Singleton {NOT} and Doubleton {YES; NOT} Gates Act as Functionally Complete Sets in DNA-Integrated Computational Circuits
Abstract
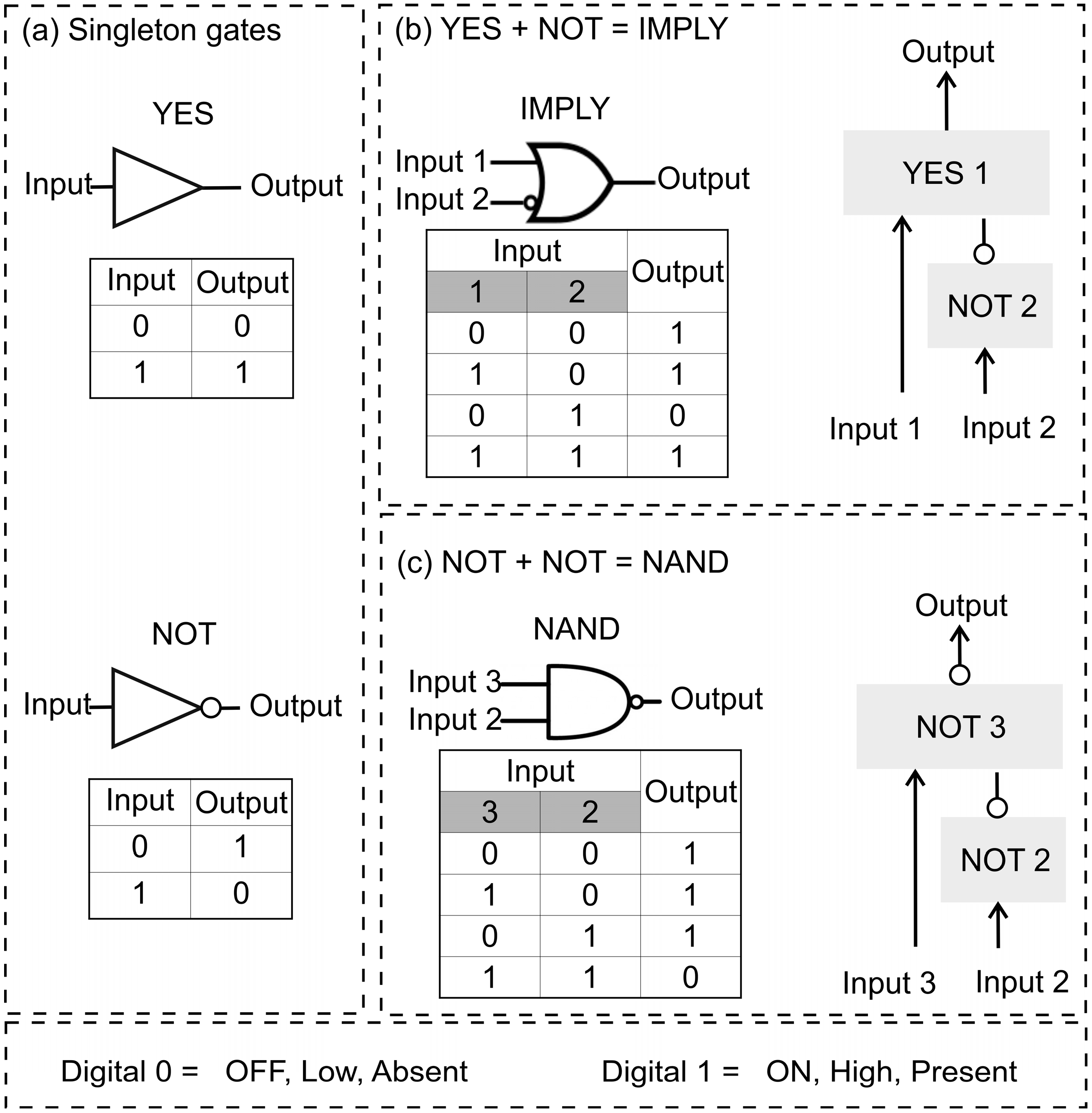
:1. Introduction
2. Materials and Methods
2.1. Materials
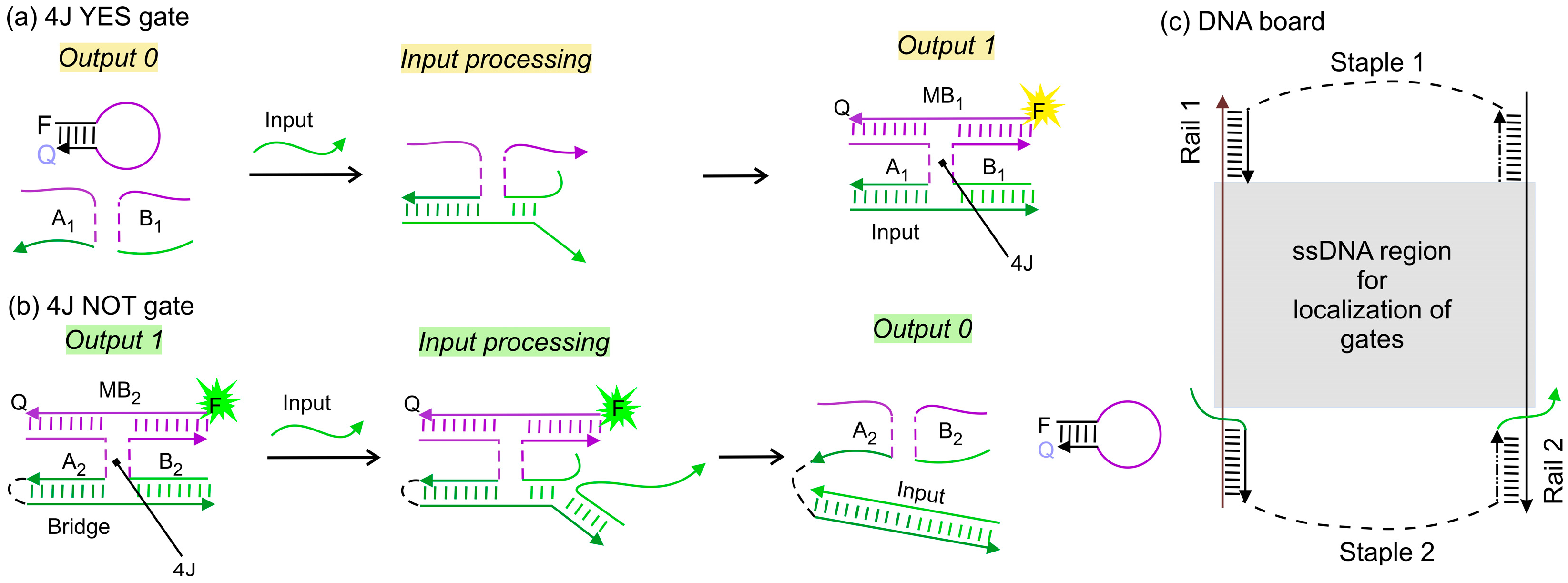
2.2. DNA Logic Gates Assembly
2.3. Fluorescence Assays
2.4. Fluorescence Data Analysis
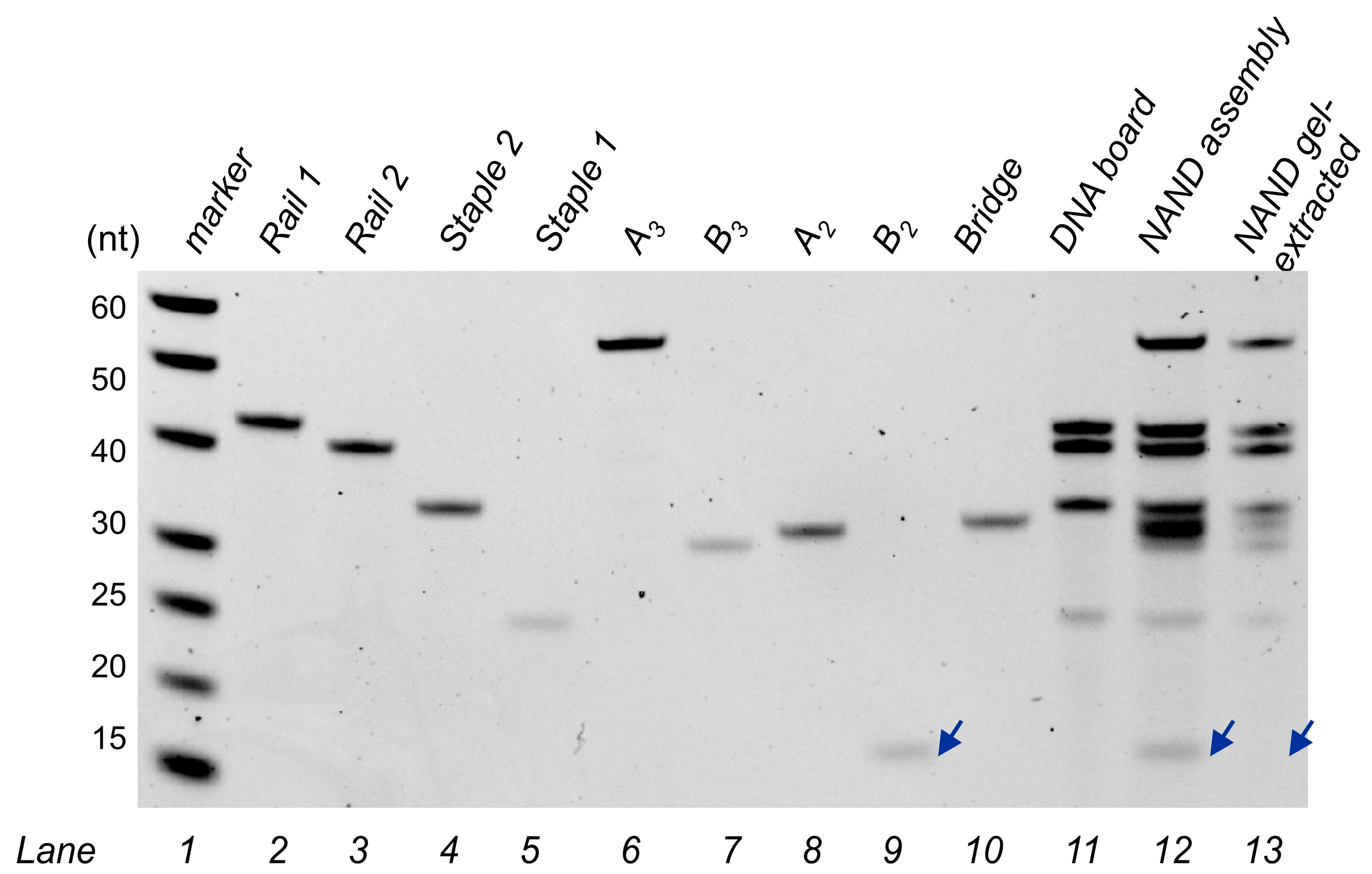
2.5. Gel Electrophoresis
2.6. Assembly Gel-Extraction
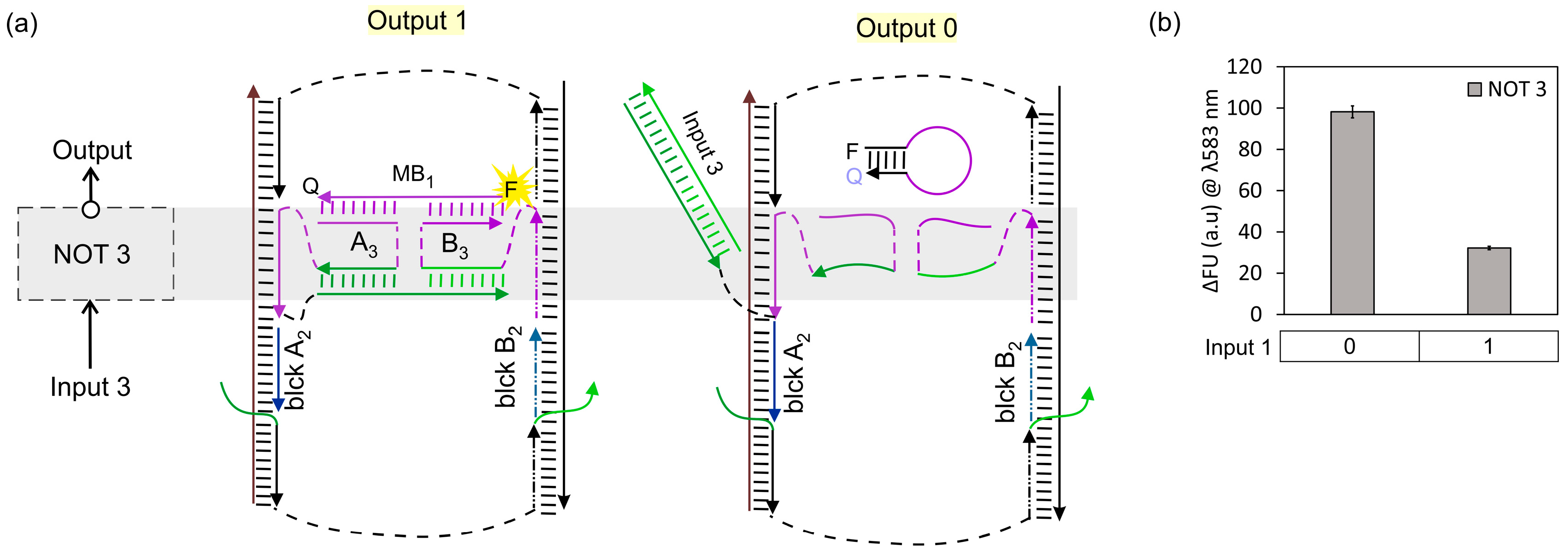
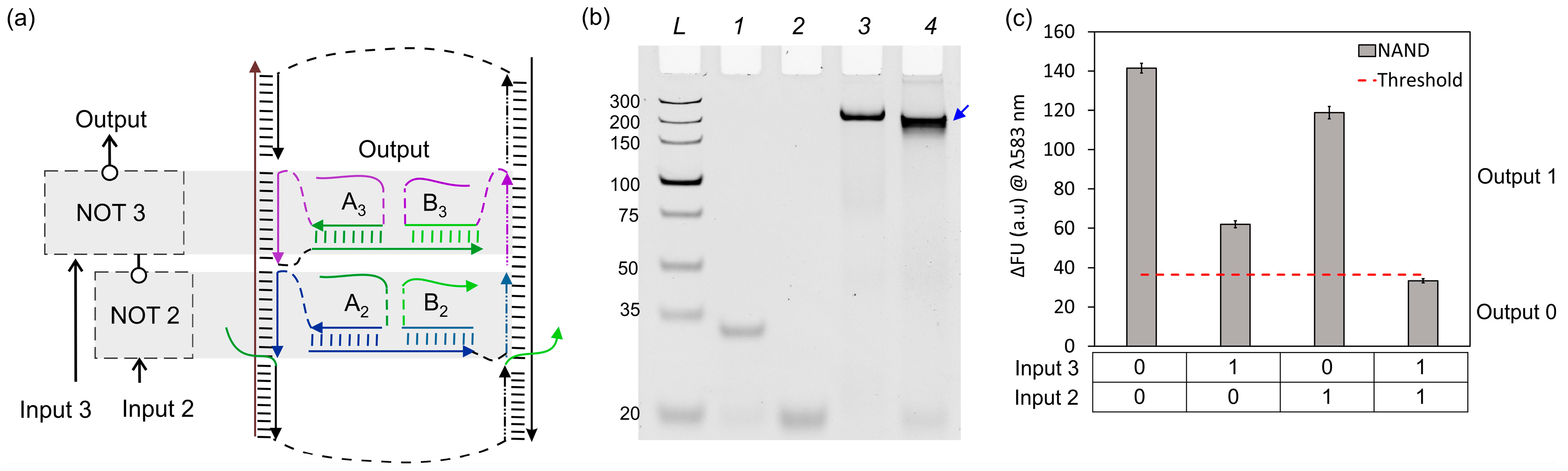
3. Results and Discussion
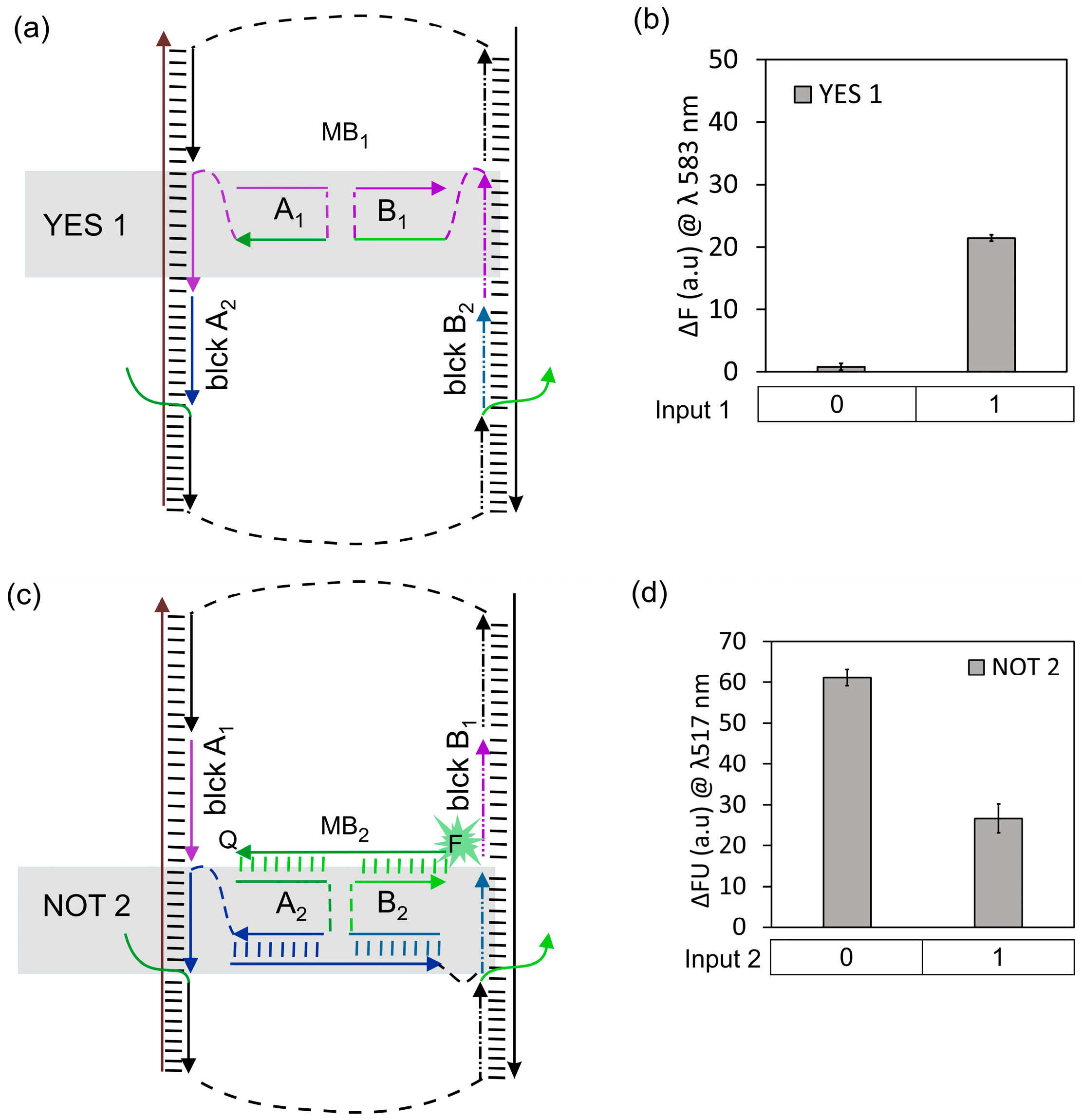
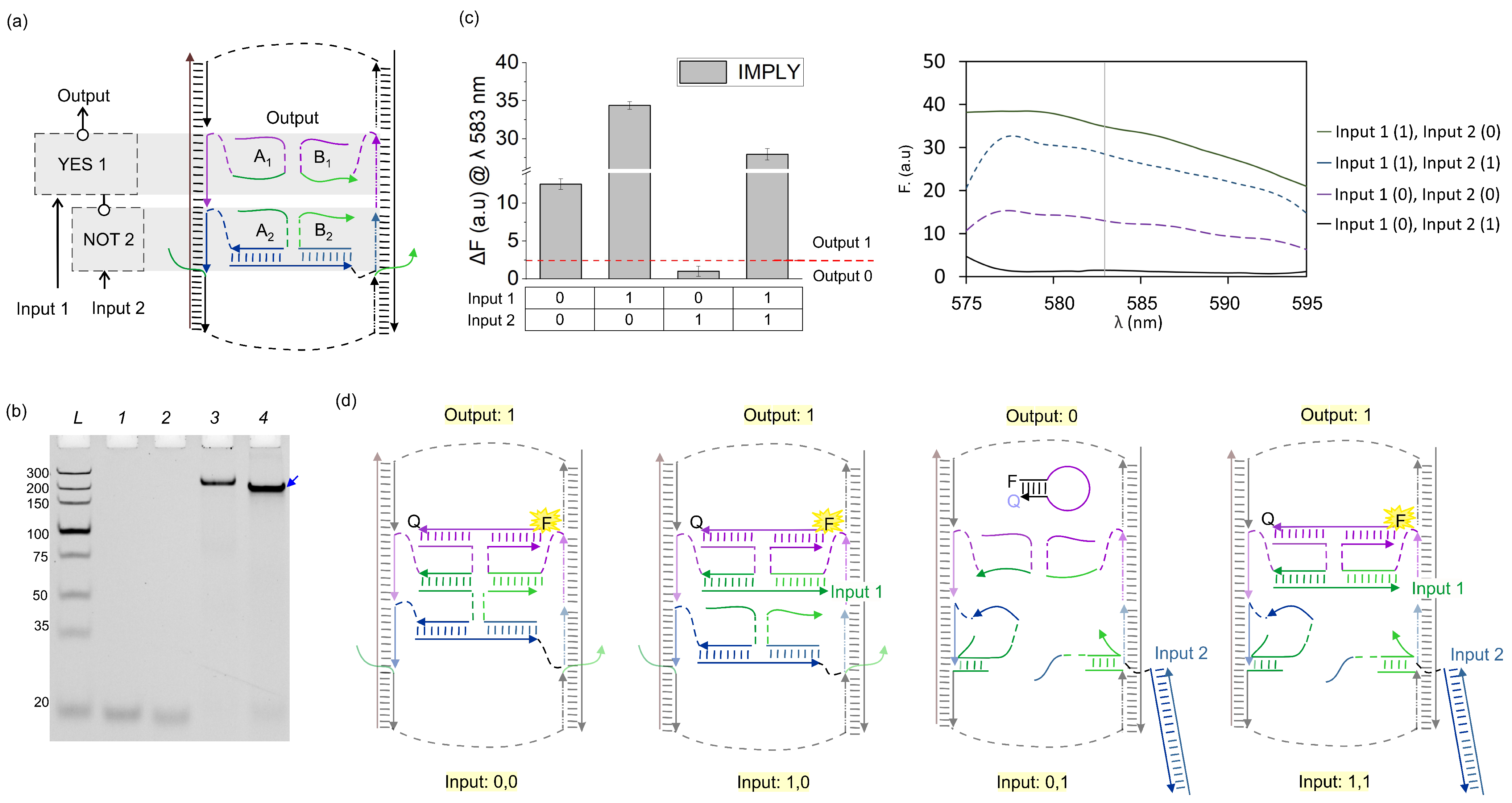
3.1. IMPLY Logic Circuit (YES + NOT)
3.2. NAND Logic Circuit (NOT + NOT)
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Malvino, A.P.; Brown, J.A. Digital Computer Electronics, 3rd ed.; Glencoe McGraw-Hill: New York, NY, USA, 1999. [Google Scholar]
