The Influence of Silver-Containing Bionanomaterials Based on Humic Ligands on Biofilm Formation in Opportunistic Pathogens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis of Humic Acid Derivatives
2.1.1. Synthesis of Phenol-Enriched Derivatives of HSs Using the Fenton Reaction
2.1.2. Phenol–Formaldehyde Copolycondensation
2.2. Synthesis and Characterization of Silver Nanoparticles
2.3. 13C NMR Spectroscopic Study of Hydroxylated Derivatives
2.4. Preparation of Microbial Cultures
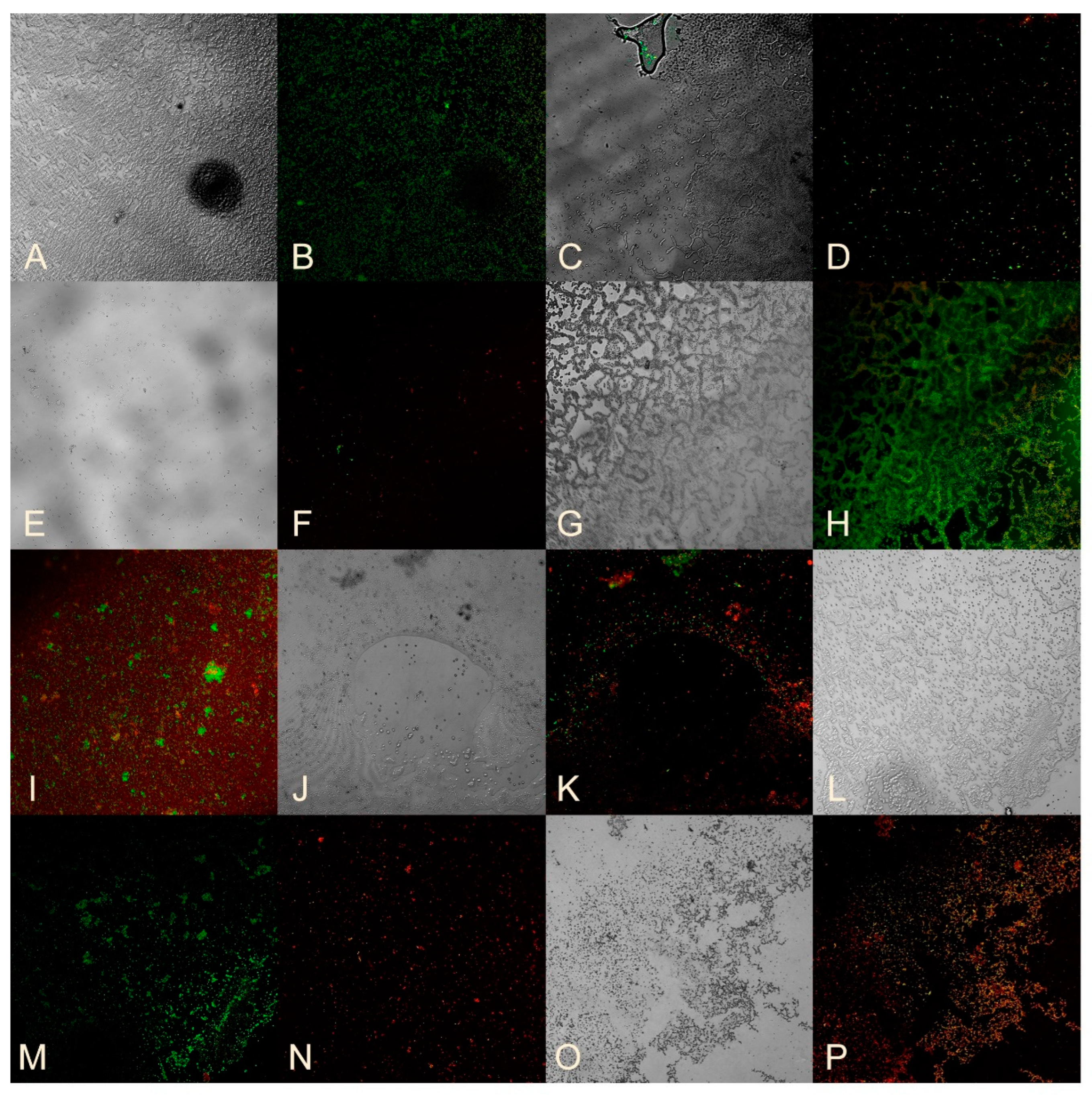
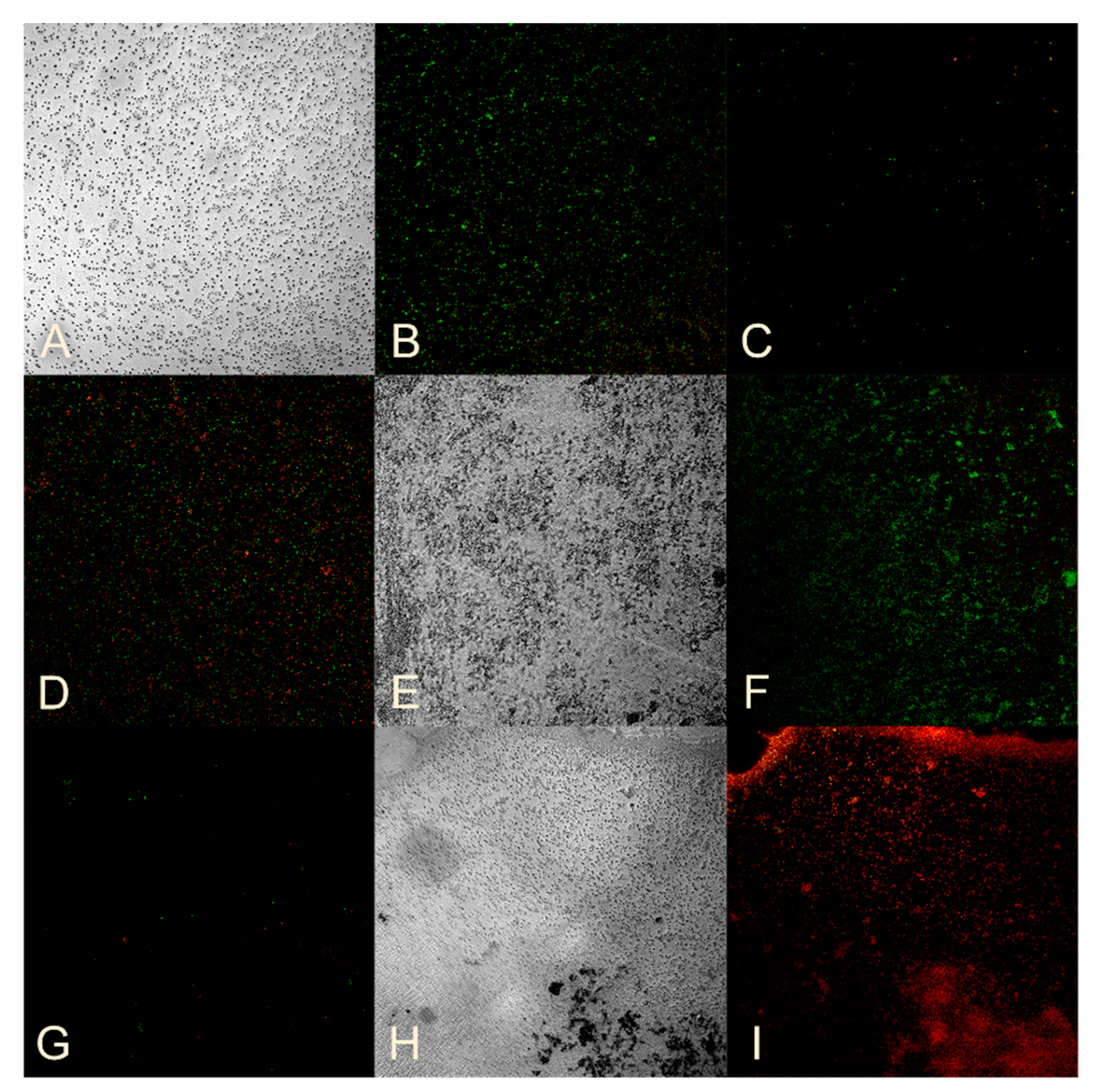
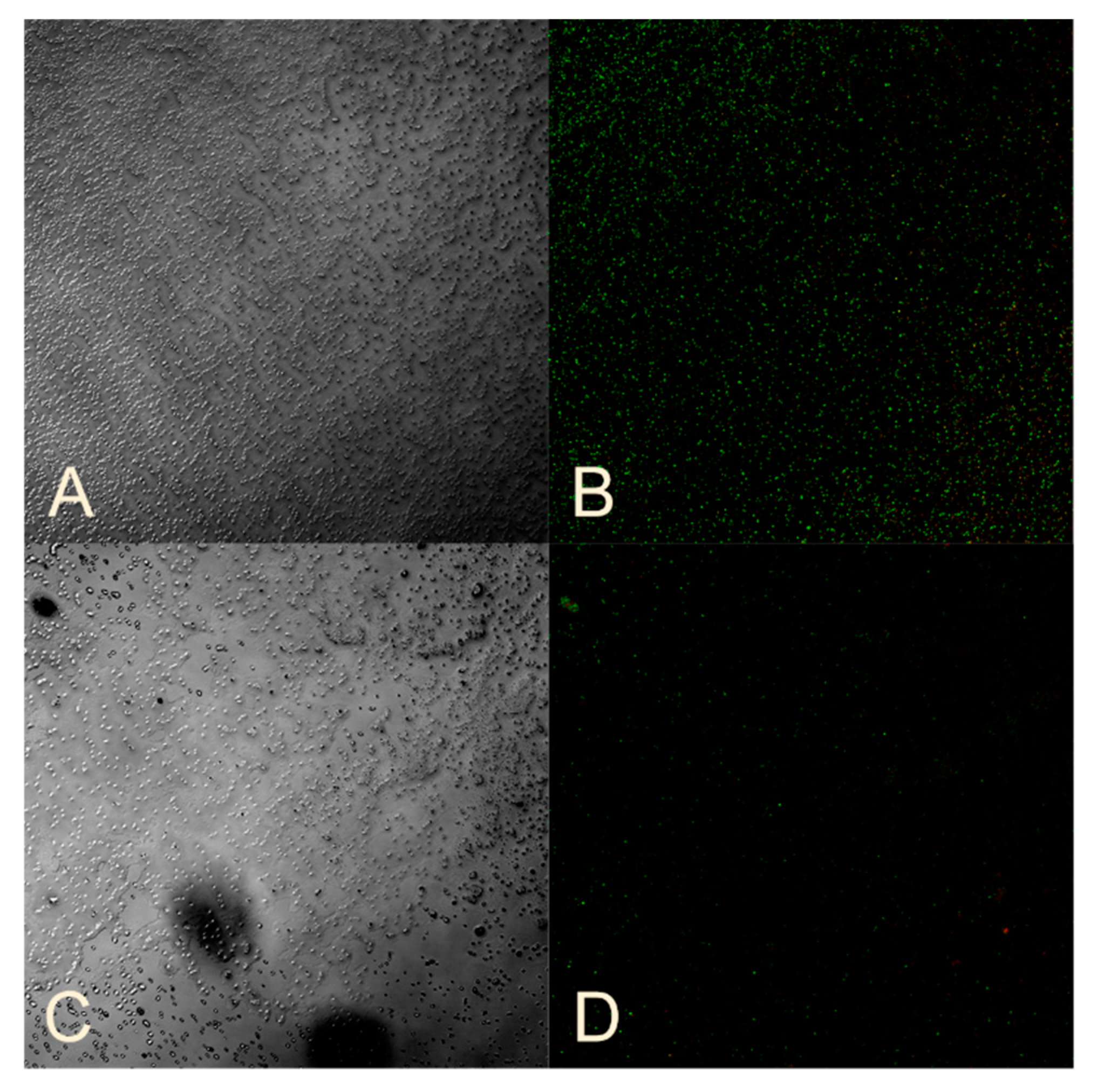
2.5. Studying the Ability of Microorganisms to Form Biofilms
2.6. Determination of the Sensitivity of Microorganisms to Nanocomposites
2.7. Study of Changes in Cell Wall Uptake of Crystal Violet Dye
2.8. Assessment of Bacterial Viability
2.9. Statistical Processing of Results
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nicolaou, K.C.; Rigol, S. A brief history of antibiotics and select advances in their synthesis. J. Antibiot. 2018, 71, 153–184. [Google Scholar] [CrossRef] [PubMed]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.R.M.; Mitra, S.; Emran, T.B.; Dhama, K.; Ripon, M.K.H.; Gajdács, M.; Sahibzada, M.U.K.; et al. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J. Infect. Public Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef] [PubMed]
- Laxminarayan, R.; Matsoso, P.; Pant, S.; Brower, C.; Røttingen, J.A.; Klugman, K.; Davies, S. Access to effective antimicrobials: A worldwide challenge. Lancet 2016, 387, 168–175. [Google Scholar] [CrossRef]
- Danilov, A.I.; Zharkova, L.P. Antimicrobial resistance: Arguments and facts. Clin. Pharmacol. Ther. 2017, 26, 6–9. [Google Scholar]
- Holmes, A.H.; Moore, L.S.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Guerin, P.J.; Piddock, L.J. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Economou, V.; Gousia, P. Agriculture and food animals as a source of antimicrobial-resistant bacteria. Infect. Drug Resist. 2015, 8, 49–61. [Google Scholar] [CrossRef]
- Rousham, E.K.; Unicomb, L.; Islam, M.A. Human, animal and environmental contributors to antibiotic resistance in low-resource settings: Integrating behavioural, epidemiological and One Health approaches. Proc. Biol. Sci. 2018, 285, 20180332. [Google Scholar] [CrossRef]
- Kovalyov, E. The Potential of the Global Agri-Food Sector: Pluralism of Assessments. World Econ. Int. Relat. 2011, 8, 3–14. [Google Scholar]
- Kozlov, R.S.; Golub, A.V. To stop the progress of antimicrobial resistance today means to give a chance of mankind surviving tomorrow. Clin. Pharmacol. Ther. 2019, 21, 310–315. [Google Scholar] [CrossRef]
- Baquero, F. Threats of antibiotic resistance: An obliged reappraisal. Int. Microbiol. 2021, 24, 499–506. [Google Scholar] [CrossRef]
- Namazova-Baranova, L.S.; Baranov, A.A. Antibiotic resistance in modern world. Pediatr. Pharmacol. 2017, 14, 341–354. [Google Scholar] [CrossRef]
- Mikhaljovaa, T.V.; Zakharovab, O.I.; Iliasova, P.V. Antimicrobial Resistance: Current Approaches and Ways to Cope (a Review). Appl. Biochem. Microbiol. 2019, 5, 99–106. [Google Scholar] [CrossRef]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef] [PubMed]
