Carbon Nanodisks Decorated with Guanidinylated Hyperbranched Polyethyleneimine Derivatives as Efficient Antibacterial Agents
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Preparation of GPEI and oxCNDs
2.3. Preparation of GPEI-Functionalized oxCNDs
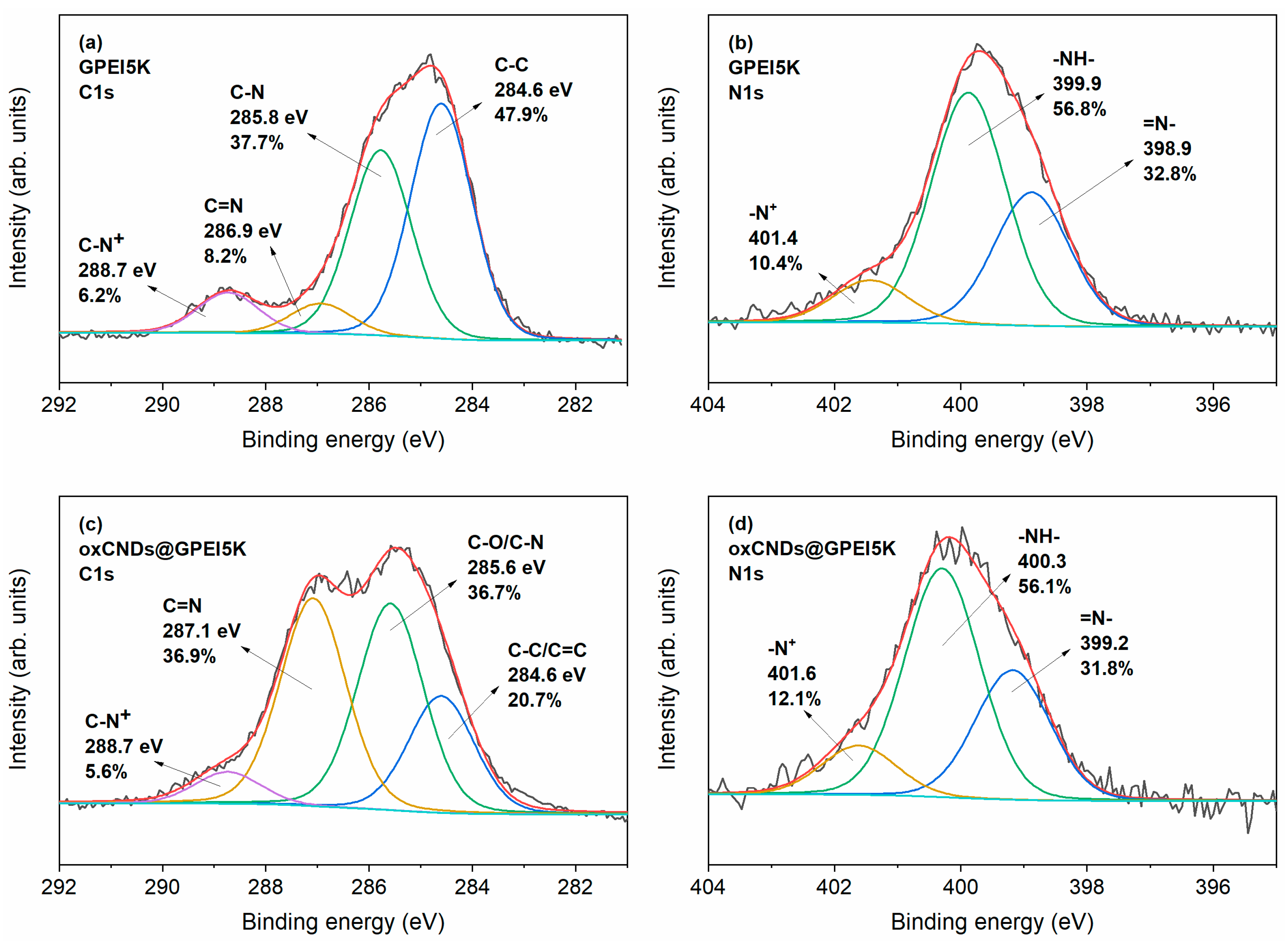
2.4. Characterization of GPEI-Functionalized oxCNDs
2.5. Assessment of the Antibacterial Properties
2.5.1. Test Microorganisms
2.5.2. Determination of Minimum Inhibitory Concentration and Minimum Bactericidal Concentration
2.5.3. Bacteria Morphology (SEM)
2.6. Evaluation of Cell Cytotoxicity
3. Results and Discussion
3.1. Synthesis and Characterization of GPEI-Functionalized oxCNDs
3.2. Aqueous Colloidal Stability of GPEI-Functionalized oxCNDs
3.3. In Vitro Viability Studies
3.4. Evaluation of Antibacterial Properties
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Makabenta, J.M.V.; Nabawy, A.; Li, C.H.; Schmidt-Malan, S.; Patel, R.; Rotello, V.M. Nanomaterial-based therapeutics for antibiotic-resistant bacterial infections. Nat. Rev. Microbiol. 2021, 19, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Barani, M.; Zeeshan, M.; Kalantar-Neyestanaki, D.; Farooq, M.; Rahdar, A.; Jha, N.K.; Sargazi, S.; Gupta, P.K.; Thakur, V.K. Nanomaterials in the Management of Gram-Negative Bacterial Infections. Nanomaterials 2021, 11, 2535. [Google Scholar] [CrossRef] [PubMed]
- Vallet-Regí, M.; González, B.; Izquierdo-Barba, I. Nanomaterials as Promising Alternative in the Infection Treatment. Int. J. Mol. Sci. 2019, 20, 3806. [Google Scholar] [CrossRef] [PubMed]
- Yaragalla, S.; Bhavitha, K.B.; Athanassiou, A. A Review on Graphene Based Materials and Their Antimicrobial Properties. Coatings 2021, 11, 1197. [Google Scholar] [CrossRef]
- Kumar, P.; Huo, P.; Zhang, R.; Liu, B. Antibacterial Properties of Graphene-Based Nanomaterials. Nanomaterials 2019, 9, 737. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Zhang, L.; Wang, Z.; Luo, Y. Mechanisms of the antimicrobial activities of graphene materials. J. Am. Chem. Soc. 2016, 138, 2064–2077. [Google Scholar] [CrossRef]
- Hegab, H.M.; ElMekawy, A.; Zou, L.; Mulcahy, D.; Saint, C.P.; Ginic-Markovic, M. The controversial antibacterial activity of graphene-based materials. Carbon 2016, 105, 362–376. [Google Scholar] [CrossRef]
- Mohammed, H.; Kumar, A.; Bekyarova, E.; Al-Hadeethi, Y.; Zhang, X.; Chen, M.; Ansari, M.S.; Cochis, A.; Rimondini, L. Antimicrobial Mechanisms and Effectiveness of Graphene and Graphene-Functionalized Biomaterials. A Scope Review. Front. Bioeng. Biotechnol. 2020, 8, 465. [Google Scholar] [CrossRef]
- Pulingam, T.; Thong, K.L.; Appaturi, J.N.; Lai, C.W.; Leo, B.F. Mechanistic actions and contributing factors affecting the antibacterial property and cytotoxicity of graphene oxide. Chemosphere 2021, 281, 130739. [Google Scholar] [CrossRef]
