Nano-Biotechnology for Bacteria Identification and Potent Anti-bacterial Properties: A Review of Current State of the Art
Abstract
:1. Introduction
2. Methods to Identify Bacteria Species
2.1. Culture Based Bacteria Identification
2.2. PCR-Based Bacterial Identification
2.3. Mass Spectrometry
2.4. Nanomaterials Based Detection
2.5. Surface Enhanced Raman Spectroscopy (SERS)
3. Methods to Destroy Bacteria
3.1. Antibiotics
3.2. Nano-Biotechnologies
3.2.1. Polymeric Nanostructures
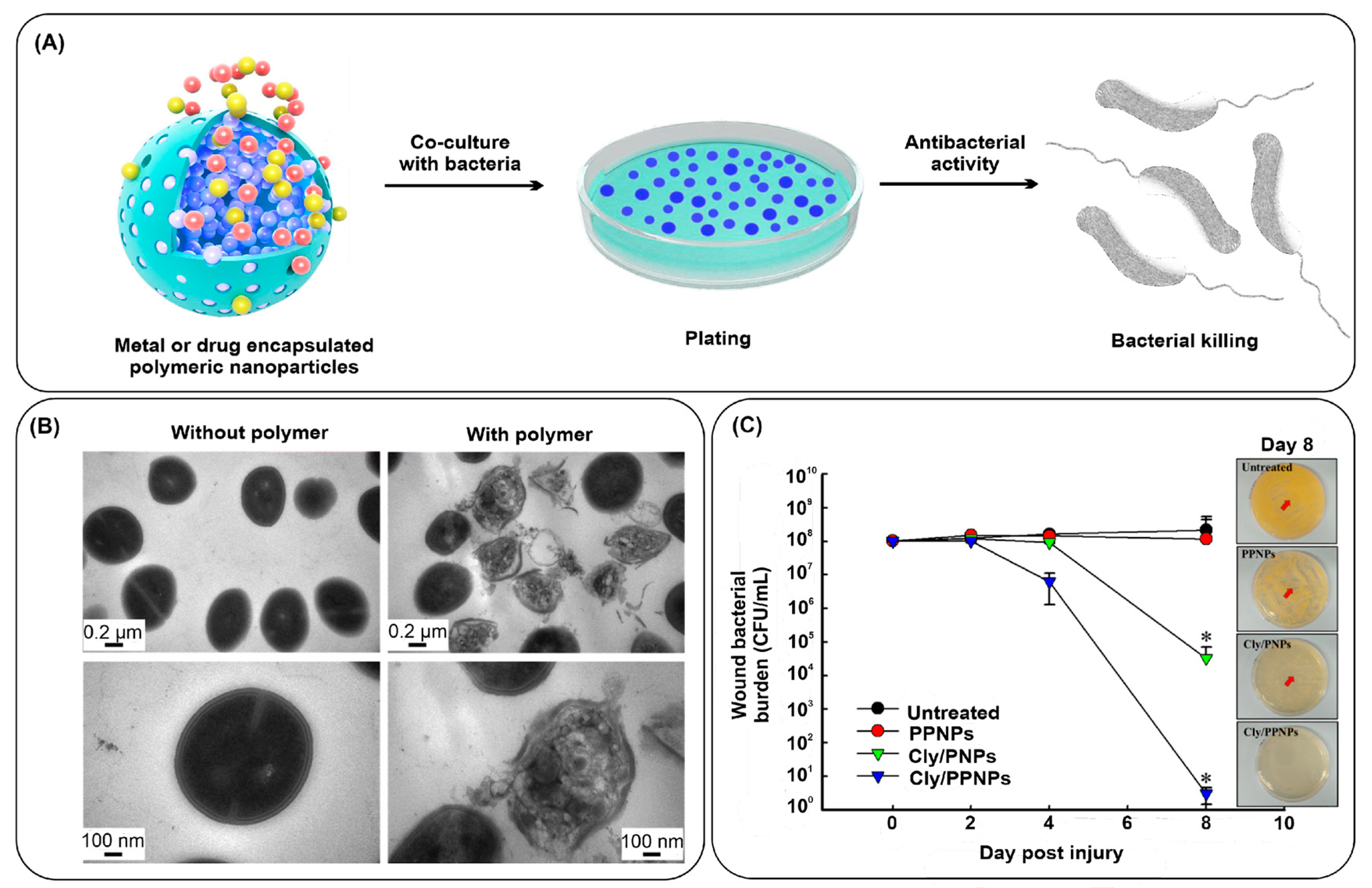
3.2.2. Noble Metal Nanoparticles
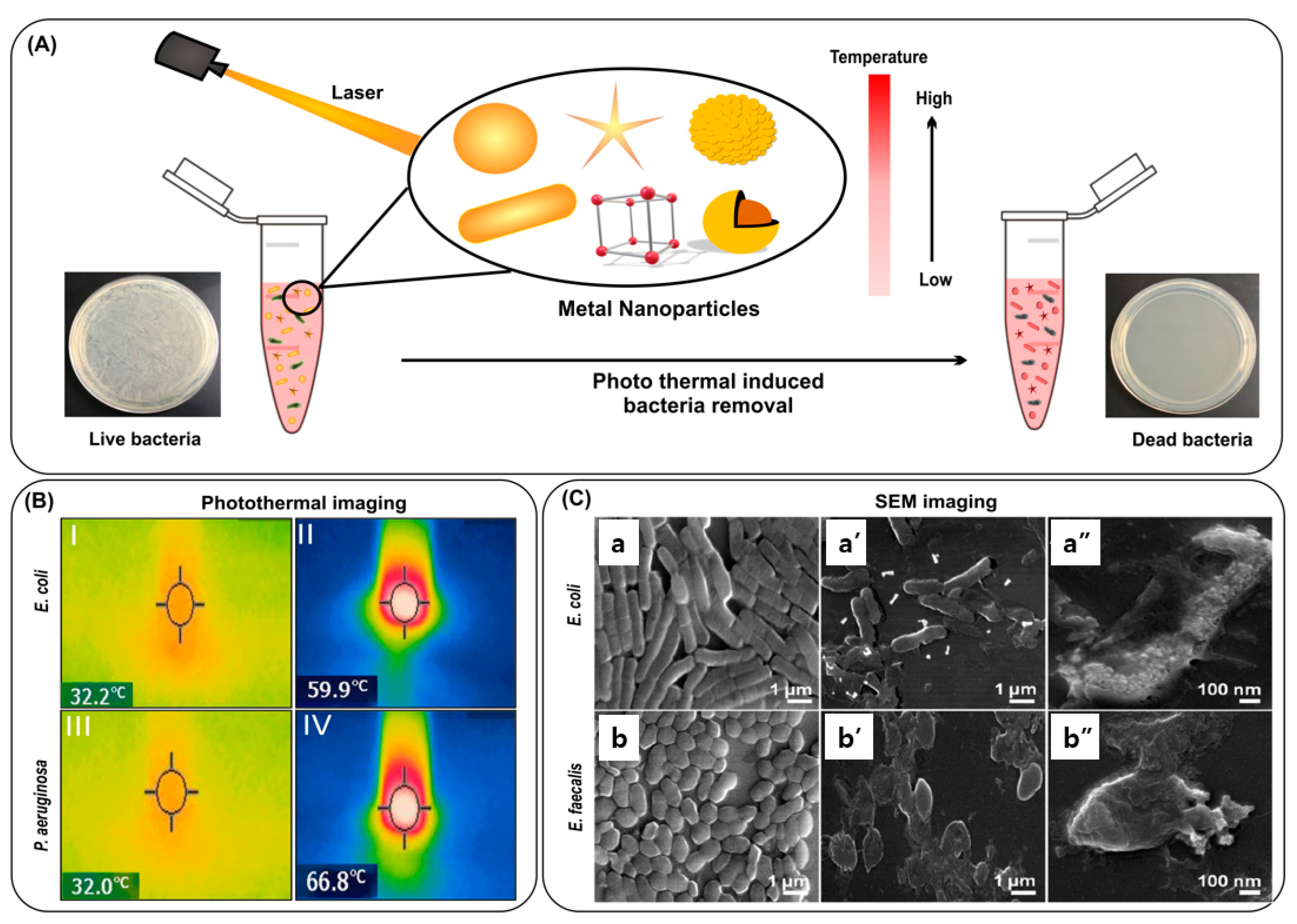
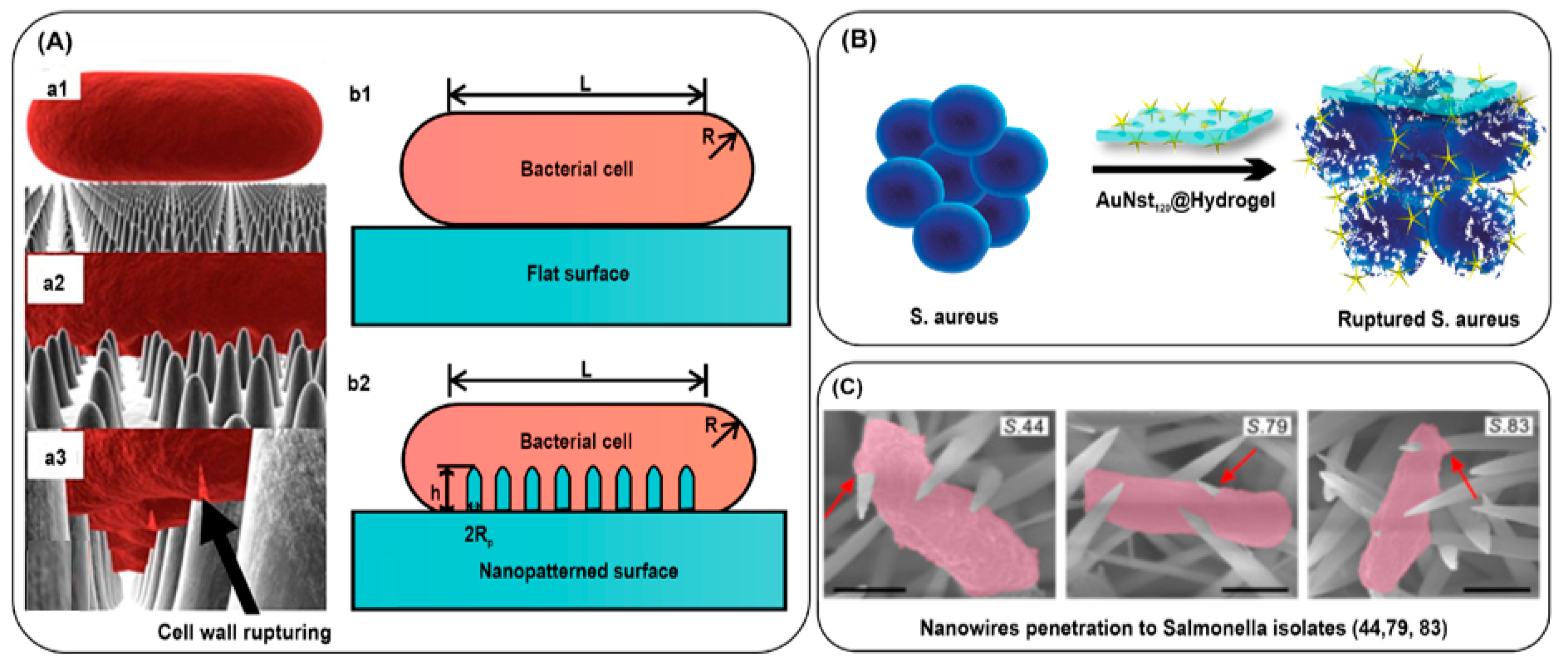
3.2.3. Bacterial Killing by Mechanical Rupture
| Mechanism of Action | Nanomaterial | Target | Scope | Limitations | Ref. |
|---|---|---|---|---|---|
| Antibiofilm activity | Mannose-functionalized chitosan nanosystems | E. coli L. monocytogenes P. aeruginosa S. aureus | Effective against both Gram-positive and Gram-negative bacteria. | The biocompatibility of the nanosystems has not been clearly explained. | [140] |
| Sustained release of antibiotics | Clindamycin-loaded polymeric particles | MRSA | Antibiotic-loaded NPs accelerate MRSA-infected wounds. | Only applicable for the treatment of topical infections. | [143] |
| Teicoplanin-encapsulated aptamer-functionalized PLGA NPs | S. aureus strains | Aptamers provide specificity against S. aureus, while PLGA NPs decrease the MIC value for teicoplanin. | The biocompatibility of the functionalized nanoparticles has not been clearly explained. | [144] | |
| PTT/PDT | Thiol chitosan-wrapped gold nanoshells | E. coli P. aeruginosa S. aureus | PTT effectively eradicates both Gram-negative and Gram-positive bacteria within 5 min. | Only applicable for the treatment of topical infections. | [150] |
| Thiol-coated gold Nanostars | E. coli S. aureus | Hyperthermia results from PTT kill 99.99% of bacteria. | The biocompatibility of the functionalized nanostars has not been clearly explained and only applicable for the eradication of bacteria from medical devices. | [151] | |
| Galactose-modified porphyrin-conjugated gold NPs | P. aeruginosa | Galactose gives the specificity against P. aeruginosa and porphyrin eliminates bacteria via PDT and PTT. | The nanomaterial was able to eradicate only 70% of bacteria (colony count method) and only applicable for the treatment of topical infections. | [154] | |
| pH-responsive gold nanoparticles | MRSA biofilm | AuNP aggregation within the biofilm enhanced the photothermal ablation of MRSA. | Only applicable for the treatment of topical infections such as wound in MRSA infection. | [156] | |
| Glycoconjugate-coated gold nanorods | E. coli P. aeruginosa | Hyperthermia results from PTT against Gram-negative bacteria. | The higher temperature rise due to the gold nanorods may affect the normal tissues if used for the treatment of bacterial infections. | [160] | |
| Gold nanorod-conjugated magnetic nanoparticles | E. coli E. faecalis | Recyclable nanocomposite for repeated photothermal effect. | No in vivo studies were performed to show the safety and efficacy of nanocomposite. | [162] | |
| Silver nanoplates | E. coli S. aureus | Anti-bacterial properties and PTT effect synergistically eradicate bacteria. | Cytotoxicity and biocompatibility must be calculated for its real time application in clinical settings. | [166] | |
| UCNP/PS (upconversion nanoparticles with photosensitizers) | E. coli S. aureus | PTT and PDT effectively eradicate both Gram-negative and Gram-positive bacteria. | The biocompatibility and cytotoxicity of the functionalized nanoparticles have not been clearly explained and applicable for only topical application. | [167] | |
| Mechanical rupture | ZnO/SiO2 nanowires | E. coli | ZnO/SiO2 nanowires inactivate 99% of E. coli inactivation. | Only effective for Gram-negative bacteria. | [179] |
| Gold nanostar-based hydrogel | E. coli P. aeruginosa S. aureus | Nanospikes of gold nanostars rupture the bacterial membrane. | Limited anti-bacterial effect in the Gram-positive bacteria S. aureus. | [181] | |
| NiCo(OH)2CO3 nanowires | Salmonella E. coli P. aeruginosa K. pneumoniae | Nanowires mechanically penetrate the bacterial cell envelope. | Only effective for Gram-negative bacteria. | [182] |
4. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Cecconi, M.; Evans, L.; Levy, M.; Rhodes, A. Sepsis and septic shock. Lancet 2018, 392, 75–87. [Google Scholar] [CrossRef]
- Rubio, I.; Osuchowski, M.F.; Shankar-Hari, M.; Skirecki, T.; Winkler, M.S.; Lachmann, G.; La Rosée, P.; Monneret, G.; Venet, F.; Bauer, M.; et al. Current gaps in sepsis immunology: New opportunities for translational research. Lancet Infect. Dis. 2019, 19, e422–e436. [Google Scholar] [CrossRef]