- Frezza, B.M.; Cockroft, S.L.; Ghadiri, M.R. Modular multi-level circuits from immobilized DNA-based logic gates. J. Am. Chem. Soc. 2007, 129, 14875–14879. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, A.N.; Russel, B. Principia Mathematica, 2nd ed.; The Syndics of the Cambridge University Press: Cambridge, UK, 1963; Volume 1. [Google Scholar]
- Borghetti, J.; Snider, G.S.; Kuekes, P.J.; Yang, J.J.; Stewart, D.R.; Williams, R.S. ‘Memristive’ Switches Enable ‘Stateful’ Logic Operations via Material Implication. Nature 2010, 464, 873–876. [Google Scholar] [CrossRef] [PubMed]
- Prezioso, M.; Riminucci, A.; Graziosi, P.; Bergenti, I.; Rakshit, R.; Cecchini, R.; Vianelli, A.; Borgatti, F.; Haag, N.; Willis, M.; et al. A Single-Device Universal Logic Gate Based on a Magnetically Enhanced Memristor. Adv. Mater. 2013, 25, 534–538. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.P.D.; Leydet, Y.; Lincheneau, C.; McClenaghan, N.D. Chemical Approaches to Nanometre-Scale Logic Gates. J. Phys. Condens. Matter 2006, 18, S1847–S1872. [Google Scholar] [CrossRef]
- Zhou, J.; Arugula, M.A.; Halámek, J.; Pita, M.; Katz, E. Enzyme-Based NAND and NOR Logic Gates with Modular Design. J. Phys. Chem. B 2009, 113, 16065–16070. [Google Scholar] [CrossRef] [PubMed]
- Katz, E. DNA- and RNA-Based Computing Systems; John Wiley & Sons: Weinheim, Germany, 2021. [Google Scholar]
- Polak, R.E.; Keung, A.J.A. Molecular Assessment of the Practical Potential of DNA-Based Computation. Curr. Opin. Biotechnol. 2023, 81, 102940. