Graphene Oxide-Based Biosensors for Liquid Biopsies in Cancer Diagnosis
Abstract
:1. Cancer and Diagnosis
2. Liquid Biopsies
3. Circulating Tumor Cells (CTCs)
4. Circulating Tumor DNA (ctDNA)
5. Exosome
6. Biomedical Diagnostic Applications of GO
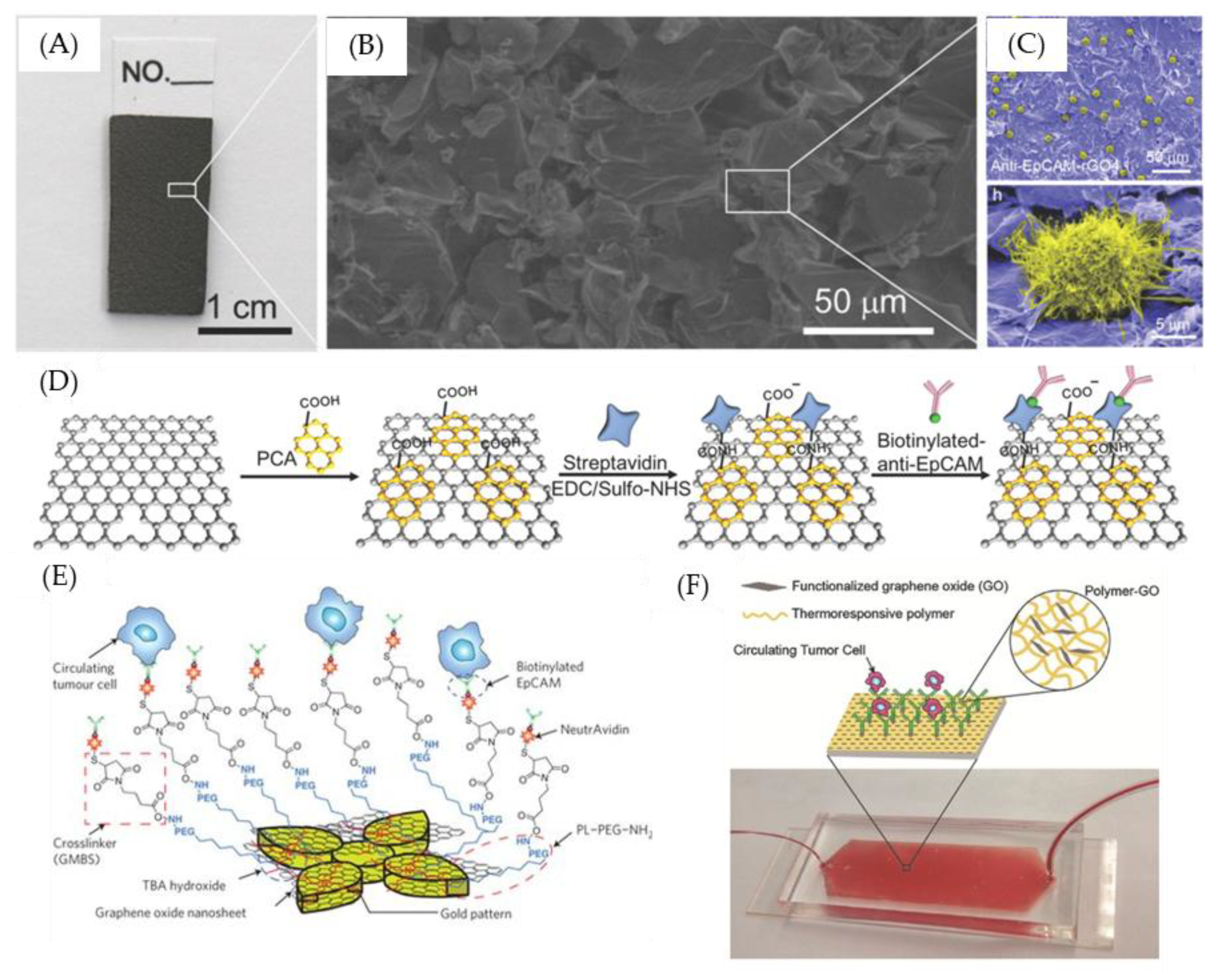
7. GO-Nanointerface for CTC Diagnosis
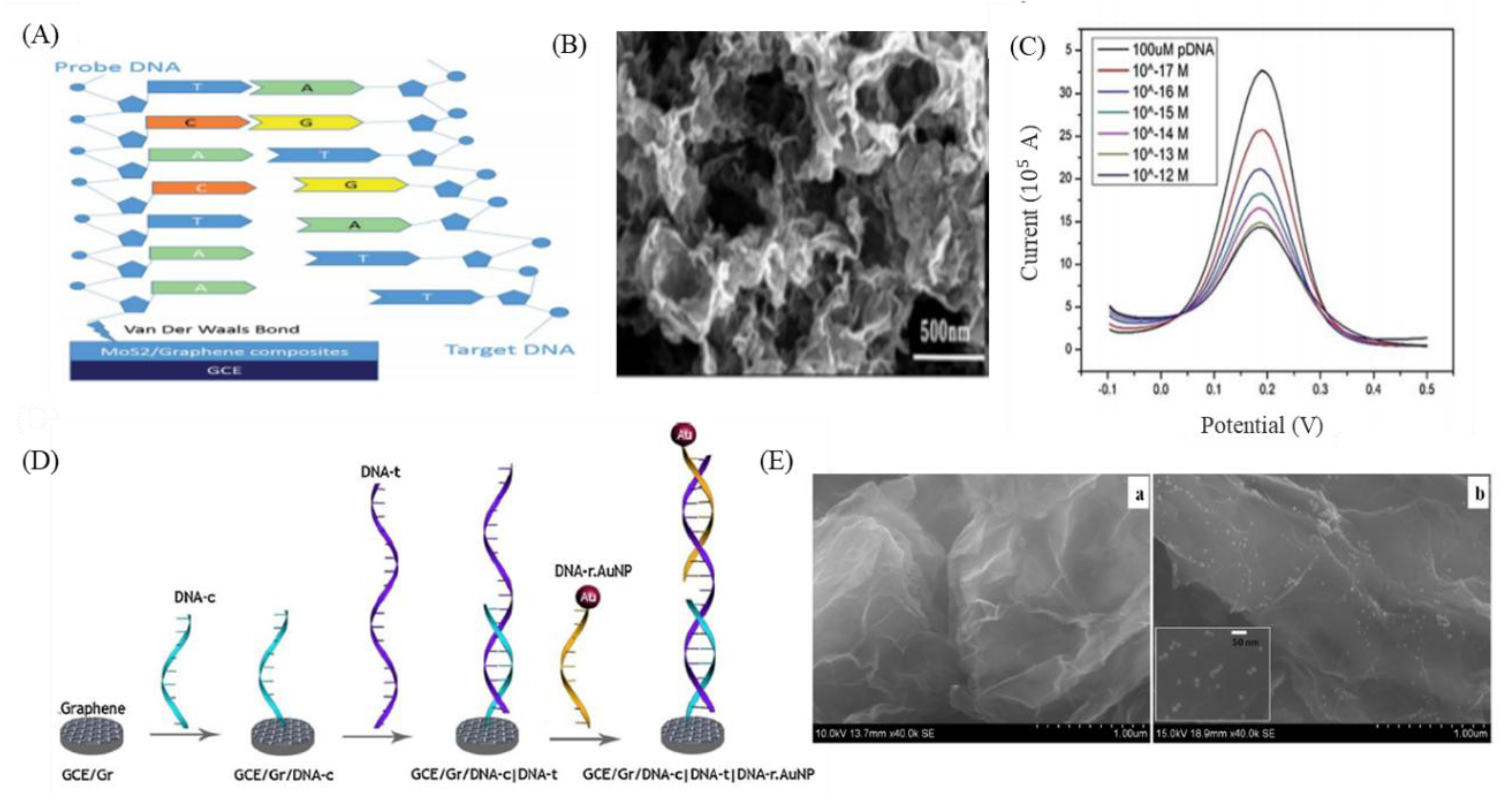
8. GO-Nanointerface for Gene Probe Diagnosis
9. GO-Nanointerface for Exosome Diagnosis
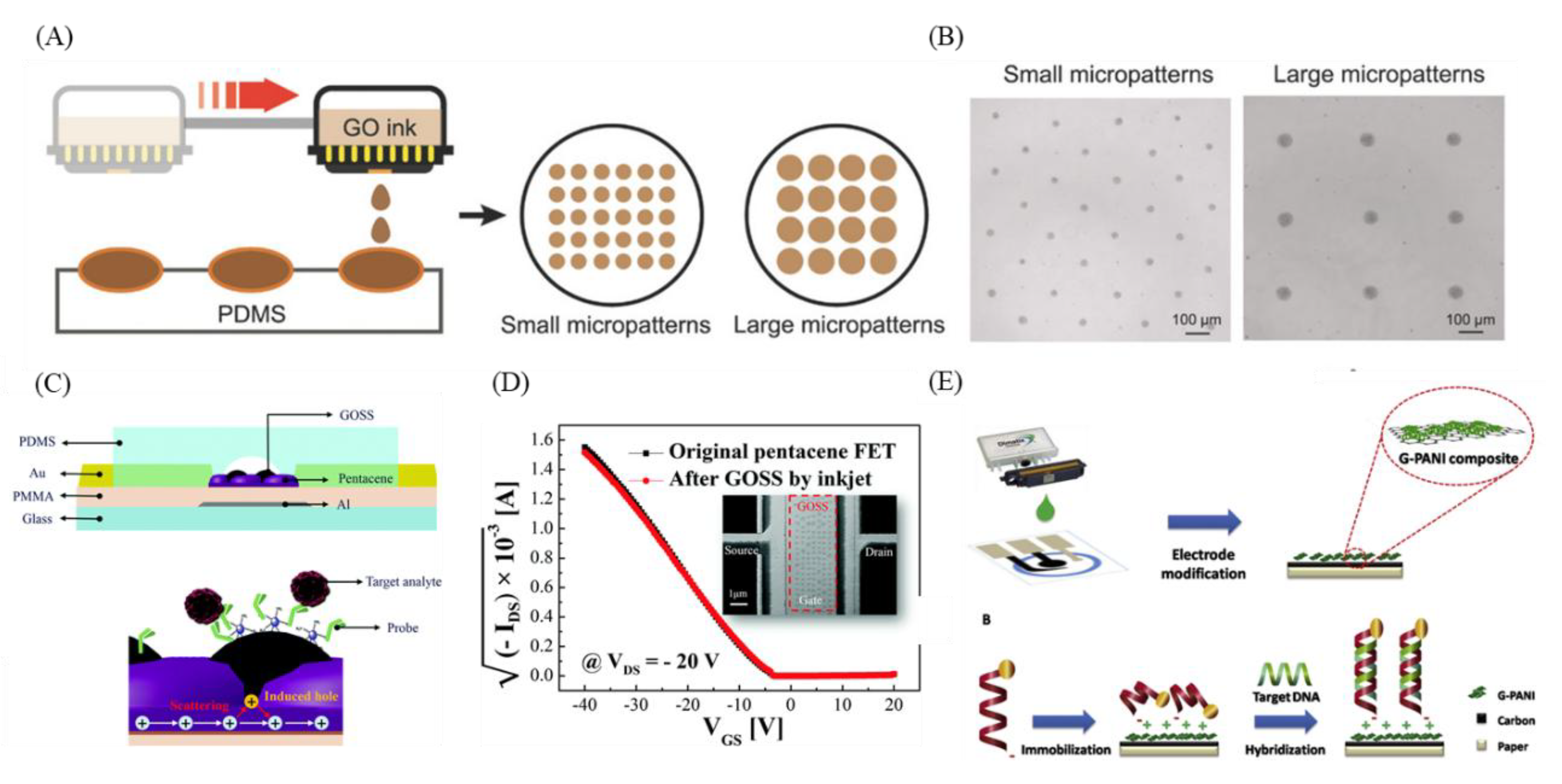
10. GO-3D Printing and Micropatterning for Diagnosis
11. GO-3D Printing and Micropatterning in CTC, ctDNA, and Exosome Diagnosis
12. Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Bouck, N. Tumor angiogenesis: The role of oncogenes and tumor suppressor genes. Cancer Cells (Cold Spring Harbor) 1990, 2, 179–185. [Google Scholar]
- Feitelson, M.A.; Arzumanyan, A.; Kulathinal, R.J.; Blain, S.W.; Holcombe, R.F.; Mahajna, J.; Marino, M.; Martinez-Chantar, M.L.; Nawroth, R.; Sanchez-Garcia, I.; et al. Sustained proliferation in cancer: Mechanisms and novel therapeutic targets. Semin. Cancer Biol. 2015, 35, S25–S54. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, J.; Rodriguez, V.; Bodey, G.P. Causes of death in cancer patients. Cancer 1974, 33, 568–573. [Google Scholar] [CrossRef]
- Edge, S.B.; Compton, C.C. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann. Surg. Oncol. 2010, 17, 1471–1474. [Google Scholar] [CrossRef] [PubMed]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; Tarpey, P. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [Green Version]
- Marusyk, A.; Polyak, K. Tumor heterogeneity: Causes and consequences. Biochim. Biophys. Acta -Rev. Cancer 2010, 1805, 105–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srinivas, P.R.; Kramer, B.S.; Srivastava, S. Trends in biomarker research for cancer detection. Lancet Oncol. 2001, 2, 698–704. [Google Scholar] [CrossRef]
- Brock, G.; Castellanos-Rizaldos, E.; Hu, L.; Coticchia, C.; Skog, J. Liquid biopsy for cancer screening, patient stratification and monitoring. Transl. Cancer Res. 2015, 4, 280–290. [Google Scholar]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef]
- De Vlaminck, I.; Martin, L.; Kertesz, M.; Patel, K.; Kowarsky, M.; Strehl, C.; Cohen, G.; Luikart, H.; Neff, N.F.; Okamoto, J.; et al. Noninvasive monitoring of infection and rejection after lung transplantation. Proc. Natl. Acad. Sci. USA 2015, 112, 13336–13341. [Google Scholar] [CrossRef] [Green Version]