- Woznica, E.A.; Inglot, M.; Woznica, R.K.; Lysenko, L. Liver dysfunction in sepsis. Adv. Clin. Exp. Med. 2018, 27, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.L.; Peters, M.J.; Alhazzani, W.; Agus, M.S.D.; Flori, H.R.; Inwald, D.P.; Nadel, S.; Schlapbach, L.J.; Tasker, R.C.; Argent, A.C.; et al. Surviving sepsis campaign international guidelines for the management of septic shock and sepsis-associated organ dysfunction in children. Intensive Care Med. 2020, 46, 10–67. [Google Scholar] [CrossRef]
- Zhao, Y.; Pu, M.; Zhang, J.; Wang, Y.; Yan, X.; Yu, L.; He, Z. Recent advancements of nanomaterial-based therapeutic strategies toward sepsis: Bacterial eradication, anti-inflammation, and immunomodulation. Nanoscale 2021, 13, 10726–10747. [Google Scholar] [CrossRef] [PubMed]
- Kardas, P.; Devine, S.; Golembesky, A.; Roberts, C. A systematic review and meta-analysis of misuse of antibiotic therapies in the community. Int. J. Antimicrob. Agents 2005, 26, 106–113. [Google Scholar] [CrossRef]
- Guo, Y.; Song, G.; Sun, M.; Wang, J.; Wang, Y. Prevalence and therapies of antibiotic-resistance in staphylococcus aureus. Front. Cell. Infect. Microbiol. 2020, 10, 107. [Google Scholar] [CrossRef]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.M.R.M.; Mitra, S.; Emran, T.B.; Dhama, K.; Ripon, M.K.H.; Gajdács, M.; Sahibzada, M.U.K.; et al. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J. Infect. Public. Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, R. Superbug Infection. World J. Pharm. Res. 2018, 7, 275–287. [Google Scholar] [CrossRef]
- Balayan, S.; Chauhan, N.; Chandra, R.; Kuchhal, N.K.; Jain, U. Recent advances in developing biosensing based platforms for neonatal sepsis. Biosens. Bioelectron. 2020, 169, 112552. [Google Scholar] [CrossRef] [PubMed]
- Surewaard, B.G.J.; Thanabalasuriar, A.; Zeng, Z.; Tkaczyk, C.; Cohen, T.S.; Bardoel, B.W.; Jorch, S.K.; Deppermann, C.; Bubeck Wardenburg, J.; Davis, R.P.; et al. α-Toxin induces platelet aggregation and liver injury during staphylococcus aureus sepsis. Cell. Host Microbe 2018, 24, 271–284.e3. [Google Scholar] [CrossRef]
- Ghaith, D.M.; Zafer, M.M.; Said, H.M.; Elanwary, S.; Elsaban, S.; Al-Agamy, M.H.; Bohol, M.F.F.; Bendary, M.M.; Al-Qahtani, A.; Al-Ahdal, M.N. Genetic diversity of carbapenem-resistant klebsiella pneumoniae causing neonatal sepsis in intensive care unit, Cairo, Egypt. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 583–591. [Google Scholar] [CrossRef]
- Bodey, G.P.; Bolivar, R.; Fainstein, V.; Jadeja, L. Infections Caused by Pseudomonas aeruginosa. Rev. Infect. Dis. 1983, 5, 279–313. [Google Scholar] [CrossRef] [PubMed]
- Askim, Å.; Mehl, A.; Paulsen, J.; DeWan, A.T.; Vestrheim, D.F.; Åsvold, B.O.; Damås, J.K.; Solligård, E. Epidemiology and outcome of sepsis in adult patients with Streptococcus pneumoniae infection in a Norwegian county 1993-2011: An observational study. BMC Infect. Dis. 2016, 16, 223. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Peng, P.; Fang, Y.; Lu, J.; Fang, M. Distribution and drug resistance of pathogenic bacteria and prognosis in patients with septicemia bloodstream infection with renal insufficiency. Infect. Drug. Resist. 2022, 15, 4109–4116. [Google Scholar] [CrossRef] [PubMed]
- Davies, H.; Pannu, K.; Edwards, J.; Pittman, M.; Mukherjee, D. Fulminant Neisseria meningitidis septicaemia with purpura fulminans requiring limb amputation. IDCases 2019, 19, e00673. [Google Scholar] [CrossRef]
- Sharma, D.; Khan, J.; Agarwal, S. Salmonella typhi as cause of neonatal sepsis: Case report and literature review. J. Matern. Fetal Neonatal Med. 2021, 34, 732–735. [Google Scholar] [CrossRef]
- Carrasquillo, M.; Dever, L.L.; Sonyey, A. Botulism-like symptoms in an immunocompetent patient with Clostridium subterminale bacteremia. IDCases 2018, 11, 80–82. [Google Scholar] [CrossRef]
- Schaefer, K.; Austhof, E.; Boyd, K.; Armstrong, A.; Hoffman, S.; Pogreba-Brown, K. Septicemia Due to Listeria monocytogenes Infection: A Systematic Review and Meta-Analysis. Foodborne Pathog. Dis. 2022, 19, 104–114. [Google Scholar] [CrossRef]
- Ono, S.; Tsujimoto, H.; Hiraki, S.; Aosasa, S. Mechanisms of sepsis-induced immunosuppression and immunological modification therapies for sepsis. Ann. Gastroenterol. Surg. 2018, 2, 351–358. [Google Scholar] [CrossRef]
- Bullock, B.; Benham, M.D. Bacterial Sepsis. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. Available online: https://www.ncbi.nlm.nih.gov/books/NBK537054/ (accessed on 21 June 2022).