- Tillotson, G.S.; Zinner, S.H. Burden of antimicrobial resistance in an era of decreasing susceptibility. Expert Rev. Anti-Infect. Ther. 2017, 15, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Huemer, M.; Shambat, S.M.; Brugger, S.D.; Zinkernagel, A.S. Antibiotic resistance and persistence-Implications for human health and treatment perspectives. EMBO Rep. 2020, 21, e51034. [Google Scholar] [CrossRef]
- Murray, C.J.L. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- WHO. Available online: https://www.who.int/publications/i/item/9789240093461 (accessed on 1 June 2024).
- Rather, M.A.; Gupta, K.; Mandal, M. Microbial biofilm: Formation, architecture, antibiotic resistance, and control strategies. Braz. J. Microbiol. 2021, 52, 1701–1718. [Google Scholar] [CrossRef]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 2019, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- Vishwanath, N.; Whitaker, C.; Allu, S.; Clippert, D.; Jouffroy, E.; Hong, J.; Stone, B.; Connolly, W.; Barrett, C.C.; Antoci, V. Silver as an Antibiotic-Independent Antimicrobial: Review of Current Formulations and Clinical Relevance. Surg. Infect. 2022, 23, 769–780. [Google Scholar] [CrossRef]
- Zhao, X.; Zhou, L.; Riaz Rajoka, M.S.; Yan, L.; Jiang, C.; Shao, D.; Zhu, J.; Shi, J.; Huang, Q.; Yang, H. Fungal silver nanoparticles: Synthesis, application and challenges. Crit. Rev. Biotechnol. 2018, 38, 817–835. [Google Scholar] [CrossRef]
- Guglielmelli, A.; D’Aquila, P.; Palermo, G.; Dell’Aglio, M.; Passarino, G.; Strangi, G.; Bellizzi, D. Role of the Human Serum Albumin Protein Corona in the Antimicrobial and Photothermal Activity of Metallic Nanoparticles against Escherichia coli Bacteria. ACS Omega 2023, 8, 31333–31343. [Google Scholar] [CrossRef]
- Hou, T.; Sana, S.S.; Kumbhakar, D.V.; Li, H.; Boya, V.K.N.; Aly Saad Aly, M.; Zhang, Z.; Pham, T.D. Synthesis of sodium-alginate capped silver nanocomposite microgel beads via bio-reduction featuring multifunctional antibacterial and apoptotic properties in human prostate cancer. J. Drug Deliv. Sci. Technol. 2023, 87, 104799. [Google Scholar] [CrossRef]
- Wiley, B.; Sun, Y.; Mayers, B.; Xia, Y. Shape-controlled synthesis of metal nanostructures: The case of silver. Chem. Eur. J. 2005, 11, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Wiley, B.; Herricks, T.; Sun, Y.; Xia, Y. Polyol synthesis of silver nanoparticles: Use of chloride and oxygen to promote the formation of single-crystal, truncated cubes and tetrahedrons. Nano Lett. 2004, 4, 1733–1739. [Google Scholar] [CrossRef]
- Lesnichaya, M.V.; Aleksandrova, G.P.; Feoktistova, L.P.; Sapozhnicov, A.N.; Fadeeva, T.V.; Sukhov, B.G.; Trofimov, B.A. Silver-containing nanocomposites based on galactomannan and carrageenan: Synthesis, structure, and antimicrobial properties. Rus. Chem. Bull. 2010, 59, 2323–2328. [Google Scholar] [CrossRef]
- Sivakumar, P.; Karthika, P.; Sivakuma, P.; Muralidharan, N.G.; Devendran, P.; Renganathan, S. Biosynthesis of silver nanoparticles from active compounds Quacetin–3-OBd-galactopyranoside containing plant extract and its antifungal application. Asian J. Pharm. Clin. Res. 2013, 6, 76–79. [Google Scholar]
- El-Shahaby, O.; El-Zayat, M.; Salih, E.; El-Sherbiny, I.M.; Reicha, F.M. Evaluation of antimicrobial activity of water infusion plant-mediated silver nanoparticles. J. Nanomed. Nanotechol. 2013, 4, 1000178. [Google Scholar]