- Duffy, M.J. Serum tumor markers in breast cancer: Are they of clinical value? Clin. Chem. 2006, 52, 345–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fazel, R.; Krumholz, H.M.; Wang, Y.; Ross, J.S.; Chen, J.; Ting, H.H.; Shah, N.D.; Nasir, K.; Einstein, A.J.; Nallamothu, B.K. Exposure to low-dose ionizing radiation from medical imaging procedures. N. Engl. J. Med. 2009, 361, 849–857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, C.-H.; Lim, S.C.; Wu, L.-C.; Wang, C.-F.; Tsai, W.-S.; Wu, H.-C.; Chang, Y.-C. Site-specific antibody modification and immobilization on a microfluidic chip to promote the capture of circulating tumor cells and microemboli. Chem. Commun. 2017, 53, 4152–4155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanderlaan, P.A.; Yamaguchi, N.; Folch, E.; Boucher, D.H.; Kent, M.S.; Gangadharan, S.P.; Majid, A.; Goldstein, M.A.; Huberman, M.S.; Kocher, O.N.; et al. Success and failure rates of tumor genotyping techniques in routine pathological samples with non-small-cell lung cancer. Lung Cancer 2014, 84, 39–44. [Google Scholar] [CrossRef] [Green Version]
- Paget, S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 1989, 8, 98–101. [Google Scholar]
- Thiery, J.P.; Sleeman, J.P. Complex networks orchestrate epithelial–mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 2006, 7, 131. [Google Scholar] [CrossRef]
- Yang, J.; Mani, S.A.; Weinberg, R.A. Exploring a new twist on tumor metastasis. Cancer Res. 2006, 66, 4549–4552. [Google Scholar] [CrossRef] [Green Version]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [Green Version]
- Alunni-Fabbroni, M.; Sandri, M.T. Circulating tumour cells in clinical practice: Methods of detection and possible characterization. Methods 2010, 50, 289–297. [Google Scholar] [CrossRef]
- Paterlini-Brechot, P.; Benali, N.L. Circulating tumor cells (CTC) detection: Clinical impact and future directions. Cancer Lett. 2007, 253, 180–204. [Google Scholar] [CrossRef]
- Sun, Y.; Haglund, T.A.; Rogers, A.J.; Ghanim, A.F.; Sethu, P. Microfluidics technologies for blood-based cancer liquid biopsies. Anal. Chim. Acta 2018, 1012, 10–29. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Sozzani, S.; Locati, M.; Allavena, P.; Sica, A. Macrophage polarization: Tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002, 23, 549–555. [Google Scholar] [CrossRef]
- Leon, S.; Shapiro, B.; Sklaroff, D.; Yaros, M. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar] [PubMed]