- Narayana Iyengar, S.; Dietvorst, J.; Ferrer-Vilanova, A.; Guirado, G.; Muñoz-Berbel, X.; Russom, A. Toward Rapid Detection of Viable Bacteria in Whole Blood for Early Sepsis Diagnostics and Susceptibility Testing. ACS Sens. 2021, 6, 3357–3366. [Google Scholar] [CrossRef] [PubMed]
- Rajapaksha, P.; Elbourne, A.; Gangadoo, S.; Brown, R.; Cozzolino, D.; Chapman, J. A review of methods for the identification of pathogenic microorganisms. Analyst 2019, 144, 396–411. [Google Scholar] [CrossRef] [PubMed]
- Hameed, S.; Xie, L.; Ying, Y. Conventional and emerging identification techniques for pathogenic bacteria in food science: A review. Trends Food Sci. Technol. 2018, 81, 61–73. [Google Scholar] [CrossRef]
- Reller, L.B.; Weinstein, M.P.; Petti, C.A. Detection and Identification of Microorganisms by Gene Amplification and Sequencing. Clin. Infect. Dis. 2007, 44, 1108–1114. [Google Scholar] [CrossRef]
- Mai, M.; Müller, I.; Maneg, D.; Lohr, B.; Haecker, A.; Haberhausen, G.; Hunfeld, K.P. Real-Time PCR-based identification of bacterial and fungal pathogens from blood samples. In Sepsis Methods in Molecular Biology; Mancini, N., Ed.; Humana Press: New York, NY, USA, 2015; Volume 1237. [Google Scholar] [CrossRef]
- Straub, J.; Paula, H.; Mayr, M.; Kasper, D.; Assadian, O.; Berger, A.; Rittenschober-Böhm, J. Diagnostic accuracy of the ROCHE septifast PCR system for the rapid identification of blood pathogens in neonatal sepsis—A prospective clinical trial. PLoS ONE 2017, 12, e0187688. [Google Scholar] [CrossRef]
- Pant, A.; Mackraj, I.; Govender, T. Advances in sepsis diagnosis and management: A paradigm shift towards nanotechnology. J. Biomed. Sci. 2021, 28, 6. [Google Scholar] [CrossRef]
- Shen, Y.; Zhang, Y.; Gao, Z.F.; Ye, Y.; Wu, Q.; Chen, H.-Y.; Xu, J.-J. Recent advances in nanotechnology for simultaneous identification of multiple pathogenic bacteria. Nano Today 2021, 38, 101121. [Google Scholar] [CrossRef]
- Galvan, D.D.; Yu, Q. Surface-enhanced raman scattering for rapid identification and characterization of antibiotic-resistant bacteria. Adv. Healthc. Mater. 2018, 7, 1701335. [Google Scholar] [CrossRef]
- Chen, Y.; Gao, Y.; Chen, Y.; Liu, L.; Mo, A.; Peng, Q. Nanomaterials-based photothermal therapy and its potentials in antibacterial treatment. J. Control. Release 2020, 328, 251–262. [Google Scholar] [CrossRef]
- Peng, H.; Chen, I.A. Rapid Colorimetric identification of bacterial species through the capture of gold nanoparticles by chimeric phages. ACS Nano 2019, 13, 1244–1252. [Google Scholar] [CrossRef]
- Priyadarshi, N.; Ambule, M.D.; Kaushal, S.; Kumar, A.; Sagar, P.; Srivastava, A.K.; Singhal, N.K. Nanoglycocluster based diagnostic platform for colorimetric identification of bacteria; A comparative study analysing the effect of AuNPs size, linker length, and glycan diversity. Biosens. Bioelectron. 2022, 201, 113969. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Guo, Y.; Fan, J.; Yang, Y.; Zuo, C.; Bai, S.; Sheng, S.; Li, J.; Xie, G. A fluorometric assay for rapid enrichment and determination of bacteria by using zirconium-metal organic frameworks as both capture surface and signal amplification tag. Microchim. Acta 2020, 187, 188. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Hu, Z.; Yang, D.; Xie, S.; Jiang, Z.; Niessner, R.; Haisch, C.; Zhou, H.; Sun, P. Bacteria identification: From powerful SERS to its advanced compatible techniques. Adv. Sci. 2020, 7, 2001739. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Zhong, Y.; Wang, M.; Zheng, X.; Zhang, J. Scale-Adaptive Deep Model for Bacterial Raman Spectra Identification. IEEE J. Biomed. Health Inform. 2022, 26, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Mobed, A.; Hasanzadeh, M.; Seidi, F. Anti-bacterial activity of gold nanocomposites as a new nanomaterial weapon to combat photogenic agents: Recent advances and challenges. RSC Adv. 2021, 11, 34688–34698. [Google Scholar] [CrossRef]
- Gibała, A.; Żeliszewska, P.; Gosiewski, T.; Krawczyk, A.; Duraczyńska, D.; Szaleniec, J.; Szaleniec, M.; Oćwieja, M. Antibacterial and antifungal properties of silver nanoparticles-effect of a surface-stabilizing agent. Biomolecules 2021, 11, 1481. [Google Scholar] [CrossRef]
- Siddiqi, K.S.; ur Rahman, A.; Tajuddin; Husen, A. Properties of zinc oxide nanoparticles and their activity against microbes. Nanoscale Res. Lett. 2018, 13, 141. [Google Scholar] [CrossRef]
- Ismail, N.A.; Amin, K.A.M.; Majid, F.A.A.; Razali, M.H. Gellan gum incorporating titanium dioxide nanoparticles biofilm as wound dressing: Physicochemical, mechanical, antibacterial properties and wound healing studies. Mater. Sci. Eng. C 2019, 103, 109770. [Google Scholar] [CrossRef]
- Bashir, F.; Irfan, M.; Ahmad, T.; Iqbal, J.; Butt, M.T.; Sadef, Y.; Umbreen, M.; Shaikh, I.A.; Moniruzzaman, M. Efficient utilization of low cost agro materials for incorporation of copper nanoparticles to scrutinize their antibacterial properties in drinking water. Environ. Technol. Innov. 2021, 21, 101228. [Google Scholar] [CrossRef]
- Yougbaré, S.; Mutalik, C.; Krisnawati, D.I.; Kristanto, H.; Jazidie, A.; Nuh, M.; Cheng, T.-M.; Kuo, T.-R. Nanomaterials for the Photothermal Killing of Bacteria. Nanomaterials 2020, 10, 1123. [Google Scholar] [CrossRef]
- Liu, H.; Xing, F.; Zhou, Y.; Yu, P.; Xu, J.; Luo, R.; Xiang, Z.; Maria Rommens, P.; Liu, M.; Ritz, U. Nanomaterials-Based Photothermal Therapies for Antibacterial Applications. Mater. Des. 2023, 233, 112231. [Google Scholar] [CrossRef]
- Xu, J.-W.; Yao, K.; Xu, Z.-K. Nanomaterials with a Photothermal Effect for Antibacterial Activities: An Overview. Nanoscale 2019, 11, 8680–8691. [Google Scholar] [CrossRef] [PubMed]
- Khorsandi, K.; Hosseinzadeh, R.; Sadat Esfahani, H.; Keyvani-Ghamsari, S.; Ur Rahman, S. Nanomaterials as drug delivery systems with antibacterial properties: Current trends and future priorities. Expert. Rev. Anti Infect. Ther. 2021, 19, 1299–1323. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Meng, H.M.; Li, Z. Near-infrared inorganic nanomaterial-based nanosystems for photothermal therapy. Nanoscale 2021, 13, 8751–8772. [Google Scholar] [CrossRef] [PubMed]
- Ferone, M.; Gowen, A.; Fanning, S.; Scannell, A.G.M. Microbial detection and identification methods: Bench top assays to omics approaches. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3106–3129. [Google Scholar] [CrossRef]
- Bonnet, M.; Lagier, J.C.; Raoult, D.; Khelaifia, S. Bacterial culture through selective and non-selective conditions: The evolution of culture media in clinical microbiology. New. Microbes New. Infect. 2020, 34, 100622. [Google Scholar] [CrossRef]
- Peri, A.M.; Stewart, A.; Hume, A.; Irwin, A.; Harris, P.N.A. New Microbiological Techniques for the Diagnosis of Bacterial Infections and Sepsis in ICU Including Point of Care. Curr. Infect. Dis. Rep. 2021, 23, 12. [Google Scholar] [CrossRef]
- Davis, C. Enumeration of probiotic strains: Review of culture-dependent and alternative techniques to quantify viable bacteria. J. Microbiol. Methods 2014, 103, 9–17. [Google Scholar] [CrossRef]
- Li, Y.; Guo, J.; Yang, H.; Li, H.; Shen, Y.; Zhang, D. Comparison of culture-negative and culture-positive sepsis or septic shock: A systematic review and meta-analysis. Crit. Care 2021, 25, 167. [Google Scholar] [CrossRef]
