Polyethylene Terephthalate Microplastics Generated from Disposable Water Bottles Induce Interferon Signaling Pathways in Mouse Lung Epithelial Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Commercially Purchased MPs
2.2. Lab-Generation of MPs
2.2.1. Preparation of Nylon MPs from Bulk Nylon Polyamide Powder
2.2.2. Preparation of Nylon MPs from the Tea Bags
2.2.3. Preparation of PETE and PP MPs
2.2.4. Endotoxin Contamination Assessment
2.3. MP Characterization
2.3.1. Raman Spectroscopy
2.3.2. Scanning Electron Microscopy (SEM)
2.4. Cell Culture
2.5. Dynamic Light Scattering (DLS) and Electrophoretic Light Scattering (ELS) Characterization of MPs
2.6. Relative Cell Survival Assessment via Trypan Blue Exclusion Assay
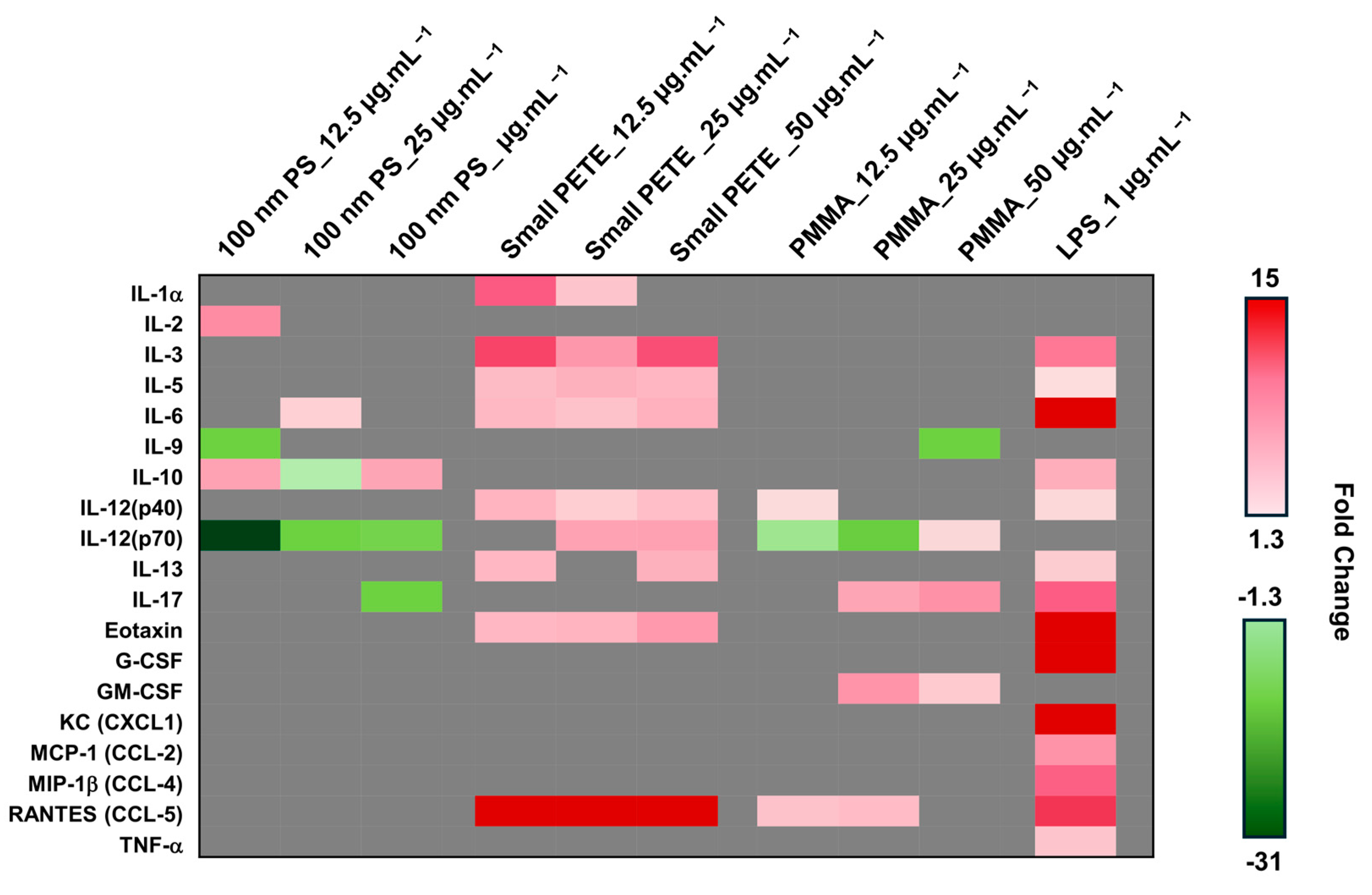
2.7. Assessment of Pro-Inflammatory Mediators by ELISA
2.7.1. Measurement of IL-1β and IL-6
2.7.2. Bio-Plex Pro Assay for the Assessment of Pro-Inflammatory Mediators
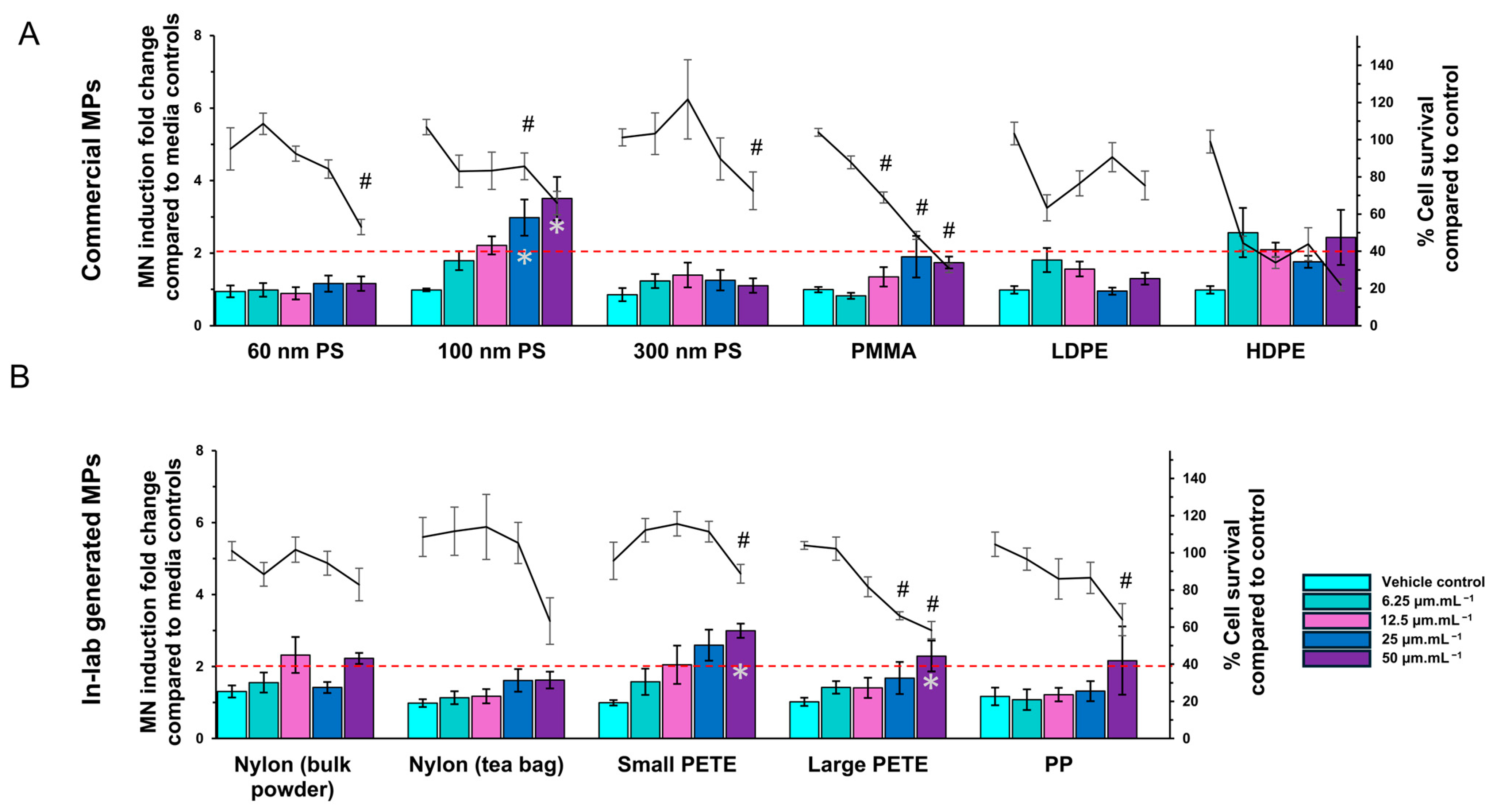
2.8. Micronucleus Assay
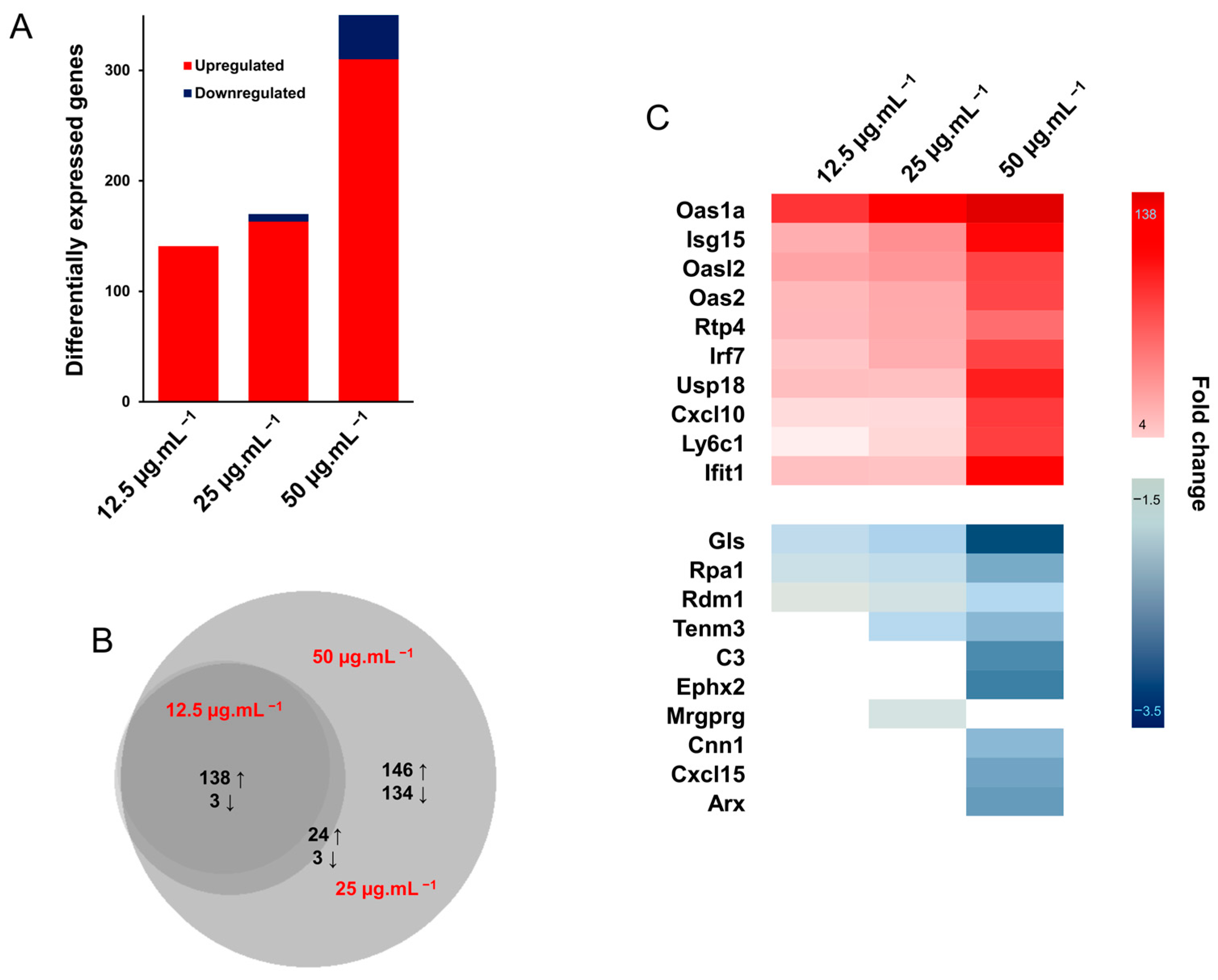
2.9. Gene Expression Analysis
2.9.1. RNA Extraction, Purification, and Integrity Analysis
2.9.2. Microarray Hybridization
2.9.3. Statistical Analysis of Microarray Data
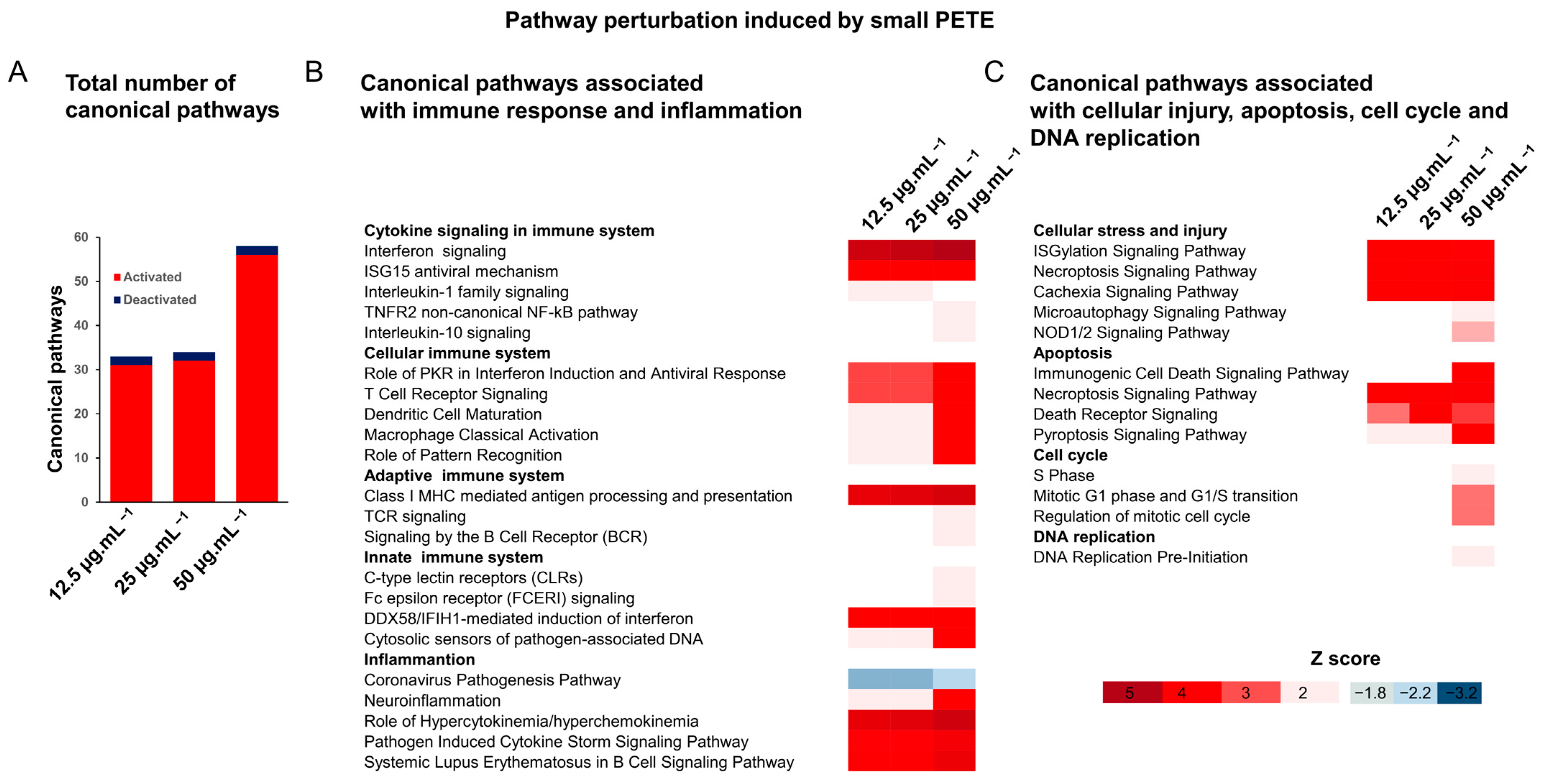
2.9.4. Pathway Enrichment Analysis
2.10. Detecting MPs in Cells
2.10.1. Immunofluorescence Labeling
2.10.2. Immunofluorescence and Enhanced Darkfield Hyperspectral Imaging (EDF-HSI)
3. Results
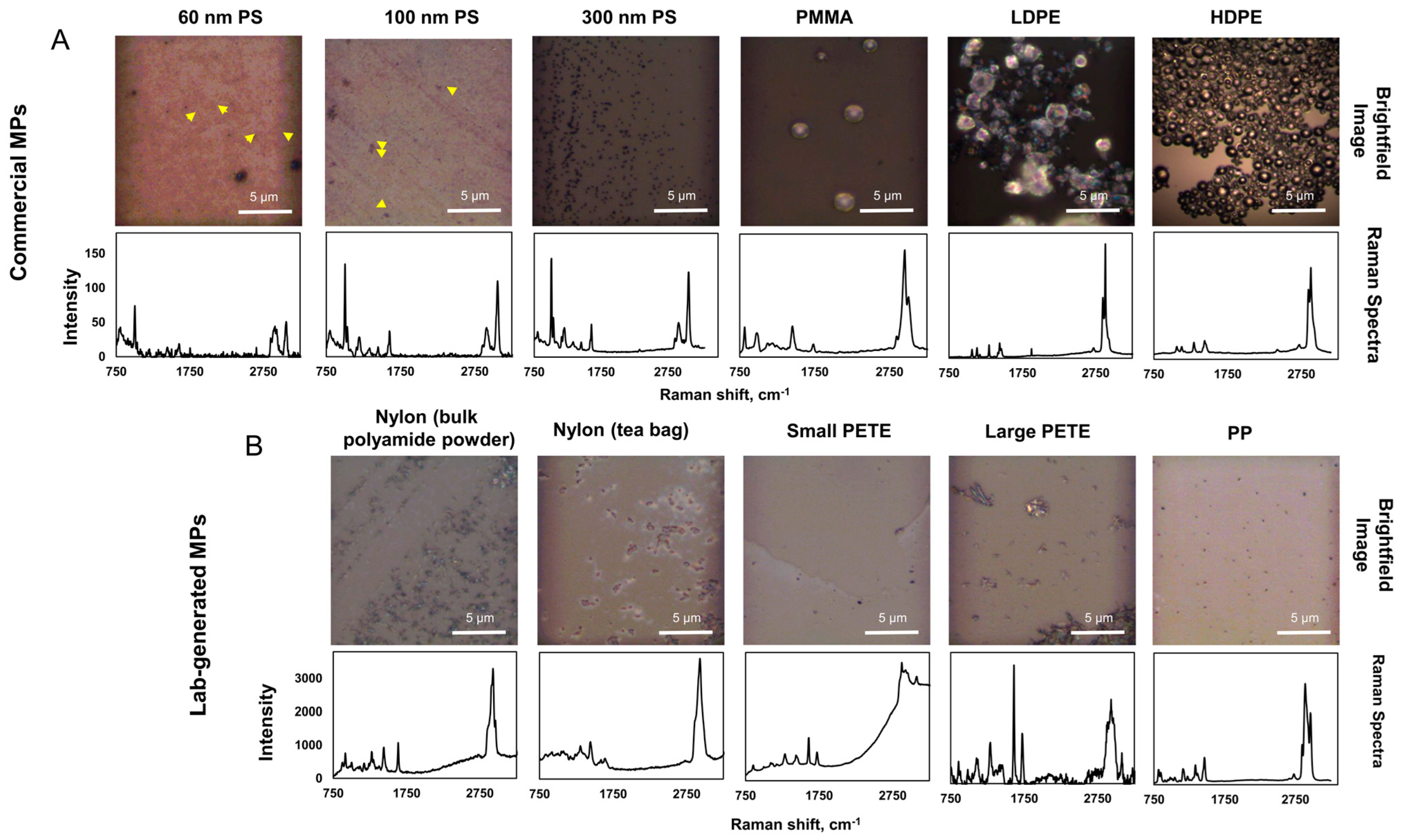
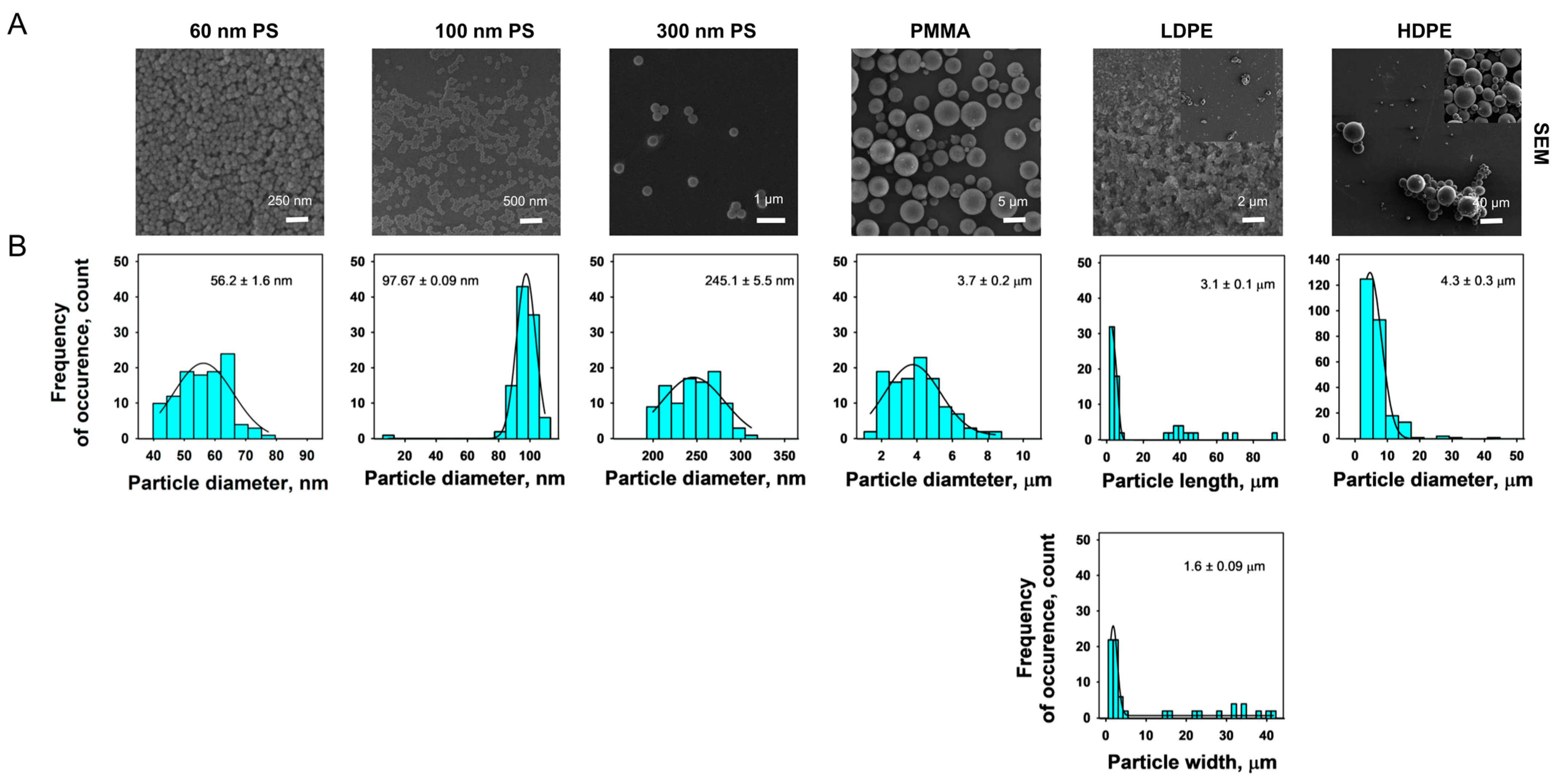
3.1. MPs Characterization
3.1.1. Lab Production of MPs
3.1.2. Chemical Identity of MPs
3.1.3. SEM
3.1.4. DLS and ELS Analysis of MPs
3.1.5. Endotoxin Assessment Results
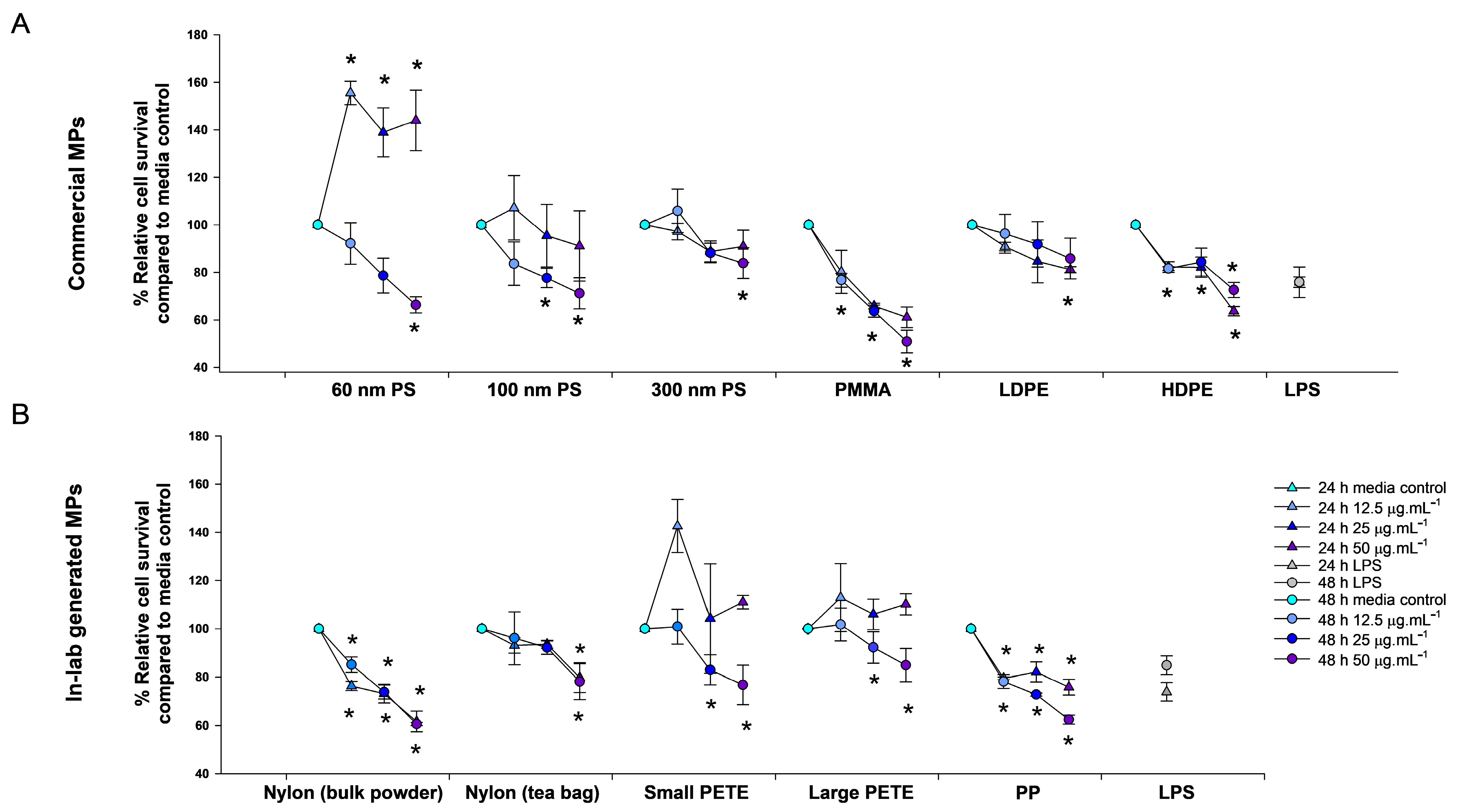
3.2. Toxicity Assessment of MPs
3.2.1. Relative Cell Survival
Commercial MPs
Lab-Generated MPs
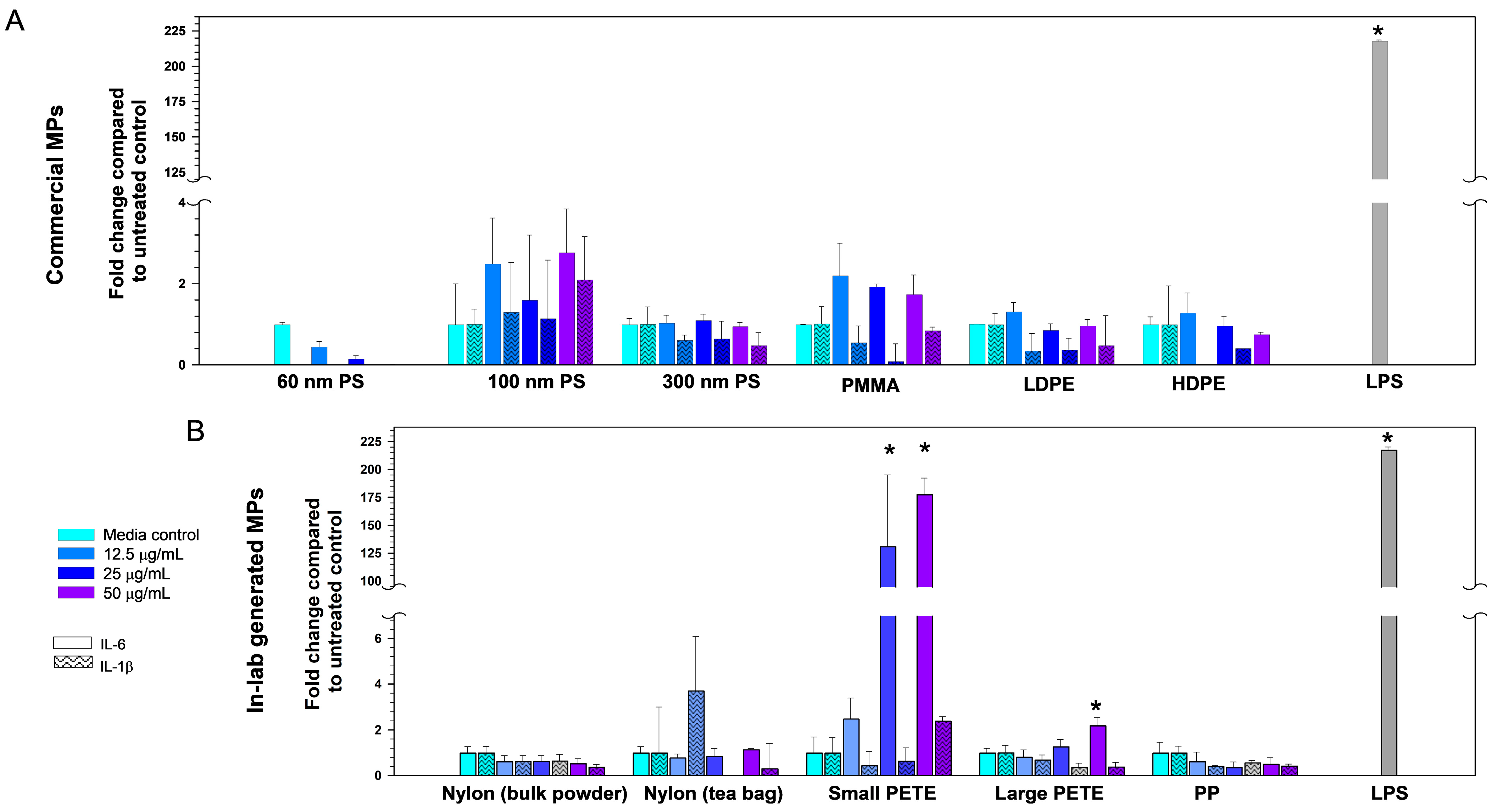
3.2.2. Pro-Inflammatory Cytokines Expression
3.2.3. MN Induction
3.2.4. Gene Expression Analysis
3.3. Cellular Uptake of MPs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wright, S.L.; Thompson, R.C.; Galloway, T.S. The physical impacts of microplastics on marine organisms: A review. Environ. Pollut. 2013, 178, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Rochman, C.M.; Cook, A.; Koelmans, A.A. Plastic debris and policy: Using current scientific understanding to invoke positive change. Environ. Toxicol. Chem. 2016, 35, 1617–1626. [Google Scholar] [CrossRef] [PubMed]
- Gregory, M.R. Plastic ‘scrubbers’ in hand cleansers: A further (and minor) source for marine pollution identified. Mar. Pollut. Bull. 1996, 32, 867–871. [Google Scholar] [CrossRef]
- Gewert, B.; Plassmann, M.M.; MacLeod, M. Pathways for degradation of plastic polymers floating in the marine environment. Environ. Sci. Process. Impacts 2015, 17, 1513–1521. [Google Scholar] [CrossRef] [PubMed]