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.; Wang, J.; Wang, E.; Dong, S. Propelling DNA Computing with Materials’ Power: Recent Advancements in Innovative DNA Logic Computing Systems and Smart Bio-Applications. Adv. Sci. 2020, 7, 2001766. [Google Scholar] [CrossRef] [PubMed]
- Gerasimova, Y.V.; Kolpashchikov, D.M. Connectable DNA logic gates: OR and XOR logics. Chem. Asian J. 2012, 7, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Gerasimova, Y.V.; Kolpashchikov, D.M. Towards a DNA Nanoprocessor: Reusable Tile-Integrated DNA Circuits. Angew. Chem. 2016, 128, 10400–10403. [Google Scholar] [CrossRef]
- Tyagi, S.; Kramer, F.R. Molecular Beacons: Probes That Fluoresce upon Hybridization. Nat. Biotechnol. 1996, 14, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, G.; Dalchau, N.; Muscat, R.A.; Phillips, A.; Seelig, G. A Spatially Localized Architecture for Fast and Modular DNA Computing. Nat. Nanotechnol. 2017, 12, 920–927. [Google Scholar] [CrossRef] [PubMed]
- Connelly, R.P.; Morozkin, E.S.; Gerasimova, Y.V. Alphanumerical Visual Display Made of DNA Logic Gates for Drug Susceptibility Testing of Pathogens. ChemBioChem 2018, 19, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Benenson, Y.; Gil, B.; Ben-Dor, U.; Adar, R.; Shapiro, E. An Autonomous Molecular Computer for Logical Control of Gene Expression. Nature 2004, 429, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Fu, Y.; Yan, J.; Zhao, B.; Dai, B.; Chao, J.; Liu, H.; He, D.; Zhang, Y.; Fan, C.; et al. Molecular Logic Gates on DNA Origami Nanostructures for MicroRNA Diagnostics. Anal. Chem. 2014, 86, 1932–1936. [Google Scholar] [CrossRef]
- You, M.; Zhu, G.; Chen, T.; Donovan, M.J.; Tan, W. Programmable and Multiparameter DNA-Based Logic Platform For Cancer Recognition and Targeted Therapy. J. Am. Chem. Soc. 2015, 137, 667–674. [Google Scholar] [CrossRef]