- Cheng, M.P.; Stenstrom, R.; Paquette, K.; Stabler, S.N.; Akhter, M.; Davidson, A.C.; Gavric, M.; Lawandi, A.; Jinah, R.; Saeed, Z.; et al. Blood culture results before and after antimicrobial administration in patients with severe manifestations of sepsis: A diagnostic study. Ann. Intern. Med. 2019, 171, 547–554. [Google Scholar] [CrossRef]
- Maugeri, G.; Lychko, I.; Sobral, R.; Roque, A.C.A. Identification and antibiotic-susceptibility profiling of infectious bacterial agents: A review of current and future trends. Biotechnol. J. 2019, 14, e1700750. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Lee, S.; Park, Y.-S. Molecular typing tools for identifying and characterizing lactic acid bacteria: A review. Food Sci. Biotechnol. 2020, 29, 1301–1318. [Google Scholar] [CrossRef]
- Sjöling, Å.; Sadeghipoorjahromi, L.; Novak, D.; Tobias, J. Identification of major diarrheagenic bacterial pathogens by multiplex PCR panels. Microbiol. Res. 2015, 172, 34–40. [Google Scholar] [CrossRef]
- Bao, M.; Zheng, Z.Z.; Sun, X.; Chen, J.; Deng, X. Enhancing PCR capacity to detect ‘Candidatus Liberibacter asiaticus’ utilizing whole genome sequence information. Plant. Dis. 2020, 104, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Ibal, J.C.; Pham, H.Q.; Park, C.E.; Shin, J.-H. Information about variations in multiple copies of bacterial 16S rRNA genes may aid in species identification. PLoS ONE 2019, 14, e0212090. [Google Scholar] [CrossRef]
- Trung, N.T.; Thau, N.S.; Bang, M.H.; Song, L.H. PCR-based Sepsis@Quick test is superior in comparison with blood culture for identification of sepsis-causative pathogens. Sci. Rep. 2019, 9, 13663. [Google Scholar] [CrossRef]
- Dark, P.; Blackwood, B.; Gates, S.; McAuley, D.; Perkins, G.D.; McMullan, R.; Wilson, C.; Graham, D.; Timms, K.; Warhurst, G. Accuracy of LightCycler® SeptiFast for the identification and identification of pathogens in the blood of patients with suspected sepsis: A systematic review and meta-analysis. Intensive Care Med. 2015, 41, 21–33. [Google Scholar] [CrossRef]
- Zboromyrska, Y.; Cillóniz, C.; Cobos-Trigueros, N.; Almela, M.; Hurtado, J.C.; Vergara, A.; Mata, C.; Soriano, A.; Mensa, J.; Marco, F.; et al. Evaluation of the Magicplex™ sepsis real-time test for the rapid diagnosis of bloodstream infections in adults. Front. Cell. Infect. Microbiol. 2019, 9, 56. [Google Scholar] [CrossRef]
- Li, D.; Yi, J.; Han, G.; Qiao, L. MALDI-TOF Mass Spectrometry in Clinical Analysis and Research. ACS Meas. Sci. Au 2022, 2, 385–404. [Google Scholar] [CrossRef]
- Tsuchida, S.; Nakayama, T. MALDI-Based Mass Spectrometry in Clinical Testing: Focus on Bacterial Identification. Appl. Sci. 2022, 12, 2814. [Google Scholar] [CrossRef]
- Patel, R. MALDI-TOF MS for the Diagnosis of Infectious Diseases. Clin. Chem. 2015, 61, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Seng, P.; Drancourt, M.; Gouriet, F.; La Scola, B.; Fournier, P.-E.; Rolain, J.M.; Raoult, D. Ongoing revolution in bacteriology: Routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 2009, 49, 543–551. [Google Scholar] [CrossRef]
- Clark Andrew, E.; Kaleta Erin, J.; Arora, A.; Wolk Donna, M. Matrix-assisted laser desorption ionization–time of flight mass spectrometry: A fundamental shift in the routine practice of clinical microbiology. Clin. Microbiol. Rev. 2013, 26, 547–603. [Google Scholar] [CrossRef] [PubMed]
- Edwards-Jones, V.; Claydon, M.A.; Evason, D.J.; Walker, J.; Fox, A.J.; Gordan, D.B. Rapid discrimination between methicillin-sensitive and methicillin-resistant staphylococcus aureus by intact cell mass spectrometry. J. Med. Microbiol. 2000, 49, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.-Y.; Chiang-Ni, C.; Teng, S.-H. Current status of MALDI-TOF mass spectrometry in clinical microbiology. J. Food Drug. Anal. 2019, 27, 404–414. [Google Scholar] [CrossRef]
- Buchan Blake, W.; Riebe Katherine, M.; Ledeboer Nathan, A. Comparison of the MALDI biotyper system using sepsityper specimen processing to routine microbiological methods for identification of bacteria from positive blood culture bottles. J. Clin. Microbiol. 2012, 50, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.S.; Sterling, S.A.; To, H.; Seals, S.R.; Jones, A.E. Diagnostic performance of matrix-assisted laser desorption ionisation time-of-flight mass spectrometry in blood bacterial infections: A systematic review and meta-analysis. Infect. Dis. 2016, 48, 530–536. [Google Scholar] [CrossRef]
- Faron, M.L.; Buchan, B.W.; Ledeboer, N.A. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for use with positive blood cultures: Methodology, performance, and optimization. J. Clin. Microbiol. 2017, 55, 3328–3338. [Google Scholar] [CrossRef]
- Florio, W.; Baldeschi, L.; Rizzato, C.; Tavanti, A.; Ghelardi, E.; Lupetti, A. Identification of antibiotic-resistance by MALDI-TOF mass spectrometry: An expanding area. Front. Cell. Infect. Microbiol. 2020, 10, 572909. [Google Scholar] [CrossRef]
- Florio, W.; Tavanti, A.; Barnini, S.; Ghelardi, E.; Lupetti, A. Recent Advances and Ongoing Challenges in the Diagnosis of Microbial Infections by MALDI-TOF Mass Spectrometry. Front. Microbiol. 2018, 9, 1097. [Google Scholar] [CrossRef]
- Han, S.S.; Jeong, Y.S.; Choi, S.K. Current Scenario and Challenges in the Direct Identification of Microorganisms Using MALDI TOF MS. Microorganisms 2021, 9, 1917. [Google Scholar] [CrossRef] [PubMed]
- Wunschel, S.C.; Jarman, K.H.; Petersen, C.E.; Valentine, N.B.; Wahl, K.L.; Schauki, D.; Jackman, J.; Nelson, C.P.; White, E. Bacterial analysis by MALDI-TOF mass spectrometry: An inter-laboratory comparison. J. Am. Soc. Mass. Spectrom. 2005, 16, 456–462. [Google Scholar] [CrossRef] [PubMed]
- Ray, P.C.; Khan, S.A.; Singh, A.K.; Senapati, D.; Fan, Z. Nanomaterials for Targeted Detection and Photothermal Killing of Bacteria. Chem. Soc. Rev. 2012, 41, 3193–3209. [Google Scholar] [CrossRef]
- Qui, M.; Zheng, M.; Zhang, J.; Yang, X.; Zhang, W.; Man, C.; Zhao, Q.; Jiang, Y. Recent Advances on Emerging Biosensing Technologies and on-site Analytical Devices for the Detection of Drug Resistant Foodborne Pathogens. Trends Anal. Chem. 2023, 167, 117258. [Google Scholar] [CrossRef]
- Jin, R.; Zeng, C.; Zhou, M.; Chen, Y. Atomically Precise Colloidal Metal Nanoclusters and Nanoparticles: Fundamentals and Opportunities. Chem. Rev. 2016, 116, 10346–10413. [Google Scholar] [CrossRef]
- Zhang, J.; Kolhatkar, G.; Ruediger, A. Localized Surface Plasmon Resonance Shift and Its Application in Scanning Near-Field Optical Microscopy. J. Mater. Chem. C Mater. 2021, 9, 6960–6969. [Google Scholar] [CrossRef]
- Simone, G.; de Ruijter, P. Plasmon Resonance Excitation Enhances Raman Emission and Amplifies the Molecular Vibration on Au (111) Film. Appl. Surf. Sci. 2020, 530, 147207. [Google Scholar] [CrossRef]
- Tavakkoli Yaraki, M.; Tan, Y.N. Recent Advances in Metallic Nanobiosensors Development: Colorimetric, Dynamic Light Scattering and Fluorescence Detection. Sens. Int. 2020, 1, 100049. [Google Scholar] [CrossRef]