- Cao, G.; Cai, Z. Getting Health Hazards of Inhaled Nano/Microplastics into Focus: Expectations and Challenges. Environ. Sci. Technol. 2023, 57, 3461–3463. [Google Scholar] [CrossRef] [PubMed]
- Kelly, F.J.; Fussell, J.C. Toxicity of airborne particles—Established evidence, knowledge gaps and emerging areas of importance. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2020, 378, 20190322. [Google Scholar] [CrossRef] [PubMed]
- Alqahtani, S.; Alqahtani, S.; Saquib, Q.; Mohiddin, F. Toxicological impact of microplastics and nanoplastics on humans: Understanding the mechanistic aspect of the interaction. Front. Toxicol. 2023, 5, 1193386. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Huang, W.; Fang, M.; Liao, Z.; Wang, Y.; Xu, L.; Mu, Q.; Shi, C.; Lu, C.; Deng, H.; et al. Airborne Microplastic Concentrations in Five Megacities of Northern and Southeast China. Environ. Sci. Technol. 2021, 55, 12871–12881. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Wang, J.; Peng, J.; Tan, Z.; Zhan, Z.; Tan, X.; Chen, Q. Characteristic of microplastics in the atmospheric fallout from Dongguan city, China: Preliminary research and first evidence. Environ. Sci. Pollut. Res. 2017, 24, 24928–24935. [Google Scholar] [CrossRef]
- Dris, R.; Gasperi, J.; Rocher, V.; Saad, M.; Renault, N.; Tassin, B. Microplastic contamination in an urban area: A case study in Greater Paris. Environ. Chem. 2015, 12, 592–599. [Google Scholar] [CrossRef]
- Liao, Z.; Ji, X.; Ma, Y.; Lv, B.; Huang, W.; Zhu, X.; Fang, M.; Wang, Q.; Wang, X.; Dahlgren, R.; et al. Airborne microplastics in indoor and outdoor environments of a coastal city in Eastern China. J. Hazard. Mater. 2021, 417, 126007. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Wang, X.; Fang, T.; Xu, P.; Zhu, L.; Li, D. Source and Potential Risk Assessment of Suspended Atmospheric Microplastics in Shanghai. Sci. Total. Environ. 2019, 675, 462. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Shao, L.; Li, Y.; Jones, T.; Cao, Y.; Yang, C.X.; Zhang, M.; Santosh, M.; Feng, X.; BéruBé, K. Microplastic atmospheric dustfall pollution in urban environment: Evidence from the types, distribution, and probable sources in Beijing, China. Sci. Total Environ. 2022, 838, 155989. [Google Scholar] [CrossRef] [PubMed]
- Dris, R.; Gasperi, J.; Mirande, C.; Mandin, C.; Guerrouache, M.; Langlois, V.; Tassin, B. A first overview of textile fibers, including microplastics, in indoor and outdoor environments. Environ. Pollut. 2017, 221, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Dris, R.; Gasperi, J.; Saad, M.; Mirande, C.; Tassin, B. Synthetic fibers in atmospheric fallout: A source of microplastics in the environment? Mar. Pollut. Bull. 2016, 104, 290–293. [Google Scholar] [CrossRef]