- Stroun, M.; Anker, P.; Maurice, P.; Lyautey, J.; Lederrey, C.; Beljanski, M. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology 1989, 46, 318–322. [Google Scholar] [CrossRef]
- Wan, J.C.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef]
- Dressman, D.; Yan, H.; Traverso, G.; Kinzler, K.W.; Vogelstein, B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc. Natl. Acad. Sci. USA 2003, 100, 8817–8822. [Google Scholar] [CrossRef] [Green Version]
- Newton, C.; Graham, A.; Heptinstall, L.; Powell, S.; Summers, C.; Kalsheker, N.; Smith, J.; Markham, A. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res. 1989, 17, 2503–2516. [Google Scholar] [CrossRef]
- Freidin, M.B.; Freydina, D.V.; Leung, M.; Fernandez, A.M.; Nicholson, A.G.; Lim, E. Circulating tumor DNA outperforms circulating tumor cells for KRAS mutation detection in thoracic malignancies. Clin. Chem. 2015, 61, 1299–1304. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.Y.; Ugozzoli, L.; Pal, B.K.; Wallace, R.B. Allele-specific enzymatic amplification of beta-globin genomic DNA for diagnosis of sickle cell anemia. Proc. Natl. Acad. Sci. USA 1989, 86, 2757–2760. [Google Scholar] [CrossRef] [Green Version]
- Vogelstein, B.; Kinzler, K.W. Digital pcr. Proc. Natl. Acad. Sci. USA 1999, 96, 9236–9241. [Google Scholar] [CrossRef] [Green Version]
- Alix-Panabières, C.; Pantel, K. Clinical prospects of liquid biopsies. Nat. Biomed. Eng. 2017, 1. [Google Scholar] [CrossRef]
- Zheng, D.; Ye, X.; Zhang, M.; Sun, Y.; Wang, J.; Ni, J.; Zhang, H.; Zhang, L.; Luo, J.; Zhang, J. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci. Rep. 2016, 6, 20913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, M.; Dawson, S.-J.; Tsui, D.W.; Gale, D.; Forshew, T.; Piskorz, A.M.; Parkinson, C.; Chin, S.-F.; Kingsbury, Z.; Wong, A.S. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013, 497, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Heitzer, E.; Ulz, P.; Belic, J.; Gutschi, S.; Quehenberger, F.; Fischereder, K.; Benezeder, T.; Auer, M.; Pischler, C.; Mannweiler, S. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 2013, 5, 30. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Yuen, S.T.; Xu, J.; Lee, S.P.; Yan, H.H.; Shi, S.T.; Siu, H.C.; Deng, S.; Chu, K.M.; Law, S. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat. Genet. 2014, 46, 573–582. [Google Scholar] [CrossRef]