- Jazayeri, M.H.; Aghaie, T.; Avan, A.; Vatankhah, A.; Ghaffari, M.R.S. Colorimetric Detection Based on Gold Nano Particles (GNPs): An Easy, Fast, Inexpensive, Low-Cost and Short Time Method in Detection of Analytes (Protein, DNA, and Ion). Sens. Biosensing Res. 2018, 20, 1–8. [Google Scholar] [CrossRef]
- Miranda, O.R.; Li, X.; Garcia-Gonzalez, L.; Zhu, Z.-J.; Yan, B.; Bunz, U.H.F.; Rotello, V.M. Colorimetric Bacteria Sensing Using a Supramolecular Enzyme–Nanoparticle Biosensor. J. Am. Chem. Soc. 2011, 133, 9650–9653. [Google Scholar] [CrossRef]
- Yang, J.; Wang, X.; Sun, Y.; Chen, B.; Hu, F.; Guo, C.; Yang, T. Recent Advances in Colorimetric Sensors Based on Gold Nanoparticles for Pathogen Detection. Biosensors 2023, 13, 29. [Google Scholar] [CrossRef]
- Li, B.; Li, X.; Dong, Y.; Wang, B.; Li, D.; Shi, Y.; Wu, Y. Colorimetric Sensor Array Based on Gold Nanoparticles with Diverse Surface Charges for Microorganisms Identification. Anal. Chem. 2017, 89, 10639–10643. [Google Scholar] [CrossRef]
- Yuan, P.; Ding, X.; Yang, Y.Y.; Xu, Q.-H. Metal Nanoparticles for Diagnosis and Therapy of Bacterial Infection. Adv. Health Mater. 2018, 7, 1701392. [Google Scholar] [CrossRef] [PubMed]
- Yin, M.; Wu, C.; Li, H.; Jia, Z.; Deng, Q.; Wang, S.; Zhang, Y. Simultaneous Sensing of Seven Pathogenic Bacteria by Guanidine-Functionalized Upconversion Fluorescent Nanoparticles. ACS Omega 2019, 4, 8953–8959. [Google Scholar] [CrossRef]
- Phillips, R.L.; Miranda, O.R.; You, C.-C.; Rotello, V.M.; Bunz, U.H.F. Rapid and Efficient Identification of Bacteria Using Gold-Nanoparticle–Poly(Para-Phenyleneethynylene) Constructs. Angew. Chem. Int. Ed. 2008, 47, 2590–2594. [Google Scholar] [CrossRef]
- Zheng, S.; Yang, X.; Zhang, B.; Cheng, S.; Han, H.; Jin, Q.; Wang, C.; Xiao, R. Sensitive Detection of Escherichia Coli O157:H7 and Salmonella Typhimurium in Food Samples Using Two-Channel Fluorescence Lateral Flow Assay with Liquid Si@quantum Dot. Food Chem. 2021, 363, 130400. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Wang, H.; Fu, F.; Li, L.; Li, J.; Li, G.; Song, Y.; Swihart, M.T.; Song, E. Dual-Recognition Förster Resonance Energy Transfer Based Platform for One-Step Sensitive Detection of Pathogenic Bacteria Using Fluorescent Vancomycin–Gold Nanoclusters and Aptamer–Gold Nanoparticles. Anal. Chem. 2017, 89, 4085–4090. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wu, H.; Yang, Y.; Yan, R.; Zhao, Y.; Wang, Y.; Chen, A.; Shao, S.; Jiang, P.; Li, Y.-Q. Bacterial Species-Identifiable Magnetic Nanosystems for Early Sepsis Diagnosis and Extracorporeal Photodynamic Blood Disinfection. Nanoscale 2018, 10, 132–141. [Google Scholar] [CrossRef]
- Herrmann, I.K.; Urner, M.; Graf, S.; Schumacher, C.M.; Roth-Z’graggen, B.; Hasler, M.; Stark, W.J.; Beck-Schimmer, B. Endotoxin Removal by Magnetic Separation-Based Blood Purification. Adv. Healthc. Mater. 2013, 2, 829–835. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-J.; Jeong, K.J.; Hashimoto, M.; Kwon, A.H.; Rwei, A.; Shankarappa, S.A.; Tsui, J.H.; Kohane, D.S. Synthetic Ligand-Coated Magnetic Nanoparticles for Microfluidic Bacterial Separation from Blood. Nano Lett. 2014, 14, 1–5. [Google Scholar] [CrossRef]
- Shi, Z.; Jin, L.; He, C.; Li, Y.; Jiang, C.; Wang, H.; Zhang, J.; Wang, J.; Zhao, W.; Zhao, C. Hemocompatible Magnetic Particles with Broad-Spectrum Bacteria Capture Capability for Blood Purification. J. Colloid. Interface Sci. 2020, 576, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Langer, J.; Jimenez de Aberasturi, D.; Aizpurua, J.; Alvarez-Puebla, R.A.; Auguié, B.; Baumberg, J.J.; Bazan, G.C.; Bell, S.E.J.; Boisen, A.; Brolo, A.G.; et al. Present and future of surface-enhanced Raman scattering. ACS Nano 2020, 14, 28–117. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Zhang, D.; Yang, K.; Zhang, X.; Zhu, Y.G. Perspective on surface-enhanced Raman spectroscopic investigation of microbial world. Anal. Chem. 2019, 91, 15345–15354. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Sun, Y.; Li, X.; Wang, L.; Xu, Y.; He, L.; Li, G. Recent advances in dual recognition-based surface enhanced Raman scattering for pathogenic bacteria identification: A review. Anal. Chim. Acta 2021, 1157, 338279. [Google Scholar] [CrossRef]
- Usman, M.; Tang, J.-W.; Li, F.; Lai, J.-X.; Liu, Q.-H.; Liu, W.; Wang, L. Recent advances in surface enhanced Raman spectroscopy for bacterial pathogen identifications. J. Adv. Res. 2023, 51, 91–107. [Google Scholar] [CrossRef] [PubMed]
- Harz, M.; Rosch, P.; Peschke, K.D.; Ronneberger, O.; Burkhardt, H.; Popp, J. Micro-Raman spectroscopic identification of bacterial cells of the genus staphylococcus and dependence on their cultivation conditions. Analyst 2005, 130, 1543–1550. [Google Scholar] [CrossRef]
- Ciloglu, F.U.; Hora, M.; Gundogdu, A.; Kahraman, M.; Tokmakci, M.; Aydin, O. SERS-based sensor with a machine learning based effective feature extraction technique for fast identification of colistin-resistant Klebsiella pneumoniae. Anal. Chim. Acta 2022, 1221, 340094. [Google Scholar] [CrossRef]
- Ciloglu, F.U.; Caliskan, A.; Saridag, A.M.; Kilic, I.H.; Tokmakci, M.; Kahraman, M.; Aydin, O. Drug-resistant Staphylococcus aureus bacteria identification by combining surface-enhanced Raman spectroscopy (SERS) and deep learning techniques. Sci. Rep. 2021, 11, 18444. [Google Scholar] [CrossRef]
- Liang, W.; Jia-Wei, T.; Fen, L.; Muhammad, U.; Chang-Yu, W.; Qing-Hua, L.; Hai-Quan, K.; Wei, L.; Bing, G. Identification of Bacterial Pathogens at Genus and Species Levels through Combination of Raman Spectrometry and Deep-Learning Algorithms. Microbiol. Spectr. 2022, 10, e02580-22. [Google Scholar] [CrossRef]
- Lussier, F.; Thibault, V.; Charron, B.; Wallace, G.Q.; Masson, J.-F. Deep learning and artificial intelligence methods for Raman and surface-enhanced Raman scattering. TrAC Trends Anal. Chem. 2020, 124, 115796. [Google Scholar] [CrossRef]
- Ho, C.S.; Jean, N.; Hogan, C.A.; Blackmon, L.; Jeffrey, S.S.; Holodniy, M.; Banaei, N.; Saleh, A.A.E.; Ermon, S.; Dionne, J. Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning. Nat. Commun. 2019, 10, 4927. [Google Scholar] [CrossRef] [PubMed]
- Maruthamuthu, M.K.; Raffiee, A.H.; De Oliveira, D.M.; Ardekani, A.A.-O.; Verma, M.A.-O. Raman spectra-based deep learning: A tool to identify microbial contamination. Microbiol. Open. 2020, 00, e112. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Lin, Q.; Zhang, J.; Young, G.M.; Jiang, C.; Zhong, Y.; Zhang, J. Rapid identification of pathogens by using surface-enhanced Raman spectroscopy and multi-scale convolutional neural network. Anal. Bioanal. Chem. 2021, 413, 3801–3811. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.; Kang, S.; Wang, W.; Huang, Q.; Kim, I.; Vikesland, P.J. Lectin-Modified Bacterial Cellulose Nanocrystals Decorated with Au Nanoparticles for Selective Detection of Bacteria Using Surface-Enhanced Raman Scattering Coupled with Machine Learning. ACS Appl. Nano Mater. 2022, 5, 259–268. [Google Scholar] [CrossRef]
- Thrift, W.J.; Ronaghi, S.; Samad, M.; Wei, H.; Nguyen, D.G.; Cabuslay, A.S.; Groome, C.E.; Santiago, P.J.; Baldi, P.; Hochbaum, A.I.; et al. Deep Learning Analysis of Vibrational Spectra of Bacterial Lysate for Rapid Antimicrobial Susceptibility Testing. ACS Nano 2020, 14, 15336–15348. [Google Scholar] [CrossRef]