- Abbasi, S.; Keshavarzi, B.; Moore, F.; Turner, A.; Kelly, F.J.; Dominguez, A.O.; Jaafarzadeh, N. Distribution and potential health impacts of microplastics and microrubbers in air and street dusts from Asaluyeh County, Iran. Environ. Pollut. 2019, 244, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.; Fischer, E.K. Microplastic abundance in atmospheric deposition within the Metropolitan area of Hamburg, Germany. Sci. Total Environ. 2019, 685, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Rahman, L.; Mallach, G.; Kulka, R.; Halappanavar, S. Microplastics and nanoplastics science: Collecting and characterizing airborne microplastics in fine particulate matter. Nanotoxicology 2021, 15, 1253–1278. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, M.; Mützel, S.; Primpke, S.; Tekman, M.B.; Trachsel, J.; Gerdts, G. White and wonderful? Microplastics prevail in snow from the Alps to the Arctic. Sci. Adv. 2019, 5, eaax1157. [Google Scholar] [CrossRef] [PubMed]
- Karbalaei, S.; Hanachi, P.; Walker, T.R.; Cole, M. Occurrence, sources, human health impacts and mitigation of microplastic pollution. Environ. Sci. Pollut. Res. 2018, 25, 36046–36063. [Google Scholar] [CrossRef]
- Cox, K.D.; Covernton, G.A.; Davies, H.L.; Dower, J.F.; Juanes, F.; Dudas, S.E. Human Consumption of Microplastics. Environ. Sci. Technol. 2019, 53, 7068–7074. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, S.; Duan, Z.; Wang, L. Pulmonary toxicology assessment of polyethylene terephthalate nanoplastic particles in vitro. Environ. Int. 2022, 162, 107177. [Google Scholar] [CrossRef] [PubMed]
- Ambrosini, R.; Azzoni, R.S.; Pittino, F.; Diolaiuti, G.; Franzetti, A.; Parolini, M. First evidence of microplastic contamination in the supraglacial debris of an alpine glacier. Environ. Pollut. 2019, 253, 297–301. [Google Scholar] [CrossRef] [PubMed]
- Revel, M.; Châtel, A.; Mouneyrac, C. Micro (nano) plastics: A threat to human health? Curr. Opin. Environ. Sci. Health 2018, 1, 17–23. [Google Scholar] [CrossRef]
- Rytting, E.; Nguyen, J.; Wang, X.; Kissel, T. Biodegradable polymeric nanocarriers for pulmonary drug delivery. Expert Opin. Drug Deliv. 2008, 5, 629–639. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Huang, X.; Bi, R.; Guo, Q.; Yu, X.; Zeng, Q.; Huang, Z.; Liu, T.; Wu, H.; Chen, Y.; et al. Detection and Analysis of Microplastics in Human Sputum. Environ. Sci. Technol. 2022, 56, 2476–2486. [Google Scholar] [CrossRef] [PubMed]
- Baeza-Martínez, C.; Olmos, S.; González-Pleiter, M.; López-Castellanos, J.; García-Pachón, E.; Masiá-Canuto, M.; Hernández-Blasco, L.; Bayo, J. First evidence of microplastics isolated in European citizens’ lower airway. J. Hazard. Mater. 2022, 438, 129439. [Google Scholar] [CrossRef]