- Shu, Y.; Wu, X.; Tong, X.; Wang, X.; Chang, Z.; Mao, Y.; Chen, X.; Sun, J.; Wang, Z.; Hong, Z. Circulating tumor DNA mutation profiling by targeted next generation sequencing provides guidance for personalized treatments in multiple cancer types. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Forshew, T.; Murtaza, M.; Parkinson, C.; Gale, D.; Tsui, D.W.; Kaper, F.; Dawson, S.-J.; Piskorz, A.M.; Jimenez-Linan, M.; Bentley, D. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med. 2012, 4, 136ra68. [Google Scholar] [CrossRef]
- Levy, B.; Hu, Z.I.; Cordova, K.N.; Close, S.; Lee, K.; Becker, D. Clinical utility of liquid diagnostic platforms in non-small cell lung cancer. Oncology 2016, 21, 1121–1130. [Google Scholar] [CrossRef] [Green Version]
- Johnstone, R. Maturation of reticulocytes: Formation of exosomes as a mechanism for shedding membrane proteins. Biochem. Cell Biol. 1992, 70, 179–190. [Google Scholar] [CrossRef]
- Pan, B.-T.; Johnstone, R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef]
- Pan, B.-T.; Teng, K.; Wu, C.; Adam, M.; Johnstone, R.M. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J. Cell Biol. 1985, 101, 942–948. [Google Scholar] [CrossRef] [Green Version]
- Tkach, M.; Théry, C. Communication by extracellular vesicles: Where we are and where we need to go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Keller, S.; Sanderson, M.P.; Stoeck, A.; Altevogt, P. Exosomes: From biogenesis and secretion to biological function. Immunol. Lett. 2006, 107, 102–108. [Google Scholar] [CrossRef]
- Meckes, D.G. Exosomal communication goes viral. J. Virol. 2015, 89, 5200–5203. [Google Scholar] [CrossRef] [Green Version]
- Barteneva, N.S.; Maltsev, N.; Vorobjev, I.A. Microvesicles and intercellular communication in the context of parasitism. Front. Cell. Infect. Microbiol. 2013, 3. [Google Scholar] [CrossRef] [Green Version]
- Kucharzewska, P.; Belting, M. Emerging roles of extracellular vesicles in the adaptive response of tumour cells to microenvironmental stress. J. Extracell. Vesicles 2013, 2. [Google Scholar] [CrossRef]
- Andaloussi, S.E.; Mäger, I.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef]
- De Toro, J.; Herschlik, L.; Waldner, C.; Mongini, C. Emerging roles of exosomes in normal and pathological conditions: New insights for diagnosis and therapeutic applications. Front. Immunol. 2015, 6, 203. [Google Scholar] [CrossRef] [Green Version]