- Pham, V.T.; Truong, V.K.; Quinn, M.D.; Notley, S.M.; Guo, Y.; Baulin, V.A.; Al Kobaisi, M.; Crawford, R.J.; Ivanova, E.P. Graphene induces formation of pores that kill spherical and rod-shaped bacteria. ACS Nano 2015, 9, 8458–8467. [Google Scholar] [CrossRef]
- Akhavan, O.; Ghaderi, E. Toxicity of graphene and graphene oxide nanowalls against bacteria. ACS Nano 2010, 4, 5731–5736. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zeng, T.H.; Hofmann, M.; Burcombe, E.; Wei, J.; Jiang, R.; Kong, J.; Chen, Y. Antibacterial activity of graphite, graphite oxide, graphene oxide, and reduced graphene oxide: Membrane and oxidative stress. ACS Nano 2011, 5, 6971–6980. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.H.; Chen, G.Y.; Xia, M.Y.; Xie, Y.; Chi, Y.Q.; He, Z.Y.; Zhang, C.L.; Zhang, T.; Chen, Q.M.; Peng, Q. Understanding the Sheet Size-Antibacterial Activity Relationship of Graphene Oxide and the Nano-Bio Interaction-Based Physical Mechanisms. Colloids Surf. B Biointerfaces 2020, 191, 111009. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Hu, M.; Zeng, T.H.; Wu, R.; Jiang, R.; Wei, J.; Wang, L.; Kong, J.; Chen, Y. Lateral Dimension-Dependent Antibacterial Activity of Graphene Oxide Sheets. Langmuir 2012, 28, 12364–12372. [Google Scholar] [CrossRef] [PubMed]
- Majidi, H.J.; Babaei, A.; Bafrani, Z.A.; Shahrampour, D.; Zabihi, E.; Jafari, S.M. Investigating the best strategy to diminish the toxicity and enhance the antibacterial activity of graphene oxide by chitosan addition. Carbohydr. Polym. 2019, 225, 115220. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Gao, Y.; Sun, Z.; Chang, D.; Gao, G.; Dong, A. Synthesis, Characterization, and Bactericidal Evaluation of Chitosan/Guanidine Functionalized Graphene Oxide Composites. Molecules 2016, 22, 12. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.M.; Tria, M.C.R.; Vergara, R.A.M.V.; Ahmed, F.; Advincula, R.C.; Rodrigues, D.F. Antimicrobial graphene polymer (PVK-GO) nanocomposite films. ChemComm 2011, 47, 8892. [Google Scholar] [CrossRef]
- Some, S.; Ho, S.-M.; Dua, P.; Hwang, E.; Shin, Y.H.; Yoo, H.; Kang, J.-S.; Lee, D.-K.; Lee, H. Dual Functions of Highly Potent Graphene Derivative–Poly-l-Lysine Composites To Inhibit Bacteria and Support Human Cells. ACS Nano 2012, 6, 7151–7161. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Sun, S.; Dong, A.; Hao, Y.; Shi, S.; Sun, Z.; Gao, G.; Chen, Y. Developing of a novel antibacterial agent by functionalization of graphene oxide with guanidine polymer with enhanced antibacterial activity. Appl. Surf. Sci. 2015, 355, 446–452. [Google Scholar] [CrossRef]
- Fan, Z.; Po, K.H.L.; Wong, K.K.; Chen, S.; Lau, S.P. Polyethylenimine-Modified Graphene Oxide as a Novel Antibacterial Agent and Its Synergistic Effect with Daptomycin for Methicillin-Resistant Staphylococcus aureus. ACS Appl. Nano Mater. 2018, 1, 1811–1818. [Google Scholar] [CrossRef]
