preactivation (1 mM L-Phe) | No L-Phe Preactivation | ||
| 0 | 6095 ± 372 | 1317 ± 117 | 4.6 |
| 0.3 | 6643 ± 14 | 1643 ± 28 * | 4.0 |
| 0.6 | 6966 ± 60 * | 2075 ± 131 * | 3.4 |
| 1.2 | 6861 ± 339 * | 2178 ± 43 * | 3.2 |
| 2 | 6104 ± 298 | 2011 ± 176 * | 3.0 |
| [N1112(OH)][C4F9SO3] (% v/v) | ΔTm (-L-Phe) | ΔTm (+L-Phe) | ΔTmL-Phe | |||
|---|---|---|---|---|---|---|
| ΔTm1 | ΔTm2 | ΔTm1 | ΔTm2 | ΔTm1 L-Phe | ΔTm2 L-Phe | |
| 0 | - | - | - | - | 7.5 | 2.0 |
| 0.3 | −8.1 | −4.5 | −5.5 | −1.8 | 10.1 | 4.8 |
| 0.6 | −9.4 | −6.0 | −9.4 | −3.3 | 7.5 | 4.7 |
| 1.2 | −11.8 | −8.7 | −19.0 | −6.5 | 0.25 | 4.3 |
| 2 | −15.8 | −12.0 | −24.3 | −10.2 | −1.1 | 3.8 |
| Kobs (min−1) | ||
|---|---|---|
| [N1112(OH)][C4F9SO3] (% v/v) | No L-Phe | 1 mM L-Phe |
| 0 | 0.279 | 0.197 |
| 0.6 | 0.053 | 0.281 |
| [N1112(OH)][C4F9SO3] (% v/v) | α-Helix | β-Sheet | Random coil |
|---|---|---|---|
| 0 | 0.39 | 0.09 | 0.52 |
| 0.3 | 0.40 | 0.14 | 0.46 |
| 0.6 | 0.38 | 0.08 | 0.54 |
| 1.2 | 0.40 | 0.11 | 0.49 |
| 2 | 0.38 | 0.07 | 0.55 |
| [N1112(OH)][C4F9SO3] (% v/v) | No L-Phe | 1 mM L-Phe | ||||||
|---|---|---|---|---|---|---|---|---|
| Rg (Å) | VP (Å3) | Dmax (Å) | MM (kDa) | Rg (Å) | VP (Å3) | Dmax (Å) | MM (kDa) | |
| 0 | 53.4 ± 1.1 | 459,446 | 168 | 287 | 61.7 ± 5.3 | 741,198 | 187 | 463 |
| 0.3 | 50.4 ± 1.2 | 354,904 | 137 | 221 | 56.7 ± 2.9 | 617,827 | 164 | 386 |
| 0.6 | 46.3 ± 1.8 | 122,885 | 117 | 77 | 58.7 ± 3.6 | 556,597 | 158 | 348 |
| 1.2 | - | - | - | - | 58.9 ± 4.3 | 476,911 | 143 | 298 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alves, M.M.S.; Leandro, P.; Mertens, H.D.T.; Pereiro, A.B.; Archer, M. Impact of Fluorinated Ionic Liquids on Human Phenylalanine Hydroxylase—A Potential Drug Delivery System. Nanomaterials 2022, 12, 893. https://doi.org/10.3390/nano12060893
Alves MMS, Leandro P, Mertens HDT, Pereiro AB, Archer M. Impact of Fluorinated Ionic Liquids on Human Phenylalanine Hydroxylase—A Potential Drug Delivery System. Nanomaterials. 2022; 12(6):893. https://doi.org/10.3390/nano12060893
Chicago/Turabian StyleAlves, Márcia M. S., Paula Leandro, Haydyn D. T. Mertens, Ana B. Pereiro, and Margarida Archer. 2022. "Impact of Fluorinated Ionic Liquids on Human Phenylalanine Hydroxylase—A Potential Drug Delivery System" Nanomaterials 12, no. 6: 893. https://doi.org/10.3390/nano12060893