- Soung, Y.H.; Ford, S.; Zhang, V.; Chung, J. Exosomes in cancer diagnostics. Cancers 2017, 9, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greening, D.W.; Xu, R.; Ji, H.; Tauro, B.J.; Simpson, R.J. A protocol for exosome isolation and characterization: Evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. In Proteomic Profiling; Springer: Berlin/Heidelberg, Germany, 2015; pp. 179–209. [Google Scholar]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30. [Google Scholar] [CrossRef]
- Alvarez, M.L.; Khosroheidari, M.; Ravi, R.K.; DiStefano, J.K. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 2012, 82, 1024–1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tauro, B.J.; Greening, D.W.; Mathias, R.A.; Ji, H.; Mathivanan, S.; Scott, A.M.; Simpson, R.J. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 2012, 56, 293–304. [Google Scholar] [CrossRef] [PubMed]
- McDonald, J.C.; Duffy, D.C.; Anderson, J.R.; Chiu, D.T.; Wu, H.; Schueller, O.J.; Whitesides, G.M. Fabrication of microfluidic systems in poly (dimethylsiloxane). Electrophor. Int. J. 2000, 21, 27–40. [Google Scholar] [CrossRef]
- Kim, H.; Namgung, R.; Singha, K.; Oh, I.K.; Kim, W.J. Graphene oxide-polyethylenimine nanoconstruct as a gene delivery vector and bioimaging tool. Bioconjug. Chem. 2011, 22, 2558–2567. [Google Scholar] [CrossRef]
- Chen, G.Y.; Pang, D.W.P.; Hwang, S.M.; Tuan, H.Y.; Hu, Y.C. A graphene-based platform for induced pluripotent stem cells culture and differentiation. Biomaterials 2012, 33, 418–427. [Google Scholar] [CrossRef]
- Zhang, L.; Lu, Z.; Zhao, Q.; Huang, J.; Shen, H.; Zhang, Z. Enhanced chemotherapy efficacy by sequential delivery of siRNA and anticancer drugs using PEI-grafted graphene oxide. Small 2011, 7, 460–464. [Google Scholar] [CrossRef]
- Zhang, L.; Xia, J.; Zhao, Q.; Liu, L.; Zhang, Z. Functional graphene oxide as a nanocarrier for controlled loading and targeted delivery of mixed anticancer drugs. Small 2010, 6, 537–544. [Google Scholar] [CrossRef]
- Yang, K.; Wan, J.; Zhang, S.; Tian, B.; Zhang, Y.; Liu, Z. The influence of surface chemistry and size of nanoscale graphene oxide on photothermal therapy of cancer using ultra-low laser power. Biomaterials 2012, 33, 2206–2214. [Google Scholar] [CrossRef]