- Krishnan, A.; Dujardinm, E.; Treacy MM, J.; Hugdahl, J.; Lynum, S.; Ebbesen, T.W. Graphitic cones and the nucleation of curved carbon surfaces. Nature 1997, 388, 451–454. [Google Scholar] [CrossRef]
- Lynum, S.; Hugdahl, J.; Hox, K.; Hildrum, R.; Nordvik, M. Micro-Domain Graphitic Materials and Method for Producing the Same. U.S. Patent 7,462,343, 9 December 2008. [Google Scholar]
- Zhang, W.; Dubois, M.; Guerin, K.; Bonnet, P.; Petit, E.; Delpuech, N.; Albertini, D.; Masin, F.; Hamwi, A. Effect of graphitization on fluorination of carbon nanocones and nanodiscs. Carbon 2009, 47, 2763–2775. [Google Scholar] [CrossRef]
- Garberg, T.; Naess, S.N.; Helgesen, G.; Knudsen, K.D.; Kopstad, G.; Elgsaeter, A. A transmission electron microscope and electron diffraction study of carbon nanodisks. Carbon 2008, 46, 1535–1543. [Google Scholar] [CrossRef]
- Staudenmaier, L. Verfahren zur Darstellung der Graphitsäure. Berichte Der Dtsch. Chem. Ges. 1898, 31, 1481–1499. [Google Scholar] [CrossRef]
- Sali, S.; Mackey, H.R.; Abdala, A.A. Effect of graphene oxide synthesis method on properties and performance of polysulfone-graphene oxide mixed matrix membranes. Nanomaterials 2019, 9, 769. [Google Scholar] [CrossRef] [PubMed]
- Poh, H.L.; Šaněk, F.; Ambrosi, A.; Zhao, G.; Sofer, Z.; Pumera, M. Graphenes prepared by Staudenmaier, Hofmann and Hummers methods with consequent thermal exfoliation exhibit very different electrochemical properties. Nanoscale 2012, 4, 3515–3522. [Google Scholar] [CrossRef] [PubMed]
- Zygouri, P.; Tsoufis, T.; Kouloumpis, A.; Patila, M.; Potsi, G.; Sevastos, A.A.; Sideratou, Z.; Katsaros, F.; Charalambopoulou, G.; Stamatis, H.; et al. Synthesis, characterization and assessment of hydrophilic oxidized carbon nanodiscs in bio-related applications. RSC Adv. 2018, 8, 122–131. [Google Scholar] [CrossRef]
- Lin, C.-T.; Lee, C.-Y.; Chiu, H.-T.; Chin, T.-S. Graphene Structure in Carbon Nanocones and Nanodiscs. Langmuir 2007, 23, 12806–12810. [Google Scholar] [CrossRef] [PubMed]
- Osváth, Z.; Vértesy, Z.; Lábár, J.; Nemes-Incze, P.; Horváth, Z.E.; Biró, L.P. Substrate-induced strain in carbon nanodisks. Thin Solid Films 2014, 565, 111–115. [Google Scholar] [CrossRef]
- Lalwani, G.; D’Agati, M.; Khan, A.M.; Sitharaman, B. Toxicology of graphene-based nanomaterials. Adv. Drug Deliv Rev. 2016, 105, 109–144. [Google Scholar] [CrossRef]
- Madannejad, R.; Shoaie, N.; Jahanpeyma, F.; Darvishi, M.H.; Azimzadeh, M.; Javadi, H. Toxicity of carbon-based nanomaterials: Reviewing recent reports in medical and biological systems. Chem. Biol. Interact. 2019, 307, 206–222. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Zhang, X.; Sun, L.; Wei, Y.; Wei, X. Cellular Toxicity and Immunological Effects of Carbon-based Nanomaterials. Part Fibre Toxicol. 2019, 16, 18. [Google Scholar] [CrossRef]