- Jenner, L.C.; Rotchell, J.M.; Bennett, R.T.; Cowen, M.; Tentzeris, V.; Sadofsky, L.R. Detection of microplastics in human lung tissue using mFTIR spectroscopy. Sci. Total Environ. 2022, 831, 154907. [Google Scholar] [CrossRef] [PubMed]
- Law, B.D.; Bunn, W.B.; Hesterberg, T.W. Solubility of polymeric organic fibers and manmade vitreous fibers in gambles solution. Inhal. Toxicol. 1990, 2, 321–339. [Google Scholar] [CrossRef]
- Gasperi, J.; Wright, S.L.; Dris, R.; Collard, F.; Mandin, C.; Guerrouache, M.; Langlois, V.; Kelly, F.J.; Tassin, B. Microplastics in air: Are we breathing it in? Curr. Opin. Environ. Sci. Health 2018, 1, 1–5. [Google Scholar] [CrossRef]
- Marfella, R.; Prattichizzo, F.; Sardu, C.; Fulgenzi, G.; Graciotti, L.; Spadoni, T.; D’Onofrio, N.; Scisciola, L.; La Grotta, R.; Frigé, C.; et al. Microplastics and Nanoplastics in Atheromas and Cardiovascular Events. N. Engl. J. Med. 2024, 390, 900–910. [Google Scholar] [CrossRef]
- Lougheed, M.D.; Roos, J.O.; Waddell, W.R.; Munt, P.W. Desquamative interstitial pneumonitis and diffuse alveolar damage in textile workers: Potential role of mycotoxins. Chest 1995, 108, 1196–1200. [Google Scholar] [CrossRef]
- Kern, D.G.; Crausman, R.S.; Durand, K.T.; Nayer, A.; Kuhn, C., III. Flock Worker’s Lung: Chronic Interstitial Lung Disease in the Nylon Flocking Industry. Ann. Intern. Med. 1998, 129, 261–272. [Google Scholar] [CrossRef]
- Wan, Z.; Wang, C.; Zhou, J.; Shen, M.; Wang, X.; Fu, Z.; Jin, Y. Effects of polystyrene microplastics on the composition of the microbiome and metabolism in larval zebrafish. Chemosphere 2019, 217, 646–658. [Google Scholar] [CrossRef]
- Chen, L.; Hu, C.; Lai, N.L.-S.; Zhang, W.; Hua, J.; Lam, P.K.; Lam, J.C.; Zhou, B. Acute exposure to PBDEs at an environmentally realistic concentration causes abrupt changes in the gut microbiota and host health of zebrafish. Environ. Pollut. 2018, 240, 17–26. [Google Scholar] [CrossRef]
- Qiao, R.; Deng, Y.; Zhang, S.; Wolosker, M.B.; Zhu, Q.; Ren, H.; Zhang, Y. Accumulation of different shapes of microplastics initiates intestinal injury and gut microbiota dysbiosis in the gut of zebrafish. Chemosphere 2019, 236, 124334. [Google Scholar] [CrossRef]
- Auguste, M.; Lasa, A.; Balbi, T.; Pallavicini, A.; Vezzulli, L.; Canesi, L. Impact of nanoplastics on hemolymph immune parameters and microbiota composition in Mytilus galloprovincialis. Mar. Environ. Res. 2020, 159, 105017. [Google Scholar] [CrossRef]
- Li, X.; Zhang, T.; Lv, W.; Wang, H.; Chen, H.; Xu, Q.; Cai, H.; Dai, J. Intratracheal administration of polystyrene microplastics induces pulmonary fibrosis by activating oxidative stress and Wnt/β-catenin signaling pathway in mice. Ecotoxicol. Environ. Saf. 2022, 232, 113238. [Google Scholar] [CrossRef]