- Bobbu, P.L.; Netala, V.R.; Aishwarya, S.; Reddy, I.R.M.; Kotakadi, V.S.; Tartte, V. Rapid synthesis of silver nanoparticles using aqueous leaf extract of Achyranthes aspera and study of their antimicrobial and free radical scavenging activities. Int. J. Pharm. Pharm. Sci. 2016, 8, 341–346. [Google Scholar]
- Zykova, M.V.; Volikov, A.B.; Buyko, E.E.; Bratishko, K.A.; Ivanov, V.V.; Konstantinov, A.I.; Logvinova, L.A.; Mihalyov, D.A.; Sobolev, N.A.; Zhirkova, A.M.; et al. Enhanced Antioxidant Activity and Reduced Cytotoxicity of Silver Nanoparticles Stabilized by Different Humic Materials. Polymers 2023, 15, 3386. [Google Scholar] [CrossRef]
- Zykova, M.V.; Zhang, Y.; Lysenko, I.V.; Arutyunyan, D.A.; Azarkina, L.A.; Mikhalev, D.A.; Belousov, M.V.; Perminova, I.V. Restoring antibiotic sensitivity to lincomycin in compositions with nanosilver and humic substance. Bull. Sib. Med. 2024, 23, 55–64. [Google Scholar] [CrossRef]
- Perminova, I.V. From green chemistry and nature-like technologies towards ecoadaptive chemistry and technology. Pure Appl. Chem. 2019, 91, 851–864. [Google Scholar] [CrossRef]
- Pena-Mendes, E.M.; Havel, J.; Patoska, J. Humic substances-compounds of still unknown structure: Applications in agriculture, industry, environment, and biomedicine. J. Appl. Biomed. 2005, 3, 13–24. [Google Scholar] [CrossRef]
- Vašková, J.; Stupák, M.; Vidová Ugurbaş, M.; Žatko, D.; Vaško, L. Therapeutic Efficiency of Humic Acids in Intoxications. Life 2023, 13, 971. [Google Scholar] [CrossRef] [PubMed]
- Van Rensburg, C.E. The antiinflammatory properties of humic substances: A mini review. Phytother. Res. 2015, 29, 791–795. [Google Scholar] [CrossRef] [PubMed]
- Zykova, M.V.; Veretennikova, E.E.; Logvinova, L.A.; Romanenko, S.V.; Bratishko, K.A.; Belousov, M.V.; Brazovsky, K.S.; Yusubov, M.S.; Lyapkov, A.A.; Danilets, M.G.; et al. New artificial network model to estimate biological activity of peat humic acids. Environ. Res. 2020, 191, 109999. [Google Scholar] [CrossRef]
- Zhernov, Y.V.; Kremb, S.; Helfer, M.; Schindler, M.; Harir, M.; Mueller, C.; Hertkorn, N.; Avvakumova, N.P.; Konstantinov, A.I.; Brack-Werner, R.; et al. Supramolecular Combinations of Fractionated Humic Polyanions as Potent and Cost-Effective Microbicides with Polymodal anti-HIV-Activities and Low Cytotoxicity. New J. Chem. 2017, 41, 212–224. [Google Scholar] [CrossRef]
- Zykova, M.V.; Brazovskii, K.S.; Bratishko, K.A.; Buyko, E.E.; Logvinova, L.A.; Romanenko, S.V.; Konstantinov, A.I.; Krivoshchekov, S.V.; Perminova, I.V.; Belousov, M.V. Quantitative Structure-Activity Relationship, Ontology-Based Model of the Antioxidant and Cell Protective Activity of Peat Humic Acids. Polymers 2022, 14, 3293. [Google Scholar] [CrossRef]
- Zykova, M.V.; Bratishko, K.A.; Buyko, E.E.; Azarkina, L.A.; Ivanov, V.V.; Mihalyov, D.A.; Trofimova, E.S.; Danilets, M.G.; Ligacheva, A.A.; Konstantinov, A.I.; et al. Coal-Derived Humic Substances: Insight into Chemical Structure Parameters and Biomedical Properties. Molecules 2024, 29, 1530. [Google Scholar] [CrossRef]
- Zhang, Y.; Larionov, K.S.; Zhang, S.; Sobolev, N.A.; Konstantinov, A.I.; Volkov, D.S.; Suslova, E.V.; Chernov, V.E.; Poloskov, A.I.; Glushakov, R.I.; et al. Humic Polyelectrolytes Facilitate Rapid Microwave Synthesis of Silver Nanoparticles Suitable for Wound-Healing Applications. Polymers 2024, 16, 587. [Google Scholar] [CrossRef]