- Hu, W.; Peng, C.; Luo, W.; Lv, M.; Li, X.; Li, D.; Huang, Q.; Fan, C. Graphene-Based Antibacterial Paper. ACS Nano 2010, 4, 4317–4323. [Google Scholar] [CrossRef] [PubMed]
- Lyra, K.-M.; Kaminari, A.; Panagiotaki, K.N.; Spyrou, K.; Papageorgiou, S.; Sakellis, E.; Katsaros, F.K.; Sideratou, Z. Multi-Walled Carbon Nanotubes Decorated with Guanidinylated Dendritic Molecular Transporters: An Efficient Platform for the Selective Anticancer Activity of Doxorubicin. Pharmaceutics 2021, 13, 858. [Google Scholar] [CrossRef]
- Heliopoulos, N.S.; Kythreoti, G.; Lyra, K.M.; Panagiotaki, K.N.; Papavasiliou, A.; Sakellis, E.; Papageorgiou, S.; Kouloumpis, A.; Gournis, D.; Katsaros, F.K.; et al. Cytotoxicity Effects of Water-Soluble Multi-Walled Carbon Nanotubes Decorated with Quaternized Hyperbranched Poly(ethyleneimine) Derivatives on Autotrophic and Heterotrophic Gram-Negative Bacteria. Pharmaceuticals 2020, 13, 293. [Google Scholar] [CrossRef] [PubMed]
- Rundlof, T.; Mathiasson, M.; Bekiroglu, S.; Hakkarainen, B.; Bowden, T.; Arvidsson, T. Survey and qualification of internal standards for quantification by 1H NMR spectroscopy. J. Pharm. Biomed. Anal. 2010, 52, 645–651. [Google Scholar] [CrossRef]
- Bouhdid, S.; Abrini, J.; Zhiri, A.; Espuny, M.J.; Manresa, A. Investigation of functional and morphological changes in Pseudomonas aeruginosa and Staphylococcus aureus cells induced by Origanum compactum essential oil. J. Appl. Microbiol. 2009, 106, 1558–1568. [Google Scholar] [CrossRef]
- Kalchayanand, N.; Dunne, P.; Sikes, A.; Ray, B. Viability loss and morphology change of foodborne pathogens following exposure to hydrostatic pressures in the presence and absence of bacteriocins. Int. J. Food Microbiol. 2004, 91, 91–98. [Google Scholar] [CrossRef]
- Sideratou, Z.; Agathokleous, M.; Theodossiou, T.A.; Tsiourvas, D. Functionalized Hyperbranched Polyethylenimines as Thermosensitive Drug Delivery Nanocarriers with Controlled Transition Temperatures. Biomacromolecules 2018, 19, 315–328. [Google Scholar] [CrossRef]
- Tsoufis, T.; Katsaros, F.; Sideratou, Z.; Kooi, B.J.; Karakassides, M.A.; Siozios, A. Intercalation Study of Low-Molecular-Weight Hyperbranched Polyethyleneimine into Graphite Oxide. Eur. J. Chem. 2014, 20, 8129–8137. [Google Scholar] [CrossRef]
- Tsoufis, T.; Katsaros, F.; Sideratou, Z.; Romanos, G.; Ivashenko, O.; Rudolf, P.; Kooi, B.J.; Papageorgiou, S.; Karakassides, M.A. Tailor-made graphite oxide–DAB poly(propylene imine) dendrimer intercalated hybrids and their potential for efficient CO2 adsorption. Chem. Commun. 2014, 50, 10967–10970. [Google Scholar] [CrossRef] [PubMed]
- Spyrou, K.; Calvaresi, M.; Diamanti, E.K.; Tsoufis, T.; Gournis, D.; Rudolf, P.; Zerbetto, F. Graphite Oxide and Aromatic Amines: Size Matters. Adv. Funct. Mat. 2015, 25, 263–269. [Google Scholar] [CrossRef]