- Jin, Y.X.; Lu, L.; Tu, W.Q.; Luo, T.; Fu, Z.W. Impacts of Polystyrene Microplastic on the Gut Barrier, Microbiota and Metabolism of Mice. Sci. Total. Environ. 2019, 649, 308. [Google Scholar] [CrossRef]
- Yang, S.; Zhang, T.; Ge, Y.; Yin, L.; Pu, Y.; Liang, G. Inhalation exposure to polystyrene nanoplastics induces chronic obstructive pulmonary disease-like lung injury in mice through multi-dimensional assessment. Environ. Pollut. 2024, 347, 123633. [Google Scholar] [CrossRef]
- Danso, I.K.; Woo, J.H.; Lee, K. Pulmonary Toxicity of Polystyrene, Polypropylene, and Polyvinyl Chloride Microplastics in Mice. Molecules 2022, 27, 7926. [Google Scholar] [CrossRef]
- Lim, D.; Jeong, J.; Song, K.S.; Sung, J.H.; Oh, S.M.; Choi, J. Inhalation toxicity of polystyrene micro(nano)plastics using modified OECD TG 412. Chemosphere 2021, 262, 128330. [Google Scholar] [CrossRef]
- Liu, S.; Li, H.; Wang, J.; Wu, B.; Guo, X. Polystyrene microplastics aggravate inflammatory damage in mice with intestinal immune imbalance. Sci. Total. Environ. 2022, 833, 155198. [Google Scholar] [CrossRef]
- Lee, A.G.; Kang, S.; Yoon, H.J.; Im, S.; Oh, S.J.; Pak, Y.K. Polystyrene Microplastics Exacerbate Systemic Inflammation in High-Fat Diet-Induced Obesity. Int. J. Mol. Sci. 2023, 24, 12421. [Google Scholar] [CrossRef]
- Lu, L.; Wan, Z.; Luo, T.; Fu, Z.; Jin, Y. Polystyrene microplastics induce gut microbiota dysbiosis and hepatic lipid metabolism disorder in mice. Sci. Total Environ. 2018, 631–632, 449–458. [Google Scholar] [CrossRef]
- Li, Z.; Zhu, S.; Liu, Q.; Wei, J.; Jin, Y.; Wang, X.; Zhang, L. Polystyrene microplastics cause cardiac fibrosis by activating Wnt/β-catenin signaling pathway and promoting cardiomyocyte apoptosis in rats. Environ. Pollut. 2020, 265, 115025. [Google Scholar] [CrossRef]
- Shi, Q.; Tang, J.; Wang, L.; Liu, R.; Giesy, J.P. Combined cytotoxicity of polystyrene nanoplastics and phthalate esters on human lung epithelial A549 cells and its mechanism. Ecotoxicol. Environ. Saf. 2021, 213, 112041. [Google Scholar] [CrossRef]
- Dong, C.D.; Chen, C.W.; Chen, Y.C.; Chen, H.H.; Lee, J.S.; Lin, C.H. Polystyrene microplastic particles: In vitro pulmonary toxicity assessment. J. Hazard. Mater. 2020, 385, 121575. [Google Scholar] [CrossRef]
- Rubio, L.; Barguilla, I.; Domenech, J.; Marcos, R.; Hernáíndez, A. Biological effects, including oxidative stress and genotoxic damage, of polystyrene nanoparticles in different human hematopoietic cell lines. J. Hazard. Mater. 2020, 398, 122900. [Google Scholar] [CrossRef]
- Poma, A.; Vecchiotti, G.; Colafarina, S.; Zarivi, O.; Aloisi, M.; Arrizza, L.; Chichiriccò, G.; Di Carlo, P. In Vitro Genotoxicity of Polystyrene Nanoparticles on the Human Fibroblast Hs27 Cell Line. Nanomaterials 2019, 9, 1299. [Google Scholar] [CrossRef]