- Nivetha, M.; Sujatha, S. Phytochemical analysis, antibacterial, antifungal and antiinflammatory activity of humic acid. Int. J. Res. Anal. Rev. 2019, 6, 853–858. [Google Scholar]
- Kulikova, N.A.; Perminova, I.V. Interactions between humic substances and microorganisms and their implications for nature-like bioremediation technologies. Molecules 2021, 26, 2706. [Google Scholar] [CrossRef] [PubMed]
- de Lacerda Coriolano, D.; de Souza, J.B.; Bueno, E.V.; Medeiros, S.M.d.F.R.d.S.; Cavalcanti, I.D.L.; Cavalcanti, I.M.F. Antibacterial and antibiofilm potential of silver nanoparticles against antibiotic-sensitive and multidrug-resistant Pseudomonas aeruginosa strains. Braz. J. Microbiol. 2021, 52, 267–278. [Google Scholar] [CrossRef]
- Bruna, T.; Maldonado-Bravo, F.; Jara, P.; Caro, N. Silver nanoparticles and their antibacterial applications. Int. J. Mol. Sci. 2021, 22, 7202. [Google Scholar] [CrossRef] [PubMed]
- Yin, I.X.; Zhang, J.; Zhao, I.S.; Mei, M.L.; Li, Q.; Chu, C.H. The Antibacterial Mechanism of Silver Nanoparticles and Its Application in Dentistry. Int. J. Nanomed. 2020, 15, 2555–2562. [Google Scholar] [CrossRef]
- Slavin, Y.N.; Asnis, J.; Hnfeli, U.O.; Bach, H. Metal nanoparticles: Understanding the mechanisms behind antibacterial activity. J. Nanobiotechnol. 2017, 15, 65. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, S.; Nazam, N.; Rizvi, S.M.D.; Ahmad, K.; Baig, M.H.; Lee, E.J.; Choi, I. Mechanistic insights into the antimicrobial actions of metallic nanoparticles and their implications for multidrug resistance. Int. J. Mol. Sci. 2019, 20, 2468. [Google Scholar] [CrossRef]
- Volikov, A.B.; Mareev, N.V.; Konstantinov, A.I.; Molodykh, A.A.; Melnikova, S.V.; Bazhanova, A.E.; Gasanov, M.E.; Nikolaev, E.N.; Zherebker, A.Y.; Volkov, D.S.; et al. Directed Synthesis of Humic and Fulvic Derivatives with Enhanced Antioxidant Properties. Agronomy 2021, 11, 2047. [Google Scholar] [CrossRef]
- Perminova, I.V.; Kovalenko, A.N.; Schmitt-Kopplin, P.; Hatfield, K.; Hertkorn, N.; Belyaeva, E.Y.; Petrosyan, V.S. Design of quinonoid-enriched humic materials with enhanced redox properties. Environ. Sci. Technol. 2005, 39, 8518–8524. [Google Scholar] [CrossRef]
- Kaur, H.; Kaur, H.; Sharma, A. Study of SPR peak shifting of silver nanoparticles with change in surrounding medium. Mater. Today Proc. 2021, 37, 3574–3576. [Google Scholar] [CrossRef]
- Hertkorn, N.; Permin, A.; Perminova, I.; Kovalevskii, D.; Yudov, M.; Petrosyan, V.; Kettrup, A. Comparative analysis of partial structures of a peat humic and fulvic acid using one- and two-dimensional nuclear magnetic resonance spectroscopy. J. Environ. Qual. 2002, 31, 375–387. [Google Scholar]
- Andrews, J.M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001, 48, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Hajfathalian, M.; de Vries, C.R.; Hsu, J.C.; Amirshaghaghi, A.; Dong, Y.C.; Ren, Z.; Liu, Y.; Huang, Y.; Li, Y.; Knight, S.A.B.; et al. Theranostic gold-in-gold cage nanoparticles enable photothermal ablation and photoacoustic imaging in biofilm-associated infection models. J. Clin. Investig. 2023, 133, 1–15. [Google Scholar] [CrossRef]
- Kingery, W.L.; Simpson, A.J.; Hayes, M.H.B.; Locke, M.; Hicks, R. The application of multidimensional NMR to the study of soil humic substances. Soil. Sci. 2000, 165, 483–494. [Google Scholar] [CrossRef]