- Song, X.; Reif, J. Nucleic Acid Databases and Molecular-Scale Computing. ACS Nano 2019, 13, 6256–6268. [Google Scholar] [CrossRef]

| Name | Comments | Sequence |
|---|---|---|
| DNA Board | ||
| Rail 1 | CCT ATC GTG TT TTG TCG CTGA CCA TC GTA TCG CTT CGT CTATG | |
| Rail 2 | CTGAG TGAAT GAG CT CTA CA C TGC AGT ACC AC CGT TAG TCA | |
| Staple 1 | ATTCA CTCAG/iSp18//iSp18/CATAG ACG AAG | |
| Staple 2 | GACA AA CAC GAT AGG/iSp18//iSp18/TGA CTA ACG GT CCAG | |
| Blck A1 | CGA TAC GAT GG | |
| Blck B1 | TGT AGA GCTC | |
| Blck A2 | TCAG CGA CAA | |
| Blck B2 | GGT ACT GCA G | |
| YES 1 | ||
| A1 | CT TTG TTC/iSp18/A GAC AAT GTA GC/iSp18/CGATAC GATGG | |
| B1 | AGTAG AGCTC/iSp18/GAAAC CCA GC/iSp18/GAT G ATT CC | |
| NOT 2 | ||
| A2 | TA CAT TGTC T/iSp18/GGT GAAC C/iSp18/TCAG CGA CAA | |