- Schirinzi, G.F.; Pérez-Pomeda, I.; Sanchís, J.; Rossini, C.; Farré, M.; Barceló, D. Cytotoxic effects of commonly used nanomaterials and microplastics on cerebral and epithelial human cells. Environ. Res. 2017, 159, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Choi, D.; Han, S.; Choi, J.; Hong, J. An assessment of the toxicity of polypropylene microplastics in human derived cells. Sci. Total Environ. 2019, 684, 657–669. [Google Scholar] [CrossRef]
- Jeon, S.; Lee, D.K.; Jeong, J.; Yang, S.I.; Kim, J.S.; Kim, J.; Cho, W.S. The reactive oxygen species as pathogenic factors of fragmented microplastics to macrophages. Environ. Pollut. 2021, 281, 117006. [Google Scholar] [CrossRef] [PubMed]
- Busch, M.; Bredeck, G.; Waag, F.; Rahimi, K.; Ramachandran, H.; Bessel, T.; Barcikowski, S.; Herrmann, A.; Rossi, A.; Schins, R.P. Assessing the NLRP3 inflammasome activating potential of a large panel of micro-and nanoplastics in THP-1 cells. Biomolecules 2022, 12, 1095. [Google Scholar] [CrossRef] [PubMed]
- Koner, S.; Florance, I.; Mukherjee, A.; Chandrasekaran, N. Cellular response of THP-1 macrophages to polystyrene microplastics exposure. Toxicology 2023, 483, 153385. [Google Scholar] [CrossRef] [PubMed]
- Paget, V.; Dekali, S.; Kortulewski, T.; Grall, R.; Gamez, C.; Blazy, K.; Aguerre-Chariol, O.; Chevillard, S.; Braun, A.; Rat, P.; et al. Specific uptake and genotoxicity induced by polystyrene nanobeads with distinct surface chemistry on human lung epithelial cells and macrophages. PLoS ONE 2015, 10, e0123297. [Google Scholar] [CrossRef] [PubMed]
- Kokalj, A.J.; Hartmann, N.B.; Drobne, D.; Potthoff, A.; Kühnel, D. Quality of nanoplastics and microplastics ecotoxicity studies: Refining quality criteria for nanomaterial studies. J. Hazard. Mater. 2021, 415, 125751. [Google Scholar] [CrossRef] [PubMed]
- Halappanavar, S.; Mallach, G. Adverse outcome pathways and in-ávitro toxicology strategies for microplastics hazard testing. Curr. Opin. Toxicol. 2021, 28, 52–61. [Google Scholar] [CrossRef]
- Barosova, H.; Maione, A.G.; Septiadi, D.; Sharma, M.; Haeni, L.; Balog, S.; O’Connell, O.; Jackson, G.R.; Brown, D.; Clippinger, A.J.; et al. Use of EpiAlveolar Lung Model to Predict Fibrotic Potential of Multiwalled Carbon Nanotubes. ACS Nano 2020, 14, 3941–3956. [Google Scholar] [CrossRef]
- Munno, K.; De Frond, H.; O’Donnell, B.; Rochman, C.M. Increasing the accessibility for characterizing microplastics: Introducing new application-based and spectral libraries of plastic particles (SLoPP and SLoPP-E). Anal. Chem. 2020, 92, 2443–2451. [Google Scholar] [CrossRef]
- KnowItAll Spectroscopy Edition Software; John Wiley & Sons, Inc.: Hoboken, NY, USA, 2022.