- Uysal Ciloglu, F.; Saridag, A.M.; Kilic, I.H.; Tokmakci, M.; Kahraman, M.; Aydin, O. Identification of methicillin-resistant Staphylococcus aureus bacteria using surface-enhanced Raman spectroscopy and machine learning techniques. Analyst 2020, 145, 7559–7570. [Google Scholar] [CrossRef]
- Rho, E.; Kim, M.; Cho, S.H.; Choi, B.; Park, H.; Jang, H.; Jung, Y.S.; Jo, S. Separation-free bacterial identification in arbitrary media via deep neural network-based SERS analysis. Biosens. Bioelectron. 2022, 202, 113991. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.H.; Lee, H.J.; Hwang, M.J.; Kim, J.; Kim, H.; Nam, S.H.; Park, J.S.; Hwang, J.E.; Kim, E.S.; Park, Y.S.; et al. Gold-silver core-shell nanodumbbells in solution state as a highly sensitive and reproducible assay platform for bacterial genome identification. Sens. Actuators B Chem. 2021, 349, 130784. [Google Scholar] [CrossRef]
- Wang, J.; Wu, X.; Wang, C.; Shao, N.; Dong, P.; Xiao, R.; Wang, S. Magnetically assisted surface-enhanced Raman spectroscopy for the identification of staphylococcus aureus based on aptamer recognition. ACS Appl. Mater. Interfaces 2015, 7, 20919–20929. [Google Scholar] [CrossRef]
- Yu, H.; Xiao, M.; Lai, W.; Alam, M.F.; Zhang, W.; Pei, H.; Wan, Y.; Li, L. A self-calibrating surface-enhanced Raman scattering-active system for bacterial phenotype identification. Anal. Chem. 2020, 92, 4491–4497. [Google Scholar] [CrossRef] [PubMed]
- Hwang, M.J.; Jang, A.S.; Lim, D.-K. Comparative study of fluorescence and surface-enhanced Raman scattering with magnetic microparticle-based assay for target bacterial DNA identification. Sens. Actuators B Chem. 2021, 329, 129134. [Google Scholar] [CrossRef]
- Zhou, Z.; Xiao, R.; Cheng, S.; Wang, S.; Shi, L.; Wang, C.; Qi, K.; Wang, S. A universal SERS-label immunoassay for pathogen bacteria identification based on Fe3O4@Au-aptamer separation and antibody-protein A orientation recognition. Anal. Chim. Acta 2021, 1160, 338421. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Yin, Y.; Wu, H.; Hao, Z.; Li, J.; Wang, S.; Liu, Y. Integrated SERS Platform for Reliable Identification and Photothermal Elimination of Bacteria in Whole Blood Samples. Anal. Chem. 2021, 93, 1569–1577. [Google Scholar] [CrossRef]
- Lynk, T.P.; Sit, C.S.; Brosseau, C.L. Electrochemical Surface-Enhanced Raman Spectroscopy as a Platform for Bacterial Identification and Identification. Anal. Chem. 2018, 90, 12639–12646. [Google Scholar] [CrossRef]
- Doron, S.; Gorbach, S.L. Bacterial Infections: Overview. In International Encyclopedia of Public Health; Elsevier: Amsterdam, The Netherlands, 2008; pp. 273–282. [Google Scholar] [CrossRef]
- Smith, I. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin. Microbiol. Rev. 2003, 16, 3. [Google Scholar] [CrossRef]
- Nathan, A.A.-O.; Teh, C.S.J.; Jabar, K.A.; Teoh, B.T.; Tangaperumal, A.; Westerhout, C.; Zaki, R.; Eg, K.P.; Thavagnanam, S.; de Bruyne, J.A. Bacterial pneumonia and its associated factors in children from a developing country: A prospective cohort study. PLoS ONE 2020, 15, e0228056. [Google Scholar] [CrossRef] [PubMed]
- Nelson, E.J.; Harris, J.B.; Glenn Morris, J., Jr.; Calderwood, S.B.; Camilli, A. Cholera transmission: The host, pathogen and bacteriophage dynamic. Nat. Rev. Microbiol. 2009, 7, 693–702. [Google Scholar] [CrossRef]
- Gygli, S.M.; Borrell, S.; Trauner, A.; Gagneux, S. Antimicrobial resistance in Mycobacterium tuberculosis: Mechanistic and evolutionary perspectives. FEMS Microbiol. Rev. 2017, 41, 354–373. [Google Scholar] [CrossRef]
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef]
- Aminov, R.I. A brief history of the antibiotic era: Lessons learned and challenges for the future. Front. Microbiol. 2010, 1, 134. [Google Scholar] [CrossRef]
- Leitão, J.A.-O.X. New insights into antibacterial compounds: From synthesis and discovery to molecular mechanisms of action. Antibiotics 2020, 9, 471. [Google Scholar] [CrossRef] [PubMed]
- Heesemann, J. Mechanisms of resistance to beta-lactam antibiotics. Infection 1993, 21, S4–S9. [Google Scholar] [CrossRef] [PubMed]
- Etebu, E.; Arikekpar, I. Antibiotics: Classification and mechanisms of action with emphasis on molecular perspectives. Int. J. Appl. Microbiol. Biotechnol. Res. 2016, 4, 90–101. [Google Scholar]
- Hamilton-Miller, J.M. Chemistry and biology of the polyene macrolide antibiotics. Bacteriol. Rev. 1973, 37, 407. [Google Scholar] [CrossRef]
- Sánchez, A.R.; Rogers, R.S., III; Sheridan, P.J. Tetracycline and other tetracycline-derivative staining of the teeth and oral cavity. Int. J. Dermatol. 2004, 43, 709–715. [Google Scholar] [CrossRef]
- Fuoco, D. Classification framework and chemical biology of tetracycline-structure-based drugs. Antibiotics 2012, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Domagala, J.M. Structure-activity and structure-side-effect relationships for the quinolone antibacterials. J. Antimicrob. Chemother. 1994, 33, 685–706. [Google Scholar] [CrossRef] [PubMed]
- Peterson, L.R. Currently available antimicrobial agents and their potential for use as monotherapy. Clin. Microbiol. Infect. 2008, 14, 30–45. [Google Scholar] [CrossRef]
- Griffith, E.C.; Wallace, M.J.; Wu, Y.; Kumar, G.; Gajewski, S.; Jackson, P.; Phelps, G.A.; Zheng, Z.; Rock, C.O.; Lee, R.E.; et al. The structural and functional basis for recurring sulfa drug resistance mutations in staphylococcus aureus dihydropteroate synthase. Front. Microbiol. 2018, 17, 1369. [Google Scholar] [CrossRef]
- Kahne, D.; Leimkuhler, C.; Lu, A.W.; Walsh, C. Glycopeptide and lipoglycopeptide antibiotics. Chem. Rev. 2005, 105, 425–448. [Google Scholar] [CrossRef]
- Allen, N.E.; Nicas, T.I. Mechanism of action of oritavancin and related glycopeptide antibiotics. FEMS Microbiol. Rev. 2003, 26, 511–532. [Google Scholar] [CrossRef] [PubMed]
- Bozdogan, B.; Appelbaum, P.C. Oxazolidinones: Activity, mode of action, and mechanism of resistance. Int. J. Antimicrob. Agents 2004, 23, 113–119. [Google Scholar] [CrossRef]
- Shinabarger, D.L.; Marotti, K.R.; Murray, R.W.; Lin, A.H.; Melchior, E.P.; Swaney, S.M.; Dunyak, D.S.; Demyan, W.F.; Buysse, J.M. Mechanism of action of oxazolidinones: Effects of linezolid and eperezolid on translation reactions. Antimicrob. Agents Chemother. 1997, 41, 2132–2136. [Google Scholar] [CrossRef]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Spirescu, V.A.; Chircov, C.; Grumezescu, A.M.; Andronescu, E. Polymeric nanoparticles for antimicrobial therapies: An up-to-date overview. Polymers 2021, 13, 724. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Li, Y.; Yang, Y.; Fang, Z.; Chen, X.; Wang, Y.; Kang, J.; Qu, X.; Yuan, W.; Dai, K.; et al. Osteoinductivity and Antibacterial Properties of Strontium Ranelate-Loaded Poly(Lactic-co-Glycolic Acid) Microspheres With Assembled Silver and Hydroxyapatite Nanoparticles. Front. Pharmacol. 2018, 9, 368. [Google Scholar] [CrossRef]
- Ejaz, S.; Ihsan, A.; Noor, T.; Shabbir, S.; Imran, M. Mannose functionalized chitosan nanosystems for enhanced antimicrobial activity against multidrug resistant pathogens. Polym. Test. 2020, 91, 106814. [Google Scholar] [CrossRef]