- Kovalevskii, D.V.; Permin, A.B.; Perminova, I.V.; Petrosyan, V.S. Choice of the time of pulse delay for quantitative 13C NMR spectroscopy of humic substances. Bull. Mosc. Univ. (Vestnik MGU) Ser. 2 2000, 41, 39–42. [Google Scholar]
- Kuzmichev, A.S.; Bogatikov, A.A.; Dobretsov, K.G.; Zaitseva, I.V. The use of nanoparticles in the treatment of purulent wounds. Russ. Biomed. Res. 2022, 7, 36–42. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, X.; He, C.; Huang, J.; Yin, S.; Zhou, M.; Ma, L.; Zhao, W.; Qiu, L.; Cheng, C.; et al. Metal-Organic Framework/Ag-Based Hybrid Nanoagents for Rapid and Synergistic Bacterial Eradication. ACS Appl. Mater. Interfaces 2020, 12, 13698–13708. [Google Scholar] [CrossRef]
- Dovnar, R.I.; Vasil’kov, A.Y.; Dovnar, I.S.; Iaskevich, N.N. Effect of silver nanoparticles on polyantibiotic resistant pathogenic microorganisms. Surgery. East. Eur. 2022, 11, 464–474. [Google Scholar]
- Khan, I.; Saeed, K.; Khan, I. Nanoparticles: Properties, applications and toxicities. Arab. J. Chem. 2019, 12, 908–931. [Google Scholar] [CrossRef]
- Sim, S.; Wong, N.K. Nanotechnology and its use in imaging and drug delivery (Review). Biomed. Rep. 2021, 14, 42. [Google Scholar] [CrossRef]
- Dovnar, R.I.; Vasil’kov, A.Y.; Sakalova, T.M.; Naumkin, A.V.; Budnikov, A.V.; Dovnar, I.S.; Iaskevich, N.N. Silver nanoparticles: An ecological method of synthesis, properties and use against antibiotic resistant microflora. Proc. Natl. Acad. Sci. Belarus Med. Ser. 2021, 18, 351–361. [Google Scholar] [CrossRef]
- Obradovic, B.; Stojkovska, J.; Zvicer, J. Preclinical functional characterization methods of nanocomposite hydrogels containing silver nanoparticles for biomedical applications. Appl. Microbiol. Biotechnol. 2020, 104, 4643–4658. [Google Scholar]
- Balu, S.K.; Andra, S.; Kannan, S.; Vidyavathy, M.S.; Muthalagu, M. Facile synthesis of silver nanoparticles with medicinal grass and its biological assessment. Mater. Lett. 2020, 259, 126900. [Google Scholar] [CrossRef]
- Qais, F.A.; Shafiq, A.; Khan, H.M.; Husain, F.M.; Khan, R.A.; Alenazi, B.; Alsalme, A.; Ahmad, I. Antibacterial Effect of Silver Nanoparticles Synthesized Using Murraya koenigii (L.) against Multidrug-Resistant Pathogens. Bioinorg. Chem. Appl. 2019, 2019, 4649506. [Google Scholar] [CrossRef] [PubMed]
- Selim, A.; Tlhaig, M.M.; Taha, S.A.; Nasr, E.A. Antibacterial activity of silver nanoparticles against field and reference strains of Mycobacterium tuberculosis, Mycobacterium bovis and multiple-drug-resistant tuberculosis strains. Rev. Sci. Tech. Off. Int. Epizoot. 2018, 37, 823–830. [Google Scholar] [CrossRef]
- Niska, K.; Knap, N.; Kędzia, A.; Jaskiewicz, M.; Kamysz, W.; Inkielewicz-Stepniak, I. Capping Agent-Dependent Toxicity and Antimicrobial Activity of Silver Nanoparticles: An In Vitro Study. Concerns about Potential Application in Dental Practice. Int. J. Med. Sci. 2016, 13, 772–782. [Google Scholar] [CrossRef]
- Pérez-Díaz, M.A.; Boegli, L.; James, G.; Velasquillo, C.; Sánchez-Sánchez, R.; Martínez-Martínez, R.E.; Martínez-Castañón, G.A.; Martinez-Gutierrez, F. Silver nanoparticles with antimicrobial activities against Streptococcus mutans and their cytotoxic effect. Mater. Sci. Eng. C Mater. Biol. Appl. 2015, 55, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Corrêa, J.M.; Mori, M.; Sanches, H.L.; da Cruz, A.D.; Poiate, E., Jr.; Poiate, I.A. Silver nanoparticles in dental biomaterials. Int. J. Biomater. 2015, 2015, 485275. [Google Scholar] [CrossRef]