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with ImageJ. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
- Solorio-Rodriguez, S.A.; Wu, D.; Boyadzhiev, A.; Christ, C.; Williams, A.; Halappanavar, S. A Systematic Genotoxicity Assessment of a Suite of Metal Oxide Nanoparticles Reveals Their DNA Damaging and Clastogenic Potential. Nanomaterials 2024, 14, 743. [Google Scholar] [CrossRef]
- Halappanavar, S.; Nikota, J.; Wu, D.; Williams, A.; Yauk, C.L.; Stampfli, M. IL-1 receptor regulates microRNA-135b expression in a negative feedback mechanism during cigarette smoke-induced inflammation. J. Immunol. 2013, 190, 3679–3686. [Google Scholar] [CrossRef]
- Husain, M.; Saber, A.T.; Guo, C.; Jacobsen, N.R.; Jensen, K.A.; Yauk, C.L.; Williams, A.; Vogel, U.; Wallin, H.; Halappanavar, S. Pulmonary instillation of low doses of titanium dioxide nanoparticles in mice leads to particle retention and gene expression changes in the absence of inflammation. Toxicol. Appl. Pharmacol. 2013, 269, 250–262. [Google Scholar] [CrossRef]
- OECD. Test No. 487: In Vitro Mammalian Cell Micronucleus Test, OECD Guidelines for the Testing of Chemicals, Section 4; OECD Publishing: Paris, France, 2023. [Google Scholar]
- White, P.A.; Douglas, G.R.; Gingerich, J.; Parfett, C.; Shwed, P.; Seligy, V.; Soper, L.; Berndt, L.; Bayley, J.; Wagner, S.; et al. Development and characterization of a stable epithelial cell line from Muta Mouse lung. Environ. Mol. Mutagen. 2003, 42, 166–184. [Google Scholar] [CrossRef]
- Fortin, A.M.; Long, A.S.; Williams, A.; Meier, M.J.; Cox, J.; Pinsonnault, C.; Yauk, C.L.; White, P.A. Application of a new approach methodology (NAM)-based strategy for genotoxicity assessment of data-poor compounds. Front. Toxicol. 2023, 5, 1098432. [Google Scholar] [CrossRef]
- Kerr, M.K. Design considerations for efficient and effective microarray studies. Biometrics 2003, 59, 822–828. [Google Scholar] [CrossRef]
- Kerr, M.K.; Churchill, G.A. Statistical design and the analysis of gene expression microarray data. Genet. Res. 2001, 77, 123–128. [Google Scholar] [CrossRef]
- Boyadzhiev, A.; Wu, D.; Avramescu, M.L.; Williams, A.; Rasmussen, P.; Halappanavar, S. Toxicity of Metal Oxide Nanoparticles: Looking through the Lens of Toxicogenomics. Int. J. Mol. Sci. 2024, 25, 529. [Google Scholar] [CrossRef]
- Cleveland, W.S. Robust locally weighted regression and smoothing scatterplots. J. Am. Stat. Assoc. 1979, 74, 829–836. [Google Scholar] [CrossRef]
- Wu, H.; Kerr, M.K.; Cui, X.; Churchill, G.A. MAANOVA: A software package for the analysis of spotted cDNA microarray experiments. In The Analysis of Gene Expression Data: Methods and Software; Springer: Berlin/Heidelberg, Germany, 2003; pp. 313–341. [Google Scholar]
- Cui, X.; Hwang, J.G.; Qiu, J.; Blades, N.J.; Churchill, G.A. Improved statistical tests for differential gene expression by shrinking variance components estimates. Biostatistics 2005, 6, 59–75. [Google Scholar] [CrossRef]
- Zar, J. Biostatistical Analysis; Upper Saddle River: Bergen, NJ, USA, 1999. [Google Scholar]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Hochberg, Y.; Benjamini, Y. More powerful procedures for multiple significance testing. Stat. Med. 1990, 9, 811–818. [Google Scholar] [CrossRef]
- Cohen, J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Hernandez, L.M.; Xu, E.G.; Larsson, H.C.E.; Tahara, R.; Maisuria, V.B.; Tufenkji, N. Plastic Teabags Release Billions of Microparticles and Nanoparticles into Tea. Environ. Sci. Technol. 2019, 53, 12300–12310. [Google Scholar] [CrossRef]
- Nisticò, R. Polyethylene terephthalate (PET) in the packaging industry. Polym. Test. 2020, 90, 106707. [Google Scholar] [CrossRef]
- Alsabri, A.; Tahir, F.; Al-Ghamdi, S.G. Environmental impacts of polypropylene (PP) production and prospects of its recycling in the GCC region. Mater. Today Proc. 2022, 56, 2245–2251. [Google Scholar] [CrossRef]
- Kihara, S.; Ashenden, A.; Kaur, M.; Glasson, J.; Ghosh, S.; van der Heijden, N.; Brooks, A.E.S.; Mata, J.P.; Holt, S.; Domigan, L.J.; et al. Cellular interactions with polystyrene nanoplastics—The role of particle size and protein corona. Biointerphases 2021, 16, 041001. [Google Scholar] [CrossRef]
- Maddah, H.A. Polypropylene as a promising plastic: A review. Am. J. Polym. Sci 2016, 6, 1–11. [Google Scholar] [CrossRef]
- Park, J.H.; Hong, S.; Kim, O.H.; Kim, C.H.; Kim, J.; Kim, J.W.; Hong, S.; Lee, H.J. Polypropylene microplastics promote metastatic features in human breast cancer. Sci. Rep. 2023, 13, 6252. [Google Scholar] [CrossRef]
- Ji, Y.; Wang, C.; Wang, Y.; Fu, L.; Man, M.; Chen, L. Realistic polyethylene terephthalate nanoplastics and the size- and surface coating-dependent toxicological impacts on zebrafish embryos. Environ. Sci. Nano 2020, 7, 2313–2324. [Google Scholar] [CrossRef]
- Alzaben, M.; Burve, R.; Loeschner, K.; Møller, P.; Roursgaard, M. Nanoplastics from ground polyethylene terephthalate food containers: Genotoxicity in human lung epithelial A549 cells. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2023, 892, 503705. [Google Scholar] [CrossRef]
- Solomon, Y.; Malkamu, B.; Berhan, A.; Eyayu, T.; Almaw, A.; Legese, B.; Woldu, B. Peripheral blood eosinophilia in adult asthmatic patients and its association with the severity of asthma. BMC Pulm. Med. 2023, 23, 96. [Google Scholar] [CrossRef]
- Sparrer, K.M.; Gack, M.U. Intracellular detection of viral nucleic acids. Curr. Opin. Microbiol. 2015, 26, 1–9. [Google Scholar] [CrossRef]
- Takeuchi, O.; Akira, S. Pattern Recognition Receptors and Inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef]
- Alijagic, A.; Hedbrant, A.; Persson, A.; Larsson, M.; Engwall, M.; Särndahl, E. NLRP3 inflammasome as a sensor of micro- and nanoplastics immunotoxicity. Mini Rev. Front. Immunol. 2023, 14, 1178434. [Google Scholar] [CrossRef]
- Lunov, O.; Syrovets, T.; Loos, C.; Nienhaus, G.U.; Mailänder, V.; Landfester, K.; Rouis, M.; Simmet, T. Amino-functionalized polystyrene nanoparticles activate the NLRP3 inflammasome in human macrophages. ACS Nano 2011, 5, 9648–9657. [Google Scholar] [CrossRef]
- Wu, Y.; Yao, Y.; Bai, H.; Shimizu, K.; Li, R.; Zhang, C. Investigation of pulmonary toxicity evaluation on mice exposed to polystyrene nanoplastics: The potential protective role of the antioxidant N-acetylcysteine. Sci. Total Environ. 2023, 855, 158851. [Google Scholar] [CrossRef]
- Chi, Q.; Xu, T.; He, Y.; Li, Z.; Tang, X.; Fan, X.; Li, S. Polystyrene nanoparticle exposure supports ROS-NLRP3 axis-dependent DNA-NET to promote liver inflammation. J. Hazard. Mater. 2022, 439, 129502. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Wang, X.; Liu, Q.; Zhou, N.; Zhu, S.; Li, Z.; Li, X.; Yao, J.; Zhang, L. The impact of polystyrene microplastics on cardiomyocytes pyroptosis through NLRP3/Caspase-1 signaling pathway and oxidative stress in Wistar rats. Environ. Toxicol. 2021, 36, 935–944. [Google Scholar] [CrossRef]
- Hou, J.; Lei, Z.; Cui, L.; Hou, Y.; Yang, L.; An, R.; Wang, Q.; Li, S.; Zhang, H.; Zhang, L. Polystyrene microplastics lead to pyroptosis and apoptosis of ovarian granulosa cells via NLRP3/Caspase-1 signaling pathway in rats. Ecotoxicol. Env. Saf. 2021, 212, 112012. [Google Scholar] [CrossRef] [PubMed]
- Shirasuna, K.; Karasawa, T.; Takahashi, M. Exogenous nanoparticles and endogenous crystalline molecules as danger signals for the NLRP3 inflammasomes. J. Cell. Physiol. 2019, 234, 5436–5450. [Google Scholar] [CrossRef] [PubMed]