- Bera, M.; Chandravati Gupta, P.; Maji, P.K. Facile One-Pot Synthesis of Graphene Oxide by Sonication Assisted Mechanochemical Approach and Its Surface Chemistry. J. Nanosci. Nanotechnol. 2018, 18, 902–912. [Google Scholar] [CrossRef] [PubMed]
- Claramunt, S.; Varea, A.; López-Díaz, D.; Velázquez, M.M.; Cornet, A.; Cirera, A. The Importance of Interbands on the Interpretation of the Raman Spectrum of Graphene Oxide. J. Phys. Chem. C 2015, 119, 10123–10129. [Google Scholar] [CrossRef]
- Alkhouzaam, A.; Abdelrazeq, H.; Khraisheh, M.; AlMomani, F.; Hameed, B.H.; Hassan, M.K.; Al-Ghouti, M.A.; Selvaraj, R. Spectral and Structural Properties of High-Quality Reduced Graphene Oxide Produced via a Simple Approach Using Tetraethylenepentamine. Nanomaterials 2022, 12, 1240. [Google Scholar] [CrossRef] [PubMed]
- Zafar, Z.; Ni, Z.H.; Wu, X.; Shi, Z.X.; Nan, H.Y.; Bai, J.; Sun, L.T. Evolution of Raman spectra in nitrogen doped graphene. Carbon 2013, 61, 57–62. [Google Scholar] [CrossRef]
- Eda, G.; Chhowalla, M. Chemically Derived Graphene Oxide: Towards Large-Area Thin-Film Electronics and Optoelectronics. Adv. Mater. 2010, 22, 2392–2415. [Google Scholar] [CrossRef]
- Mustafa, R.; Fitian, M.; Hamilton, N.B.; Li, J.; Silva, W.R.; Punihaole, D. Molecular Insights into the Binding of Linear Polyethylenimines and Single-Stranded DNA Using Raman Spectroscopy: A Quantitative Approach. J. Phys. Chem. B 2022, 126, 8404–8414. [Google Scholar] [CrossRef]
- Liu, H.; Kuila, T.; Kim, N.H.; Ku, B.-C.; Lee, J.H. In situ synthesis of the reduced graphene oxide–polyethyleneimine composite and its gas barrier properties. J. Mater. Chem. A 2013, 1, 3739–3746. [Google Scholar] [CrossRef]
- Johra, F.T.; Lee, J.-W.; Jung, W.-G. Facile and safe graphene preparation on solution based platform. J. Ind. Eng. Chem. 2014, 20, 2883–2887. [Google Scholar] [CrossRef]
- Zhou, X.; Chen, Z.; Yan, D.; Lu, H. Deposition of Fe–Ni nanoparticles on polyethyleneimine-decorated graphene oxide and application in catalytic dehydrogenation of ammonia borane. J. Mater. Chem. 2012, 22, 13506–13516. [Google Scholar] [CrossRef]
- Rani, S.; Kumar, M.; Kumar, R.; Kumar, D.; Sharma, S.; Singh, G. Characterization and dispersibility of improved thermally stable amide functionalized graphene oxide. Mater. Res. Bull. 2014, 60, 143–149. [Google Scholar] [CrossRef]
- Bhattacharjee, S. DLS and zeta potential—What they are and what they are not? J. Control. Release 2016, 235, 337–351. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, Approved Standard, 9th ed.; CLSI document M07-A9; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- CLSI. Methods for Determining Bactericidal Activity of Antimicrobial Agents. Approved Guideline; CLSI document M26-A; Clinical and Laboratory Stan- dards Institute: Wayne, PA, USA, 1998. [Google Scholar]