| B2 | TG TTG CTC/iSp18/GCT GGG | |
| Bridge | AGGG GTT CAC CGA GCA ACA TTC/iSp9/GGT ACT GCA G | |
| NOT 3 | ||
| A3 | CT TTG TTC/iSp18/A GAC AAT G/iSp18/CGATAC GATGG/iSp18/GC TAC ATT GTCT GC TGG GTTTC | |
| B3 | AGTAG AGCTC/iSp18/AAC CCA GC/iSp18/GAT G ATT CC | |
| Inputs | ||
| Input 3 | GAAAC CCA GC AGAC AAT GTA GC | |
| Input 2 | hsa-miR-409-3p | /5′-Phos/-rGrArA rUrGrU rUrGrC rUrCrG rGrUrG rArArC rCrCrC rU |
| Input 1 | hsa-miR-221-3p | /5′-Phos/rArGrC rUrArC rArUrU rGrUrC rUrGrC rUrGrG rGrUrUrUrC |
| Molecular Beacons (MB) | ||
| MB1 | /56-TAMN/CCT GG AATCATC GAACAAAG CA CAG CCAGG-3′-BHQ2 | |
| MB2 | /56-FAM/CCAGG CCCAGC AGACAATGTA CCT GG/3BHQ_1/ | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bardales, A.C.; Vo, Q.; Kolpashchikov, D.M. Singleton {NOT} and Doubleton {YES; NOT} Gates Act as Functionally Complete Sets in DNA-Integrated Computational Circuits. Nanomaterials 2024, 14, 600. https://doi.org/10.3390/nano14070600
Bardales AC, Vo Q, Kolpashchikov DM. Singleton {NOT} and Doubleton {YES; NOT} Gates Act as Functionally Complete Sets in DNA-Integrated Computational Circuits. Nanomaterials. 2024; 14(7):600. https://doi.org/10.3390/nano14070600
Chicago/Turabian StyleBardales, Andrea C., Quynh Vo, and Dmitry M. Kolpashchikov. 2024. "Singleton {NOT} and Doubleton {YES; NOT} Gates Act as Functionally Complete Sets in DNA-Integrated Computational Circuits" Nanomaterials 14, no. 7: 600. https://doi.org/10.3390/nano14070600
APA StyleBardales, A. C., Vo, Q., & Kolpashchikov, D. M. (2024). Singleton {NOT} and Doubleton {YES; NOT} Gates Act as Functionally Complete Sets in DNA-Integrated Computational Circuits. Nanomaterials, 14(7), 600. https://doi.org/10.3390/nano14070600