- Yazdi, A.S.; Guarda, G.; Riteau, N.; Drexler, S.K.; Tardivel, A.; Couillin, I.; Tschopp, J. Nanoparticles activate the NLR pyrin domain containing 3 (Nlrp3) inflammasome and cause pulmonary inflammation through release of IL-1α and IL-1β. Proc. Natl. Acad. Sci. USA 2010, 107, 19449–19454. [Google Scholar] [CrossRef]
- Hornung, V.; Bauernfeind, F.; Halle, A.; Samstad, E.O.; Kono, H.; Rock, K.L.; Fitzgerald, K.A.; Latz, E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 2008, 9, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; McLeland, C.B.; Potter, T.M.; Stern, S.T.; Adiseshaiah, P.P. Assessing NLRP3 inflammasome activation by nanoparticles. In Characterization of Nanoparticles Intended for Drug Delivery; Humana Press: New York, NY, USA, 2018; pp. 135–147. [Google Scholar] [CrossRef]
- Baron, L.; Gombault, A.; Fanny, M.; Villeret, B.; Savigny, F.; Guillou, N.; Panek, C.; Le Bert, M.; Lagente, V.; Rassendren, F.; et al. The NLRP3 inflammasome is activated by nanoparticles through ATP, ADP and adenosine. Cell Death. Dis. 2015, 6, e1629. [Google Scholar] [CrossRef]
- Halappanavar, S.; Sharma, M.; Solorio-Rodriguez, S.; Wallin, H.; Vogel, U.; Sullivan, K.; Clippinger, A.J. Substance interaction with the pulmonary resident cell membrane components leading to pulmonary fibrosis. In OECD Series on Adverse Outcome Pathways; OECD Publishing: Paris, France, 2023; Volume 33. [Google Scholar] [CrossRef]
- Halappanavar, S.; Nymark, P.; Krug, H.F.; Clift, M.J.D.; Rothen-Rutishauser, B.; Vogel, U. Non-Animal Strategies for Toxicity Assessment of Nanoscale Materials: Role of Adverse Outcome Pathways in the Selection of Endpoints. Small 2021, 17, 2007628. [Google Scholar] [CrossRef] [PubMed]
- Bølling, A.K.; Sripada, K.; Becher, R.; Bekö, G. Phthalate exposure and allergic diseases: Review of epidemiological and experimental evidence. Environ. Int. 2020, 139, 105706. [Google Scholar] [CrossRef] [PubMed]
- Campanale, C.; Savino, I.; Pojar, I.; Massarelli, C.; Uricchio, V.F. A Practical Overview of Methodologies for Sampling and Analysis of Microplastics in Riverine Environments. Sustainability 2020, 12, 6755. [Google Scholar] [CrossRef]
- Ma, L.; Wu, Z.; Lu, Z.; Yan, L.; Dong, X.; Dai, Z.; Sun, R.; Hong, P.; Zhou, C.; Li, C. Differences in toxicity induced by the various polymer types of nanoplastics on HepG2 cells. Sci. Total Environ. 2024, 918, 170664. [Google Scholar] [CrossRef]
- Martínez, A.; Barbosa, A. Chemical reactivity theory to analyze possible toxicity of microplastics: Polyethylene and polyester as examples. PLoS ONE 2024, 19, e0285515. [Google Scholar] [CrossRef]
- Jakubowska, M.; Białowąs, M.; Stankevičiūtė, M.; Chomiczewska, A.; Pažusienė, J.; Jonko-Sobuś, K.; Hallmann, A.; Urban-Malinga, B. Effects of chronic exposure to microplastics of different polymer types on early life stages of sea trout Salmo trutta. Sci. Total. Environ. 2020, 740, 139922. [Google Scholar] [CrossRef]
- Nymark, P.; Karlsson, H.L.; Halappanavar, S.; Vogel, U. Adverse Outcome Pathway Development for Assessment of Lung Carcinogenicity by Nanoparticles. Front. Toxicol. 2021, 3, 653386. [Google Scholar] [CrossRef] [PubMed]
- Babonaitė, M.; Čepulis, M.; Kazlauskaitė, J.; Lazutka, J.R. Evaluation of In Vitro Genotoxicity of Polystyrene Nanoparticles in Human Peripheral Blood Mononuclear Cells. Toxics 2023, 11, 627. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Chang, Y.; Zhang, T.; Qiao, Y.; Klobučar, G.; Li, M. Toxicological effects of polystyrene microplastics on earthworm (Eisenia fetida). Environ. Pollut. 2020, 259, 113896. [Google Scholar] [CrossRef] [PubMed]
- Panariti, A.; Miserocchi, G.; Rivolta, I. The effect of nanoparticle uptake on cellular behavior: Disrupting or enabling functions? Nanotechnol. Sci. Appl. 2012, 5, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Nagai, H.; Okazaki, Y.; Chew, S.H.; Misawa, N.; Yamashita, Y.; Akatsuka, S.; Ishihara, T.; Yamashita, K.; Yoshikawa, Y.; Yasui, H.; et al. Diameter and rigidity of multiwalled carbon nanotubes are critical factors in mesothelial injury and carcinogenesis. Proc. Natl. Acad. Sci. USA 2011, 108, E1330–E1338. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhang, M.; Zhang, Q.; Jiang, W. Graphene nanosheets damage the lysosomal and mitochondrial membranes and induce the apoptosis of RBL-2H3 cells. Sci. Total Environ. 2020, 734, 139229. [Google Scholar] [CrossRef]
- Zhou, T.; Wu, J.; Liu, Y.; Xu, A. Seawater Accelerated the Aging of Polystyrene and Enhanced Its Toxic Effects on Caenorhabditis elegans. Int. J. Mol. Sci. 2023, 24, 17219. [Google Scholar] [CrossRef]
- Choi, D.; Hwang, J.; Bang, J.; Han, S.; Kim, T.; Oh, Y.; Hwang, Y.; Choi, J.; Hong, J. In vitro toxicity from a physical perspective of polyethylene microplastics based on statistical curvature change analysis. Sci. Total Environ. 2021, 752, 142242. [Google Scholar] [CrossRef]



| MPs | Suspension Medium | Average Hydrodynamic Size, nm | PDI | Zeta Potential, mV |
|---|---|---|---|---|
| 60 nm PS | MilliQ water | 62.25 ± 0.36 | 0.0816 | −39.16 ± 0.73 |
| Cell culture media | 74.75 ± 0.89 | 0.0518 | −15.72 ± 0.91 | |
| 100 nm PS | MilliQ water | 103.88 ± 0.77 | 0.0102 | −46.88 ± 0.91 |
| Cell culture media | 124.52 ± 0.68 | 0.0682 | −14.38 ± 1.47 | |
| 300 nm PS | MilliQ water | 270.90 ± 1.72 | 0.0244 | −50.38 ± 0.44 |
| Cell culture media | 280.82 ± 2.16 | 0.0396 | −14.66 ± 1.44 | |
| PMMA | MilliQ water | 1810.33 ± 2079 | 0.66 ± 0.23 | −49.62 ± 3.48 |
| Cell culture media | 1092.98 ± 737 | 0.78 ± 0.15 | −8.47 ± 0.36 | |
| LDPE | MilliQ water | 315.1 ± 121 | 0.48 ± 0.07 | −39.4 ± 4.14 |
| Cell culture media | 342.34 ± 82.5 | 0.51 ± 0.09 | −12.98 ± 0.68 | |
| HDPE | MilliQ water | 3649.25 ± 1831 | 0.64 ± 0.25 | −47.9 ± 4.23 |
| Cell culture media | 1664.5 ± 300 | 0.48 ± 0.38 | −5.03 ± 0.45 | |
| Nylon from bulk polyamide powder | MilliQ water | 449.75 ± 11.26 | 0.44 ± 0.09 | −31.18 ± 1.38 |
| Cell culture media | 466.84 ± 15.13 | 0.49 ± 0.03 | −11.46 ± 1.15 | |
| Nylon from tea bags | MilliQ water | 698.62 ± 82.53 | 0.59 ± 0.06 | −32.06 ± 0.68 |
| Cell culture media | 223.58 ± 63.2 | 0.78 ± 0.12 | −7.08 ± 0.73 | |
| Small PETE | MilliQ water | 406 ± 5.06 | 0.28 ± 0.03 | −39.3 ± 1.4 |
| Cell culture media | 713.33 ± 27 | 0.53 ± 0.04 | −17.4 ± 1.07 | |
| Large PETE | MilliQ water | 1117.3 ± 383 | 0.61 ± 0.27 | −41.98 ± 3.49 |
| Cell culture media | 731.9 ± 107 | 0.57 ± 0.06 | −15.46 ± 0.47 | |
| PP | MilliQ water | 185.84 ± 44.86 | 0.40 ± 0.08 | −52.28 ± 11.9 |
| Cell culture media | 1809.04 ± 886.34 | 0.92 ± 0.1 | −14.94 ± 0.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© Health Canada 2024. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, L.; Williams, A.; Wu, D.; Halappanavar, S. Polyethylene Terephthalate Microplastics Generated from Disposable Water Bottles Induce Interferon Signaling Pathways in Mouse Lung Epithelial Cells. Nanomaterials 2024, 14, 1287. https://doi.org/10.3390/nano14151287
Rahman L, Williams A, Wu D, Halappanavar S. Polyethylene Terephthalate Microplastics Generated from Disposable Water Bottles Induce Interferon Signaling Pathways in Mouse Lung Epithelial Cells. Nanomaterials. 2024; 14(15):1287. https://doi.org/10.3390/nano14151287
Chicago/Turabian StyleRahman, Luna, Andrew Williams, Dongmei Wu, and Sabina Halappanavar. 2024. "Polyethylene Terephthalate Microplastics Generated from Disposable Water Bottles Induce Interferon Signaling Pathways in Mouse Lung Epithelial Cells" Nanomaterials 14, no. 15: 1287. https://doi.org/10.3390/nano14151287