- Akter, M.; Sikder, M.T.; Rahman, M.M.; Ullah, A.K.M.A.; Hossain, K.F.B.; Banik, S.; Hosokawa, T.; Saito, T.; Kurasaki, M. A systematic review on silver nanoparticles-induced cytotoxicity: Physicochemical properties and perspectives. J. Adv. Res. 2017, 9, 1–16. [Google Scholar] [CrossRef]
- Chernysheva, M.G.; Badun, G.A.; Kulikova, N.A.; Perminova, I.V. Behavior of humic substances in the liquid-liquid system directly measured using tritium label. Chemosphere 2020, 238, 124646. [Google Scholar] [CrossRef]
- Singh, S.; Datta, S.; Narayanan, K.B.; Rajnish, K.N. Bacterial exo-polysaccharides in biofilms: Role in antimicrobial resistance and treatments. J. Genet. Eng. Biotechnol. 2021, 19, 140. [Google Scholar] [CrossRef]
- Buzlama, A.V.; Chernov, I.N. Humic substances: Pharmacological properties, mechanisms of action, and prospects for use in medicine. Eksp. Klin. Farmakol. 2010, 73, 43–48. [Google Scholar] [PubMed]
- Buyko, E.E.; Zykova, M.V.; Ivanov, V.V.; Bratishko, K.A.; Ufandeev, A.A.; Grigorieva, I.O.; Tsupko, A.V.; Mikhalyov, D.A.; Perminova, I.V.; Belousov, M.V. Antioxidant Activity of Silver-containing Bionanocompositions Based on Humic Substances in Cell Culture. Drug Dev. Regist. 2021, 10, 46–53. [Google Scholar] [CrossRef]
- Trofimova, E.S.; Zykova, M.V.; Danilets, M.G.; Ligacheva, A.A.; Sherstoboev, E.Y.; Grigorieva, I.O.; Mikhalev, D.A.; Tsupko, A.V.; Logvinova, L.A.; Perminova, I.V.; et al. The effect of coal-derived humic substances and their silver-containing bionanocomposites on arginine balance in peritoneal macrophages of intact mice. Bull. Sib. Med. 2021, 20, 71–78. [Google Scholar] [CrossRef]
- Saadh, M.J. Effect of silver nanoparticles on the antibacterial activity of Levofloxacin against methicillin-resistant Staphylococcus aureus. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 5507–5510. [Google Scholar]
- Kaur, A.; Preet, S.; Kumar, V.; Kumar, R.; Kumar, R. Synergetic effect of vancomycin loaded silver nanoparticles for enhanced antibacterial activity. Colloids Surf. B Biointerfaces 2019, 176, 62–69. [Google Scholar] [CrossRef]
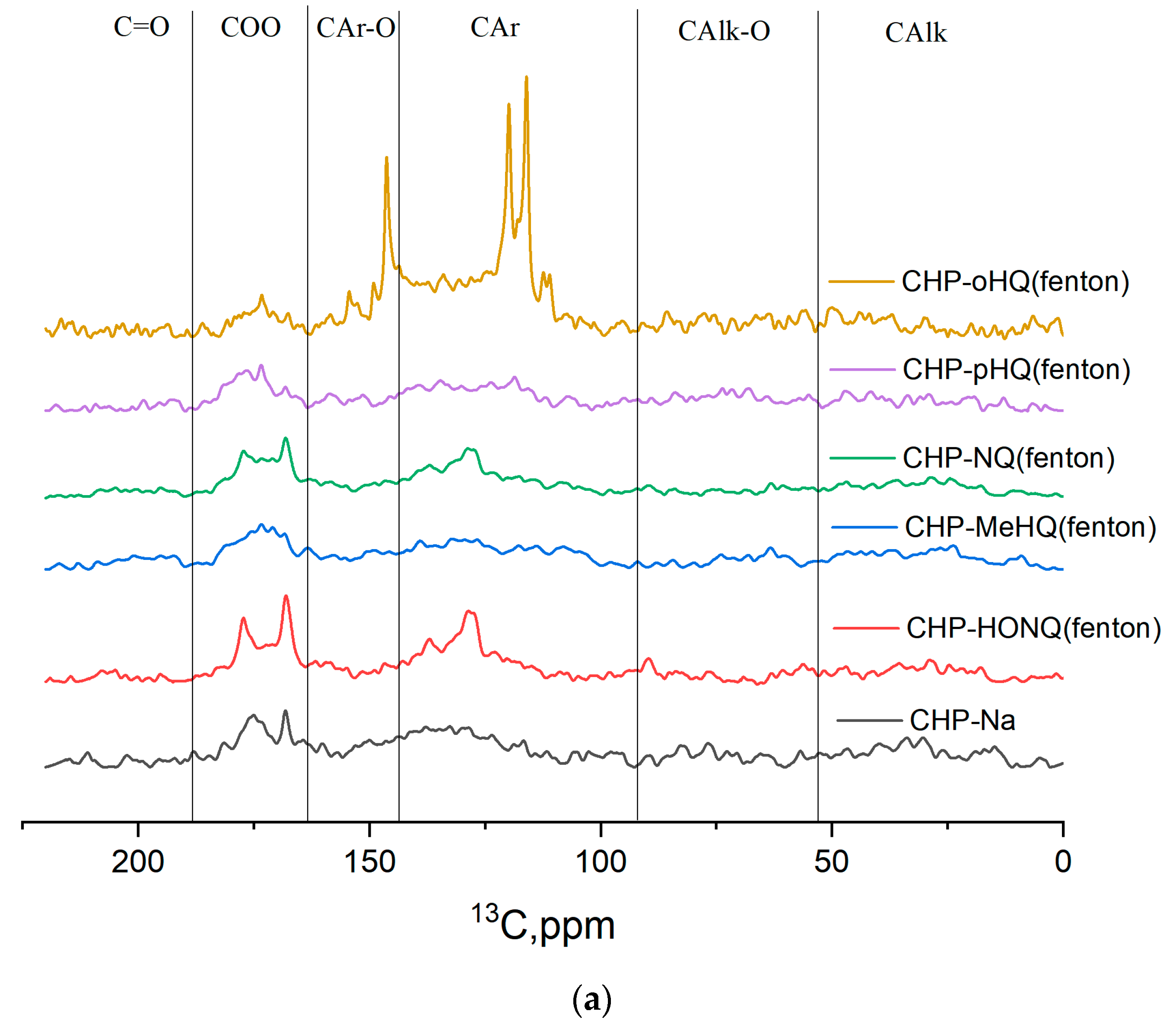


| Samples | CH | CHO | Car | CarO | COO | C=O |
|---|---|---|---|---|---|---|
| 0–47 | 47–110 | 110–145 | 145–165 | 165–185 | 185–220 | |
| CHP-Na | 17.5 | 18.7 | 34.4 | 9.8 | 20.6 | 4.7 |
| CHP-HQ-FF | 14.5 | 32.6 | 39.0 | 11.3 | 13.7 | 1.2 |
| CHP-HQ-FE | 16.7 | 31.6 | 22.4 | 10.1 | 27.6 | 4.7 |
| CHP-HONQ-FF | 11.5 | 34.9 | 34.5 | 11.8 | 17.0 | 4.2 |
| CHP-HONQ-FE | 13.3 | 24.6 | 33.9 | 9.0 | 24.0 | 4.3 |
| CHP-MeHQ-FF | 18.6 | 36.8 | 37.5 | 9.6 | 8.9 | 2.9 |
| CHP-MeHQ-FE | 19.3 | 31.3 | 25.7 | 9.8 | 18.2 | 8.0 |
| CHP-PK-FF | 15.5 | 24.3 | 40.0 | 9.9 | 15.3 | 4.2 |
| CHP-PK-FE | 9.0 | 30.1 | 49.5 | 13.4 | 6.7 | 4.0 |
| CHP-NQ-FF | 17.0 | 19.7 | 43.0 | 8.4 | 16.4 | 2.0 |
| CHP-NQ-FE | 14.6 | 44.4 | 28.7 | 7.6 | 19.0 | 4.7 |
| No. | Code | Derivative for HS-AgNPs Synthesis | Reaction for HS-AgNPs Synthesis | Ag Content, mg/100 mL |
|---|---|---|---|---|
| 1 | CHP-AgNPs | - | Template synthesis | 24.27 |
| 2 | CHP-oHQ-FE-AgNPs | o-hydroquinone | Fenton | 24.27 |
| 3 | CHP-pHQ-FE-AgNPs | p-hydroquinone | Fenton | 24.27 |
| 4 | CHP-MeHQ-FE-AgNPs | 2-methyl-1,4-hydroquinone | Fenton | 24.27 |