- Kritchenkov, A.S.; Egorov, A.R.; Artemjev, A.A.; Kritchenkov, I.S.; Volkova, O.V.; Kurliuk, A.V.; Shakola, T.V.; Rubanik, V.V.; Rubanik, V.V.; Tskhovrebov, A.G.; et al. Ultrasound-assisted catalyst-free thiol-yne click reaction in chitosan chemistry: Antibacterial and transfection activity of novel cationic chitosan derivatives and their based nanoparticles. Int. J. Biol. Macromol. 2020, 143, 143–152. [Google Scholar] [CrossRef]
- Chen, J.; Wang, F.; Liu, Q.; Du, J. Antibacterial polymeric nanostructures for biomedical applications. Chem. Commun. 2014, 50, 14482–14493. [Google Scholar] [CrossRef]
- Hasan, N.; Cao, J.; Lee, J.; Hlaing, S.P.; Oshi, M.A.; Naeem, M.; Ki, M.H.; Lee, B.L.; Jung, Y.; Yoo, J.W. Bacteria-Targeted Clindamycin Loaded Polymeric Nanoparticles: Effect of Surface Charge on Nanoparticle Adhesion to MRSA, Antibacterial Activity, and Wound Healing. Pharmaceutics 2019, 1, 236. [Google Scholar] [CrossRef]
- Ucak, S.; Sudagidan, M.; Borsa, B.A.; Mansuroğlu, B.; Ozalp, V.C. Inhibitory effects of aptamer targeted teicoplanin encapsulated PLGA nanoparticles for Staphylococcus aureus strains. World J. Microbiol. Biotechnol. 2020, 36, 69. [Google Scholar] [CrossRef]
- Deepika, M.S.; Thangam, R.; Sheena, T.S.; Vimala, R.T.V.; Sivasubramanian, S.; Jeganathan, K.; Thirumurugan, R. Dual drug loaded PLGA nanospheres for synergistic efficacy in breast cancer therapy. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 103, 109716. [Google Scholar] [CrossRef] [PubMed]
- Durak, S.; Arasoglu, T.; Ates, S.C.; Derman, S. Enhanced antibacterial and antiparasitic activity of multifunctional polymeric nanoparticles. Nanotechnology 2020, 31, 175705. [Google Scholar] [CrossRef]
- Da Costa, D.; Exbrayat-Héritier, C.; Rambaud, B.; Megy, S.; Terreux, R.; Verrier, B.; Primard, C. Surface charge modulation of rifampicin-loaded PLA nanoparticles to improve antibiotic delivery in Staphylococcus aureus biofilms. J. Nanobiotechnol. 2021, 19, 12. [Google Scholar] [CrossRef] [PubMed]
- Vrouvaki, I.; Koutra, E.; Kornaros, M.; Avgoustakis, K.; Lamari, F.N.; Hatziantoniou, S. Polymeric nanoparticles of pistacia lentiscus var. chia essential oil for cutaneous applications. Pharmaceutics 2020, 12, 353. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, M.; Chen, Q.; Guan, G.; Hu, W.; Zhao, X.; Qiao, M.; Hu, H.; Liang, Y.; Zhu, H.; et al. Gold nanorods/mesoporous silica-based nanocomposite as theranostic agents for targeting near-infrared imaging and photothermal therapy induced with laser. Int. J. Nanomed. 2015, 10, 4747–4761. [Google Scholar] [CrossRef] [PubMed]
- Manivasagan, P.; Khan, F.; Hoang, G.; Mondal, S.; Kim, H.; Hoang Minh Doan, V.; Kim, Y.M.; Oh, J. Thiol chitosan-wrapped gold nanoshells for near-infrared laser-induced photothermal destruction of antibiotic-resistant bacteria. Carbohydr. Polym. 2019, 225, 115228. [Google Scholar] [CrossRef]
- Rovati, D.; Albini, B.; Galinetto, P.; Grisoli, P.; Bassi, B.; Pallavicini, P.; Dacarro, G.; Taglietti, A. High stability thiol-coated gold nanostars monolayers with photo-thermal antibacterial activity and wettability control. Nanomaterials 2019, 9, 1288. [Google Scholar] [CrossRef]
- Kang, S.; Lee, J.; Ryu, S.; Kwon, Y.; Kim, K.H.; Jeong, D.H.; Paik, S.R.; Kim, B.S. Gold nanoparticle/graphene oxide hybrid sheets attached on mesenchymal stem cells for effective photothermal cancer therapy. Chem. Mater. 2017, 29, 3461–3476. [Google Scholar] [CrossRef]
- Mocan, L.; Tabaran, F.A.; Mocan, T.; Pop, T.; Mosteanu, O.; Agoston-Coldea, L.; Matea, C.T.; Gonciar, D.; Zdrehus, C.; Iancu, C. Laser thermal ablation of multidrug-resistant bacteria using functionalized gold nanoparticles. Int. J. Nanomed. 2017, 12, 2255–2263. [Google Scholar] [CrossRef]
- Garg, P.; Priyadarshi, N.; Ambule, M.D.; Kaur, G.; Kaul, S.; Gupta, R.; Sagar, P.; Bajaj, G.; Yadav, B.; Rishi, V.; et al. Multiepitope glycan-based laser assisted fluorescent nanocomposite with dual functionality for sensing and ablation of Pseudomonas aeruginosa. Nanoscale 2023. [Google Scholar] [CrossRef]
- Huang, W.C.; Tsai, P.J.; Chen, Y.C. Functional gold nanoparticles as photothermal agents for selective-killing of pathogenic bacteria. Nanomedicine 2007, 2, 777–787. [Google Scholar] [CrossRef]
- Hu, D.; Li, H.; Wang, B.; Ye, Z.; Lei, W.; Jia, F.; Jin, Q.; Ren, K.F.; Ji, J. Surface-adaptive gold nanoparticles with effective adherence and enhanced photothermal ablation of methicillin-resistant staphylococcus aureus biofilm. ACS Nano 2017, 11, 9330–9339. [Google Scholar] [CrossRef] [PubMed]
- Mao, W.; Son, Y.J.; Yoo, H.S. Gold nanospheres and nanorods for anti-cancer therapy: Comparative studies of fabrication, surface-decoration, and anti-cancer treatments. Nanoscale 2020, 12, 14996–15020. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Zheng, Y.; Lu, X.; Thai, T.; Lee, N.A.; Bach, U.; Gooding, J.J. Biocompatible gold nanorods: One-step surface functionalization, highly colloidal stability, and low cytotoxicity. Langmuir 2015, 31, 4973–4980. [Google Scholar] [CrossRef]
- Fernando, D.; Sulthana, S.; Vasquez, Y. Cellular uptake and cytotoxicity of varying aspect ratios of gold nanorods in HeLa cells. ACS Appl. Bio. Mater. 2020, 3, 1374–1384. [Google Scholar] [CrossRef]
- Kaushal, S.; Priyadarshi, N.; Pinnaka, A.K.; Soni, S.; Deep, A.; Singhal, N.K. Glycoconjugates coated gold nanorods based novel biosensor for optical identification and photothermal ablation of food borne bacteria. Sens. Actuators B Chem. 2019, 289, 207–215. [Google Scholar] [CrossRef]
- Lim, D.K.; Barhoumi, A.; Wylie, R.G.; Reznor, G.; Langer, R.S.; Kohane, D.S. Enhanced photothermal effect of plasmonic nanoparticles coated with reduced graphene oxide. Nano Lett. 2013, 13, 4075–4079. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, M.; Lee, S.S.; Yi, D.; Kim, K. Magnetic, optical gold nanorods for recyclable photothermal ablation of bacteria. J. Mater. Chem. B 2014, 2, 981–988. [Google Scholar] [CrossRef] [PubMed]
- Taglietti, A.; Diaz Fernandez, Y.A.; Amato, E.; Cucca, L.; Dacarro, G.; Grisoli, P.; Necchi, V.; Pallavicini, P.; Pasotti, L.; Patrini, M. Antibacterial activity of glutathione-coated silver nanoparticles against Gram positive and Gram negative bacteria. Langmuir 2012, 28, 8140–8148. [Google Scholar] [CrossRef]
- Wan, X.; Liu, M.; Ma, M.; Chen, D.; Wu, N.; Li, L.; Li, Z.; Lin, G.; Wang, X.; Xu, G. The ultrasmall biocompatible CuS@BSA nanoparticle and its photothermal effects. Front. Pharmacol. 2019, 10, 141. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wei, W.; Zhao, Y.; Dai, H. Iron oxide nanoparticles with photothermal performance and enhanced nanozyme activity for bacteria-infected wound therapy. Regen. Biomater. 2022, 9, rbac041. [Google Scholar] [CrossRef]
- D'Agostino, A.; Taglietti, A.; Desando, R.; Bini, M.; Patrini, M.; Dacarro, G.; Cucca, L.; Pallavicini, P.; Grisoli, P. Bulk surfaces coated with triangular silver nanoplates: Antibacterial action based on silver release and photo-thermal effect. Nanomaterials 2017, 7, 7. [Google Scholar] [CrossRef]
- Yin, M.; Li, Z.; Ju, E.; Wang, Z.; Dong, K.; Ren, J.; Qu, X. Multifunctional upconverting nanoparticles for near-infrared triggered and synergistic antibacterial resistance therapy. Chem. Commun. 2014, 50, 10488–10490. [Google Scholar] [CrossRef]