- Qian, L.; Xiao, H.; Zhao, G.; He, B. Synthesis of modified guanidine-based polymers and their antimicrobial activities revealed by AFM and CLSM. ACS Appl. Mater. Interfaces 2011, 3, 1895–1901. [Google Scholar] [CrossRef] [PubMed]
- Heredero-Bermejo, I.; Hernández-Ros, J.M.; Sánchez-García, L.; Maly, M.; Verdú-Expósito, C.; Soliveri, J.; De la Mata, J.F.; Copa-Patiño, J.L.; Pérez-Serrano, J.; Sánchez-Nieves, J.; et al. Ammonium and guanidine carbosilane dendrimers and dendrons as microbicides. Eur. Polym. J. 2018, 101, 159–168. [Google Scholar] [CrossRef]
- Zamperini, C.; Maccari, G.; Deodato, D.; Pasero, C.; D’Agostino, I.; Orofino, F.; De Luca, F.; Dreassi, E.; Docquier, J.D.; Botta, M. Identification, synthesis and biological activity of alkyl-guanidine oligomers as potent antibacterial agents. Sci. Rep. 2017, 7, 8251. [Google Scholar] [CrossRef] [PubMed]
- Tsogas, I.; Sideratou, Z.; Tsiourvas, D.; Theodossiou, T.A.; Paleos, C.M. Interactive transport of guanidinylated poly(propylene imine)-based dendrimers through liposomal and cellular membranes. ChemBioChem 2007, 8, 1865–1876. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lu, C.; Shi, Y.; Feng, X.; Wu, B.; Zhou, G.; Quan, G.; Pan, X.; Cai, J.; Wu, C. Structural superiority of guanidinium-rich, four-armed copolypeptides: Role of multiple peptide–membrane interactions in enhancing bacterial membrane perturbation and permeability. ACS Appl. Mater. Interfaces 2020, 12, 18363–18374. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Zhang, Z.; Ma, H.; Chen, Y. In Vitro Hemocompatibility and Toxic Mechanism of Graphene Oxide on Human Peripheral Blood T Lymphocytes and Serum Albumin. ACS Appl. Mater. Interfaces 2014, 6, 19797–19807. [Google Scholar] [CrossRef]
- Silhavy, T.J.; Kahne, D.; Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Epand, R.M.; Walker, C.; Epand, R.F.; Magarvey, N.A. Molecular mechanisms of membrane targeting antibiotics. Biochim. Biophys. Acta Biomembr. 2016, 1858, 980–987. [Google Scholar] [CrossRef] [PubMed]
- Zielonka, J.; Joseph, J.; Sikora, A.; Hardy, M.; Ouari, O.; Vasquez-Vivar, J.; Cheng, G.; Lopez, M.; Kalyanaraman, B. Mitochondria-targeted triphenylphosphonium-based compounds: Syntheses, mechanisms of action, and therapeutic and diagnostic applications. Chem. Rev. 2017, 117, 10043–10120. [Google Scholar] [CrossRef]