| 5 | CHP-NQ-FE-AgNPs | 1,4-naphthoquinone | Fenton | 24.27 |
| 6 | CHP-HONQ-FE-AgNPs | 2-hydroxy-1,4-naphthoquinone | Fenton | 24.27 |
| 7 | CHP-oHQ-FF-AgNPs | o-hydroquinone | Phenol–formaldehyde condensation | 24.27 |
| 8 | CHP-pHQ-FF-AgNPs | p-hydroquinone | Phenol–formaldehyde condensation | 24.27 |
| 9 | CHP-MeHQ-FF-AgNPs | 2-methyl-1,4-hydroquinone | Phenol–formaldehyde condensation | 24.27 |
| 10 | CHP-NQ-FF-AgNPs | 1,4-naphthoquinone | Phenol–formaldehyde condensation | 24.27 |
| 11 | CHP-HONQ-FF-AgNPs | 2-hydroxy-1,4-naphthoquinone | Phenol–formaldehyde condensation | 24.27 |
| 12 | CHP-AgNPs-MW | - | Microwave synthesis | 24.27 |
| Microorganisms | Concentration of HS-AgNPs That Suppressed the Growth of Microorganisms | ||
|---|---|---|---|
| 200 mg/L | 500 mg/L | 800 mg/L | |
| E. coli | No. 2, No. 3 | No. 2, No. 3 | No. 2, No. 3, No. 4, No. 6, No. 7 |
| MRSA | – | – | No. 12 |
| K. pneumoniae | – | – | No. 3 |
| P. aeruginosa | No. 3 | – | No. 3, No. 5, No. 6, No. 7 |
| A. baumannii | No. 2, No. 3, No. 4, No. 6 | No. 1, No. 2, No. 3, No. 4, No. 6, No. 78, No. 12 | No. 1, No. 2, No. 3, No. 4, No. 6, No. 7, No. 8, No. 10, No. 12 |
| Bacteria | Sample | Concentration of HS-AgNPs | Optical Density | p |
|---|---|---|---|---|
| E. coli (N = 3) | No. 3 | 100 | 0.164 (0.164; 0.228) | 0.513 |
| 150 | 0.141 (0.125; 0.144) | 0.049 | ||
| 200 | 0.170 (0.133; 0.181) | 0.049 | ||
| Control | 0.216 (0.195; 0.226) | |||
| A. baumannii (N = 3) | No. 3 | 100 | 0.348 (0.345; 0.348) | 0.049 |
| 150 | 0.317 (0.308; 0.326) | 0.049 | ||
| 200 | 0.316 (0.306; 0.318) | 0.049 | ||
| Control | 0.330 (0.329; 0.330) | |||
| K. pneumoniae (N = 3) | No. 3 | 700 | 0.314 (0.307; 0.316) | 0.513 |
| 750 | 0.349 (0.337; 0.365) | 0.513 | ||
| 800 | 0.293 (0.273; 0.300) | 0.513 | ||
| Control | 0.321 (0.237; 0.392) | |||
| P. aeruginosa (N = 3) | No. 3 | 700 | 0.189 (0.184; 0.190) | 0.513 |
| 750 | 0.186 (0.179; 0.193) | 0.513 | ||
| 800 | 0.178 (0.170; 0.194) | 0.513 | ||
| Control | 0.248 (0.169; 0.262) | |||
| S. aureus (N = 3) | No. 12 | 700 | 0.206 (0.205; 0.236) | 0.049 |
| 750 | 0.195 (0.192; 0.205) | 0.049 | ||
| 800 | 0,198 (0.197; 0.199) | 0.049 | ||
| Control | 0.273 (0.261; 0.284) | |||
| MRSA (N = 3) | No. 12 | 700 | 0.270 (0.261; 0.276) | 0.513 |
| 750 | 0.245 (0.241; 0.251) | 0.049 | ||
| 800 | 0.254 (0.242; 0.257) | 0.127 | ||
| Control | 0.341 (0.255; 0.372) |
| Bacteria | Material | Sample | Final Concentration of HS-AgNPs, mg/L | Optical Density | p |
|---|---|---|---|---|---|
| E. coli (N = 3) | Sputum | No. 3 | 100 | 0.129 (0.119; 0.140) | 0.275 |
| 150 | 0.111 (0.104; 0.120) | 0.049 | |||
| 200 | 0.127 (0.127; 0.132) | 0.127 | |||
| Control | 0.139 (0.130; 0.215) | ||||
| A. baumannii (N = 3) | Urine | No. 3 | 100 | 0.274 (0.255; 0.294) | 0.827 |
| 150 | 0.224 (0.213; 0.227) | 0.049 | |||
| 200 | 0.243 (0.238; 0.271) | 0.275 | |||
| Control | 0.259 (0.250; 0.347) | ||||
| K. pneumoniae (N = 3) | Urine | No. 3 | 700 | 0.166 (0.158; 0.195) | 0.049 |
| 750 | 0.206 (0.204; 0.229) | 0.049 | |||
| 800 | 0.210 (0.172; 0.215) | 0.049 | |||
| Control | 0.270 (0.244; 0.300) | ||||
| K. pneumoniae (N = 3) | Blood | No. 3 | 700 | 0.207 (0.207; 0.231) | 0.127 |
| 750 | 0.207 (0.200; 0.222) | 0.049 | |||
| 800 | 0.234 (0.223; 0.241) | 0.275 | |||
| Control | 0.260 (0.227; 0.328) |