- Jeong, C.J.; Sharker, S.M.; In, I.; Park, S.Y. Iron oxide@PEDOT-based recyclable photothermal nanoparticles with poly(vinylpyrrolidone) sulfobetaines for rapid and effective antibacterial activity. ACS Appl. Mater. Interfaces 2015, 7, 9469–9478. [Google Scholar] [CrossRef]
- Wen, W.; Song, Y.; Yan, X.; Zhu, C.; Du, D.; Wang, S.; Asiri, A.M.; Lin, Y. Recent advances in emerging 2D nanomaterials for biosensing and bioimaging applications. Mater. Today 2018, 21, 164–177. [Google Scholar] [CrossRef]
- Yin, W.; Yan, L.; Yu, J.; Tian, G.; Zhou, L.; Zheng, X.; Zhang, X.; Yong, Y.; Li, J.; Gu, Z.; et al. High-throughput synthesis of single-layer MoS2 nanosheets as a near-infrared photothermal-triggered drug delivery for effective cancer therapy. ACS Nano 2014, 8, 6922–6933. [Google Scholar] [CrossRef]
- Yin, W.; Yu, J.; Lv, F.; Yan, L.; Zheng, L.R.; Gu, Z.; Zhao, Y. Functionalized nano-MoS2 with peroxidase catalytic and near-infrared photothermal activities for safe and synergetic wound antibacterial applications. ACS Nano 2016, 10, 11000–11011. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.; Tan, L.; Lang, Q.; Ke, X.; Bai, L.; Guo, K.; Qiao, R.; Bai, S. Plasmonic molybdenum oxide nanosheets supported silver nanocubes for enhanced near-infrared antibacterial activity: Synergism of photothermal effect, silver release and photocatalytic reactions. Appl. Catal. B Environ. 2018, 224, 671–680. [Google Scholar] [CrossRef]
- Gupta, B.D.; Pathak, A.; Semwal, V. Carbon-based nanomaterials for plasmonic sensors: A review. Sensors 2019, 19, 3536. [Google Scholar] [CrossRef]
- Robinson, J.T.; Tabakman, S.M.; Liang, Y.; Wang, H.; Casalongue, H.S.; Vinh, D.; Dai, H. Ultrasmall reduced graphene oxide with high near-infrared absorbance for photothermal therapy. J. Am. Chem. Soc. 2011, 133, 6825–6831. [Google Scholar] [CrossRef]
- Turcheniuk, K.; Hage, C.H.; Spadavecchia, J.; Serrano, A.Y.; Larroulet, I.; Pesquera, A.; Zurutuza, A.; Pisfil, M.G.; Héliot, L.; Boukaert, J.; et al. Plasmonic photothermal destruction of uropathogenic E. coli with reduced graphene oxide and core/shell nanocomposites of gold nanorods/reduced graphene oxide. J. Mater. Chem. B 2015, 3, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Tripathy, A.; Sen, P.; Su, B.; Briscoe, W.H. Natural and bioinspired nanostructured bactericidal surfaces. Adv. Colloid. Interface Sci. 2017, 248, 85–104. [Google Scholar] [CrossRef]
- Dehghani, S.; Mashreghi, M.; Nezhad, A.H.N.; Karimi, J.; Hosseinpour, S.; Davoodi, A. Exploring mechano-bactericidal nature of Psalmocharias cicadas wings: An analytical nanotopology investigation based on atomic force microscopy characterization. Surf. Interfaces 2021, 26, 101407. [Google Scholar] [CrossRef]
- Ivanova, E.P.; Hasan, J.; Webb, H.K.; Truong, V.K.; Watson, G.S.; Watson, J.A.; Baulin, V.A.; Pogodin, S.; Wang, J.Y.; Tobin, M.J.; et al. Natural bactericidal surfaces: Mechanical rupture of Pseudomonas aeruginosa cells by cicada wings. Small 2012, 8, 2489–2494. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.A.-O.; Yasui, T.; Yonese, A.; Yanagida, T.; Kaji, N.A.-O.X.; Kanai, M.A.-O.; Nagashima, K.; Kawai, T.; Baba, Y. Mechanical Rupture-Based Antibacterial and Cell-Compatible ZnO/SiO(2) Nanowire Structures Formed by Bottom-Up Approaches. Micromachines 2020, 11, 610. [Google Scholar] [CrossRef] [PubMed]
- Linklater, D.P.; Baulin, V.A.-O.; Le Guével, X.; Fleury, J.A.-O.; Hanssen, E.A.-O.; Nguyen, T.H.P.; Juodkazis, S.A.-O.; Bryant, G.; Crawford, R.J.; Stoodley, P.; et al. Antibacterial Action of Nanoparticles by Lethal Stretching of Bacterial Cell Membranes. Adv. Mater. 2020, 32, e2005679. [Google Scholar] [CrossRef]
- Kaul, S.; Sagar, P.; Gupta, R.; Garg, P.; Priyadarshi, N.; Singhal, N.K. Mechanobactericidal, Gold Nanostar Hydrogel-Based Bandage for Bacteria-Infected Skin Wound Healing. ACS Appl. Mater. Interfaces 2022, 14, 44084–44097. [Google Scholar] [CrossRef]
- Liu, L.; Chen, S.; Zhang, X.; Xue, Z.; Cui, S.; Hua, X.; Yang, B.; Yan, H.; Liu, C.; Wang, J.; et al. Mechanical penetration of β-lactam–resistant Gram-negative bacteria by proGrammable nanowires. Sci. Adv. 2020, 6, eabb9593. [Google Scholar] [CrossRef]


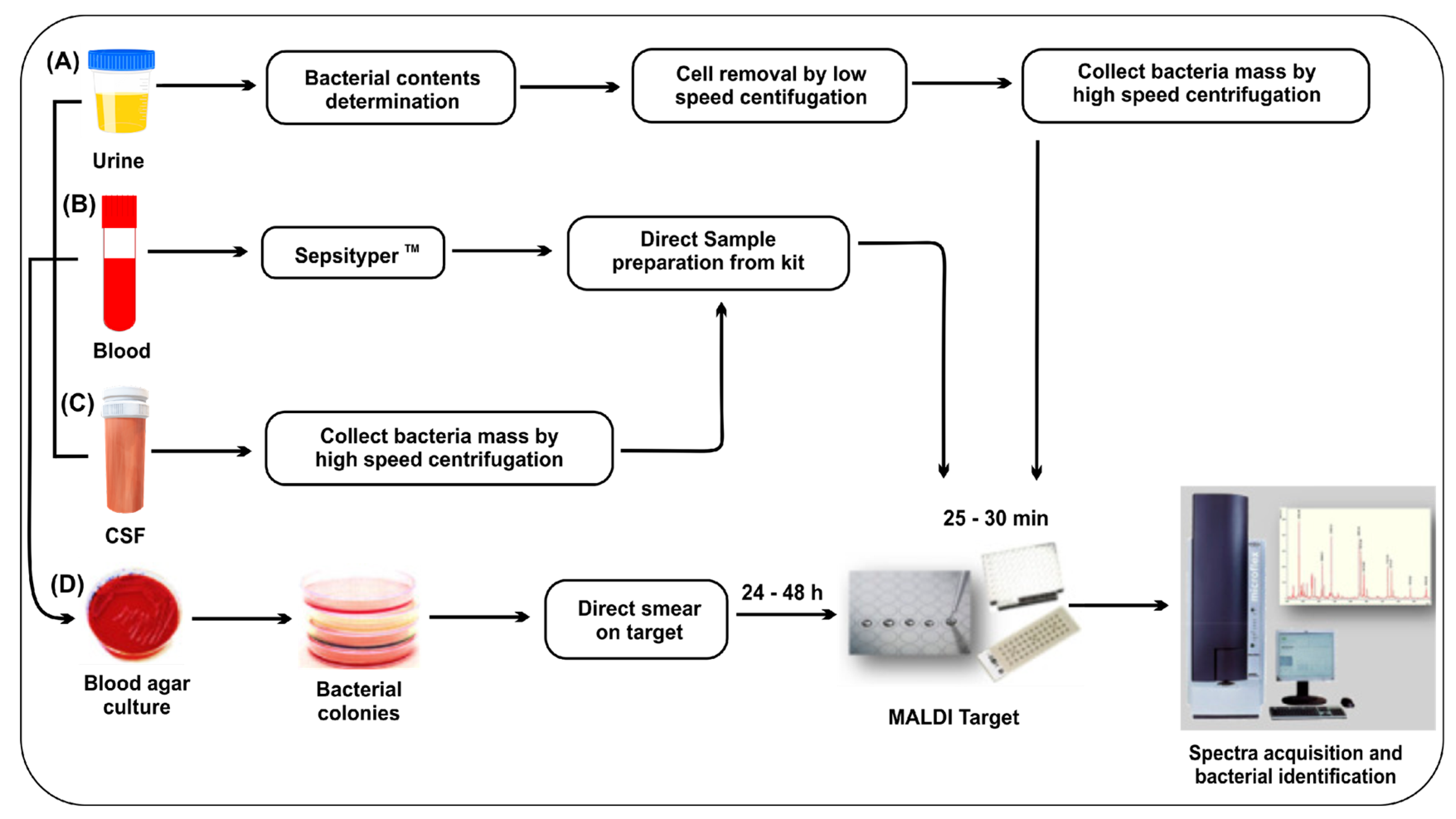
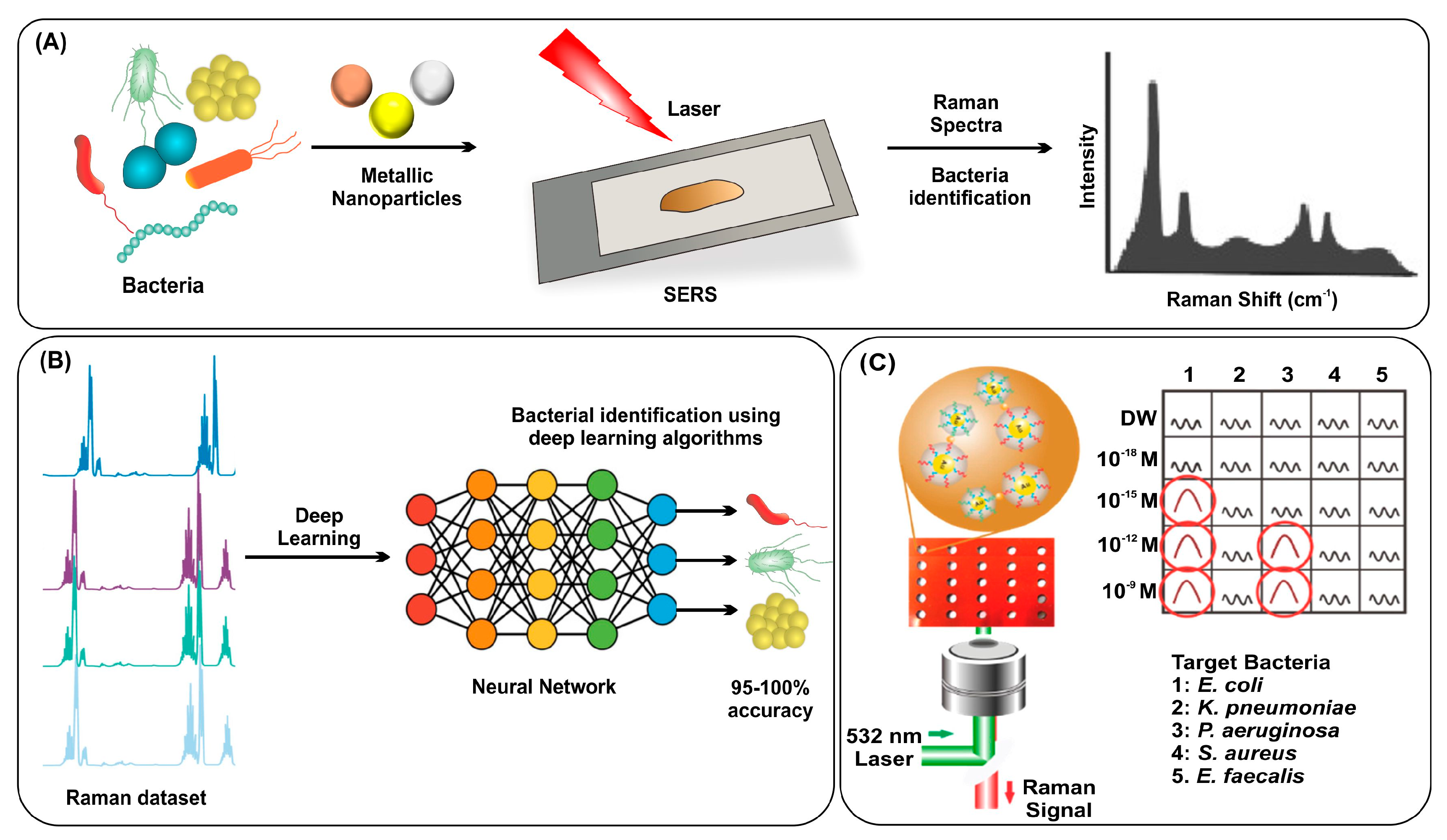
| Technique | Cost | Time | Sensitivity | Specificity | Scope | Limitations | Ref. |
|---|---|---|---|---|---|---|---|
| Culture | Low | Long | Low | High | Broad range of bacteria can be identified (both Gram-negative and Gram-positive), antibiotic susceptibility can be tested, familiar to most clinical facilities. | Time consuming, susceptible to contamination, low sensitivity for some bacterial species. | [47,48,49,50,51,52] |
| PCR | High | Short | High | High | Fast, sensitive, specific, can identify multiple bacteria simultaneously, and can quantify even small number of bacteria in real time. | False positives or negatives due to contamination, unable to differentiate among closely related bacterial strains due to complex bacterial genome sequences. | [53,54,55,56,57,58,59,60] |
| Mass spectrometry | High | Short | High | High | Sensitive, specific, and can identify broad range of bacteria directly from clinical samples, and can identify bacteria in low concentration. | Limited database coverage, expensive, requires specialized equipment and trained personnel. | [61,62,63,64,65,66,67,68,69,70,71,72,73,74] |
| Nanomaterial | Low | Short | High | High | Rapid, sensitive, specific, and can detect and quantify bacteria in real-time settings. | Sensitive to environmental factors such as temperature, pH, salinity, and non-specific aggregation in complex media. | [75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93] |
| SERS | Moderate | Short | High | High | Rapid, high sensitivity, and specificity due to unique spectral fingerprint, can identify bacteria in low concentrations, can identify broad range of bacteria in real time directly from clinical samples. | Requires specialized equipment, difficult to interpret spectra without deep-learning algorithms. | [94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaushal, S.; Priyadarshi, N.; Garg, P.; Singhal, N.K.; Lim, D.-K. Nano-Biotechnology for Bacteria Identification and Potent Anti-bacterial Properties: A Review of Current State of the Art. Nanomaterials 2023, 13, 2529. https://doi.org/10.3390/nano13182529
Kaushal S, Priyadarshi N, Garg P, Singhal NK, Lim D-K. Nano-Biotechnology for Bacteria Identification and Potent Anti-bacterial Properties: A Review of Current State of the Art. Nanomaterials. 2023; 13(18):2529. https://doi.org/10.3390/nano13182529
Chicago/Turabian StyleKaushal, Shimayali, Nitesh Priyadarshi, Priyanka Garg, Nitin Kumar Singhal, and Dong-Kwon Lim. 2023. "Nano-Biotechnology for Bacteria Identification and Potent Anti-bacterial Properties: A Review of Current State of the Art" Nanomaterials 13, no. 18: 2529. https://doi.org/10.3390/nano13182529
APA StyleKaushal, S., Priyadarshi, N., Garg, P., Singhal, N. K., & Lim, D.-K. (2023). Nano-Biotechnology for Bacteria Identification and Potent Anti-bacterial Properties: A Review of Current State of the Art. Nanomaterials, 13(18), 2529. https://doi.org/10.3390/nano13182529