- Khater, M.; Khater, S.; Gholap, H.; Patil, R.; Kulkarni, G. Comparative studies on measurement of membrane potential of bacterial cells treated with ZnO nanoparticles by Spectrofluorometry, fluorescence microscopy and flowcytometry. J. Microbiol. Methods 2020, 173, 105920. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.B. Mitochondrial membrane potential in living cells. Annu. Rev. Cell Biol. 1988, 4, 155–181. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Chi, J.; Yan, Y.; Luo, R.; Feng, X.; Zheng, Y.; Xian, D.; Li, X.; Quan, G.; Liu, D.; et al. Membrane-disruptive peptides/peptidomimetics-based therapeutics: Promising systems to combat bacteria and cancer in the drug-resistant era. Acta Pharm. Sin. B 2021, 11, 2609–2644. [Google Scholar] [CrossRef]
- Teixeira, V.; Feio, M.J.; Bastos, M. Role of lipids in the interaction of antimicrobial peptides with membranes. Prog. Lipid Res. 2012, 51, 149–177. [Google Scholar] [CrossRef]








| Sample | ||||
|---|---|---|---|---|
| CNDs | 0.15 | 0.06 | 1.03 | ∞ |
| oxCNDs | 1.56 | 1.14 | 0.93 | 1.46 |
| oxCNDs@GPEI5K | - | 0.81 | - | - |
| oxCNDs@GPEI25K | - | 0.76 | - | - |
| Sample | (001)A Peak | (001)B Peak | ||||
|---|---|---|---|---|---|---|
| 2θ (deg.) | d (Å) | A (%) | 2θ (deg.) | d (Å) | A (%) | |
| oxCNDs | 11.3 | 7.82 | 100 | - | - | - |
| oxCNDs@GPEI5K | 11.3 | 7.82 | 62.6 | 10.7 | 8.26 | 37.4 |
| oxCNDs@GPEI25K | 11.4 | 7.76 | 61.7 | 10.3 | 8.58 | 38.3 |
| Samples | E. coli | S. aureus | ||
|---|---|---|---|---|
| MIC (μg/mL) | MBC (μg/mL) | MIC (μg/mL) | MBC (μg/mL) | |
| oxCNDs | >750 | >750 | >750 | 750 |
| oxCNDs@GPEI5K | 250 | 280 | 150 | 200 |
| oxCNDs@GPEI25K | 300 | 300 | 200 | 250 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lyra, K.-M.; Tournis, I.; Subrati, M.; Spyrou, K.; Papavasiliou, A.; Athanasekou, C.; Papageorgiou, S.; Sakellis, E.; Karakassides, M.A.; Sideratou, Z. Carbon Nanodisks Decorated with Guanidinylated Hyperbranched Polyethyleneimine Derivatives as Efficient Antibacterial Agents. Nanomaterials 2024, 14, 677. https://doi.org/10.3390/nano14080677
Lyra K-M, Tournis I, Subrati M, Spyrou K, Papavasiliou A, Athanasekou C, Papageorgiou S, Sakellis E, Karakassides MA, Sideratou Z. Carbon Nanodisks Decorated with Guanidinylated Hyperbranched Polyethyleneimine Derivatives as Efficient Antibacterial Agents. Nanomaterials. 2024; 14(8):677. https://doi.org/10.3390/nano14080677
Chicago/Turabian StyleLyra, Kyriaki-Marina, Ioannis Tournis, Mohammed Subrati, Konstantinos Spyrou, Aggeliki Papavasiliou, Chrysoula Athanasekou, Sergios Papageorgiou, Elias Sakellis, Michael A. Karakassides, and Zili Sideratou. 2024. "Carbon Nanodisks Decorated with Guanidinylated Hyperbranched Polyethyleneimine Derivatives as Efficient Antibacterial Agents" Nanomaterials 14, no. 8: 677. https://doi.org/10.3390/nano14080677
APA StyleLyra, K.-M., Tournis, I., Subrati, M., Spyrou, K., Papavasiliou, A., Athanasekou, C., Papageorgiou, S., Sakellis, E., Karakassides, M. A., & Sideratou, Z. (2024). Carbon Nanodisks Decorated with Guanidinylated Hyperbranched Polyethyleneimine Derivatives as Efficient Antibacterial Agents. Nanomaterials, 14(8), 677. https://doi.org/10.3390/nano14080677