| Bacteria | Sample | Final Concentration of HS-AgNPs, mg/L | Optical Density | p |
|---|---|---|---|---|
| E. coli (N = 3) | No. 3 | 100 | 0.232 (0.204; 0.245) | 0.049 |
| 150 | 0.231 (0.224; 0.238) | 0.049 | ||
| 200 | 0.232 (0.231; 0.238) | 0.049 | ||
| Control | 0.181 (0.169; 0.193) | |||
| A. baumannii (N = 3) | No. 3 | 100 | 0.335 (0.316; 0.346) | 0.049 |
| 150 | 0.326 (0.315; 0.328) | 0.049 | ||
| 200 | 0.329 (0.327; 0.336) | 0.049 | ||
| Control | 0.232 (0.168; 0.308) | |||
| S. aureus (N = 3) | No. 12 | 700 | 0.290 (0.284; 0.305) | 0.049 |
| 750 | 0.296 (0.285; 0.300) | 0.049 | ||
| 800 | 0.292 (0.286; 0.304) | 0.049 | ||
| Control | 0.227 (0.224; 0.249) | |||
| MRSA (N = 3) | No. 12 | 700 | 0.367 (0.363; 0.373) | 0.049 |
| 750 | 0.346 (0.324; 0.350) | 0.049 | ||
| 800 | 0.370 (0.345; 0.386) | 0.049 | ||
| Control | 0.266 (0.216; 0.308) | |||
| K. pneumoniae (N = 3) | No. 3 | 700 | 0.346 (0.256; 0.351) | 0.513 |
| 750 | 0.365 (0.360; 0.373) | 0.049 | ||
| 800 | 0.337 (0.335; 0.381) | 0.049 | ||
| Control | 0.322 (0.305; 0.333) | |||
| P. aeruginosa (N = 3) | No. 3 | 700 | 0.309 (0.283; 0.310) | 0.049 |
| 750 | 0.323 (0.292; 0.330) | 0.049 | ||
| 800 | 0.266 (0.237; 0.276) | 0.513 | ||
| Control | 0.252 (0.244; 0.260) |
| Microorganism | Material | Sample | Final Concentration of HS-AgNPs, mg/L | Optical Density | p |
|---|---|---|---|---|---|
| E. coli (N = 3) | Sputum | No. 3 | 100 | 0.197 (0.181; 0.216) | 0.513 |
| 150 | 0.185 (0.179; 0.190) | 0.513 | |||
| 200 | 0.195 (0.157; 0.196) | 0.663 | |||
| Control | 0.157 (0.142; 0.235) | ||||
| A. baumannii (N = 3) | Urine | No. 3 | 100 | 0.309 (0.304; 0.314) | 0.513 |
| 150 | 0.302 (0.293; 0.319) | 0.513 | |||
| 200 | 0.293 (0.286; 0.297) | 0.512 | |||
| Control | 0.329 (0.274; 0.375) | ||||
| K. pneumoniae (N = 3) | Urine | No. 3 | 700 | 0.265 (0.236; 0.328) | 0.049 |
| 750 | 0.266 (0.247; 0.273) | 0.049 | |||
| 800 | 0.260 (0.255; 0.281) | 0.049 | |||
| Control | 0.489 (0.456; 0.553) | ||||
| K. pneumoniae (N = 3) | Blood | No. 3 | 700 | 0.249 (0.244; 0.251) | 0.049 |
| 750 | 0.271 (0.260; 0.288) | 0.513 | |||
| 800 | 0.262 (0.251; 0.279) | 0.275 | |||
| Control | 0.278 (0.269; 0.390) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zykova, M.V.; Karpova, M.R.; Zhang, Y.; Chubik, M.V.; Shunkova, D.M.; Azarkina, L.A.; Mihalyov, D.A.; Konstantinov, A.I.; Plotnikov, E.V.; Pestryakov, A.N.; et al. The Influence of Silver-Containing Bionanomaterials Based on Humic Ligands on Biofilm Formation in Opportunistic Pathogens. Nanomaterials 2024, 14, 1453. https://doi.org/10.3390/nano14171453
Zykova MV, Karpova MR, Zhang Y, Chubik MV, Shunkova DM, Azarkina LA, Mihalyov DA, Konstantinov AI, Plotnikov EV, Pestryakov AN, et al. The Influence of Silver-Containing Bionanomaterials Based on Humic Ligands on Biofilm Formation in Opportunistic Pathogens. Nanomaterials. 2024; 14(17):1453. https://doi.org/10.3390/nano14171453
Chicago/Turabian StyleZykova, Maria V., Maria R. Karpova, Yu Zhang, Marianna V. Chubik, Daria M. Shunkova, Lyudmila A. Azarkina, Dmitrii A. Mihalyov, Andrey I. Konstantinov, Evgenii V. Plotnikov, Alexey N. Pestryakov, and et al. 2024. "The Influence of Silver-Containing Bionanomaterials Based on Humic Ligands on Biofilm Formation in Opportunistic Pathogens" Nanomaterials 14, no. 17: 1453. https://doi.org/10.3390/nano14171453