- Levy, J.M.; Thorburn, A. Targeting autophagy during cancer therapy to improve clinical outcomes. Pharmacol. Ther. 2011, 131, 130–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.J.; Chee, C.E.; Huang, S.; Sinicrope, F.A. The role of autophagy in cancer: Therapeutic implications. Mol. Cancer Ther. 2011, 10, 1533–1541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Compton, O.C.; Nguyen, S.T. Graphene oxide, highly reduced graphene oxide, and graphene: Versatile building blocks for carbon-based materials. Small 2010, 6, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Murali, S.; Cai, W.; Li, X.; Suk, J.W.; Potts, J.R.; Ruoff, R.S. Graphene and graphene oxide: Synthesis, properties, and applications. Adv. Mater. 2010, 22, 3906–3924. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Kumar, R.; Singh, D.P. Graphene oxide: Strategies for synthesis, reduction and frontier applications. RSC Adv. 2016, 6, 64993–65011. [Google Scholar] [CrossRef]
- Shao, Y.; Wang, J.; Wu, H.; Liu, J.; Aksay, I.A.; Lin, Y. Graphene based electrochemical sensors and biosensors: A review. Electroanal. Int. J. Devoted Fundam. Pract. Asp. Electroanal. 2010, 22, 1027–1036. [Google Scholar] [CrossRef]
- Gao, L.; Lian, C.; Zhou, Y.; Yan, L.; Li, Q.; Zhang, C.; Chen, L.; Chen, K. Graphene oxide–DNA based sensors. Biosens. Bioelectron. 2014, 60, 22–29. [Google Scholar] [CrossRef]
- Robinson, J.T.; Perkins, F.K.; Snow, E.S.; Wei, Z.; Sheehan, P.E. Reduced graphene oxide molecular sensors. Nano Lett. 2008, 8, 3137–3140. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.; Wu, D.; Cui, Z.; Li, Y.; Zhang, Y.; Du, B.; Wei, Q. Graphene-based optical and electrochemical biosensors: A review. Anal. Lett. 2013, 46, 1–17. [Google Scholar] [CrossRef]
- Artiles, M.S.; Rout, C.S.; Fisher, T.S. Graphene-based hybrid materials and devices for biosensing. Adv. Drug Deliv. Rev. 2011, 63, 1352–1360. [Google Scholar] [CrossRef]
- Jung, J.H.; Cheon, D.S.; Liu, F.; Lee, K.B.; Seo, T.S. A graphene oxide based immuno-biosensor for pathogen detection. Angew. Chem. 2010, 49, 5708–5711. [Google Scholar] [CrossRef]
- Shi, X.; Gong, H.; Li, Y.; Wang, C.; Cheng, L.; Liu, Z. Graphene-based magnetic plasmonic nanocomposite for dual bioimaging and photothermal therapy. Biomaterials 2013, 34, 4786–4793. [Google Scholar] [CrossRef] [PubMed]
- Mei, Q.; Zhang, Z. Photoluminescent graphene oxide ink to print sensors onto microporous membranes for versatile visualization bioassays. Angew. Chem. 2012, 51, 5602–5606. [Google Scholar] [CrossRef] [PubMed]
- Ryoo, S.R.; Lee, J.; Yeo, J.; Na, H.K.; Kim, Y.K.; Jang, H.; Lee, J.H.; Han, S.W.; Lee, Y.; Kim, V.N.; et al. Quantitative and multiplexed microRNA sensing in living cells based on peptide nucleic acid and nano graphene oxide (PANGO). ACS Nano 2013, 7, 5882–5891. [Google Scholar] [CrossRef]
- Yoon, H.J.; Kim, T.H.; Zhang, Z.; Azizi, E.; Pham, T.M.; Paoletti, C.; Lin, J.; Ramnath, N.; Wicha, M.S.; Hayes, D.F.; et al. Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets. Nat. Nanotechnol. 2013, 8, 735–741. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lu, Q.; Liu, H.; Wang, J.; Zhang, P.; Liang, H.; Jiang, L.; Wang, S. Antibody-Modified Reduced Graphene Oxide Films with Extreme Sensitivity to Circulating Tumor Cells. Adv. Mater. 2015, 27, 6848–6854. [Google Scholar] [CrossRef] [PubMed]
- Trilling, A.K.; Harmsen, M.M.; Ruigrok, V.J.; Zuilhof, H.; Beekwilder, J. The effect of uniform capture molecule orientation on biosensor sensitivity: Dependence on analyte properties. Biosens. Bioelectron. 2013, 40, 219–226. [Google Scholar] [CrossRef]
- Jeon, S.; Moon, J.M.; Lee, E.S.; Kim, Y.H.; Cho, Y. An electroactive biotin-doped polypyrrole substrate that immobilizes and releases EpCAM-positive cancer cells. Angew. Chem. 2014, 53, 4597–4602. [Google Scholar] [CrossRef]
- Hou, S.; Zhao, H.; Zhao, L.; Shen, Q.; Wei, K.S.; Suh, D.Y.; Nakao, A.; Garcia, M.A.; Song, M.; Lee, T.; et al. Capture and stimulated release of circulating tumor cells on polymer-grafted silicon nanostructures. Adv. Mater. 2013, 25, 1547–1551. [Google Scholar] [CrossRef] [Green Version]
- Yoon, H.J.; Shanker, A.; Wang, Y.; Kozminsky, M.; Jin, Q.; Palanisamy, N.; Burness, M.L.; Azizi, E.; Simeone, D.M.; Wicha, M.S. Tunable Thermal-Sensitive Polymer–Graphene Oxide Composite for Efficient Capture and Release of Viable Circulating Tumor Cells. Adv. Mater. 2016, 28, 4891–4897. [Google Scholar] [CrossRef] [Green Version]
- Pumera, M. Graphene in biosensing. Mater. Today 2011, 14, 308–315. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, B.; Ding, J.; Liu, J. Fluorescent sensors using DNA-functionalized graphene oxide. Anal. Bioanal. Chem. 2014, 406, 6885–6902. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Song, B.; Li, D.; Zhu, C.; Qi, W.; Wen, Y.; Wang, L.; Song, S.; Fang, H.; Fan, C. A graphene nanoprobe for rapid, sensitive, and multicolor fluorescent DNA analysis. Adv. Funct. Mater. 2010, 20, 453–459. [Google Scholar] [CrossRef]
- Antony, J.; Grimme, S. Structures and interaction energies of stacked graphene–nucleobase complexes. Phys. Chem. Chem. Phys. 2008, 10, 2722–2729. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Zhu, C.; Ren, J.; Dong, S. A graphene-based real-time fluorescent assay of deoxyribonuclease I activity and inhibition. Anal. Chim. Acta 2012, 740, 88–92. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Z.; Weber, T.J.; Hu, D.; Lin, C.-T.; Li, J.; Lin, Y. In situ live cell sensing of multiple nucleotides exploiting DNA/RNA aptamers and graphene oxide nanosheets. Anal. Chem. 2013, 85, 6775–6782. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, Y.; Leng, F.; Huang, C.Z. Highly selective and sensitive detection of coralyne based on the binding chemistry of aptamer and graphene oxide. Talanta 2013, 112, 117–122. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, J.; Han, L.; Jin, L.; Zhu, C.; Wang, E.; Dong, S. Graphene-based aptamer logic gates and their application to multiplex detection. ACS Nano 2012, 6, 6659–6666. [Google Scholar] [CrossRef]
- Riely, G.J.; Pao, W.; Pham, D.; Li, A.R.; Rizvi, N.; Venkatraman, E.S.; Zakowski, M.F.; Kris, M.G.; Ladanyi, M.; Miller, V.A. Clinical course of patients with non–small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin. Cancer Res. 2006, 12, 839–844. [Google Scholar] [CrossRef] [Green Version]
- Eftekhari-Sis, B.; Aliabad, M.A.; Karimi, F. Graphene oxide based nano-biosensor for the detection of deletion mutation in exon 19 of EGFR gene, leading to lung cancer. Mater. Lett. 2016, 183, 441–443. [Google Scholar] [CrossRef]
- Muti, M.; Sharma, S.; Erdem, A.; Papakonstantinou, P. Electrochemical monitoring of nucleic acid hybridization by single-use graphene oxide-based sensor. Electroanalysis 2011, 23, 272–279. [Google Scholar] [CrossRef]
- Chu, Y.; Cai, B.; Ma, Y.; Zhao, M.; Ye, Z.; Huang, J. Highly sensitive electrochemical detection of circulating tumor DNA based on thin-layer MoS 2/graphene composites. RSC Adv. 2016, 6, 22673–22678. [Google Scholar] [CrossRef]
- Du, M.; Yang, T.; Jiao, K. Immobilization-free direct electrochemical detection for DNA specific sequences based on electrochemically converted gold nanoparticles/graphene composite film. J. Mater. Chem. 2010, 20, 9253–9260. [Google Scholar] [CrossRef]
- Rasheed, P.A.; Sandhyarani, N. Graphene-DNA electrochemical sensor for the sensitive detection of BRCA1 gene. Sens. Actuators B Chem. 2014, 204, 777–782. [Google Scholar] [CrossRef]
- Lu, C.H.; Li, J.; Liu, J.J.; Yang, H.H.; Chen, X.; Chen, G.N. Increasing the sensitivity and single-base mismatch selectivity of the molecular beacon using graphene oxide as the “nanoquencher”. Chem. A Eur. J. 2010, 16, 4889–4894. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Zhen, S.J.; Huang, C.Z. Graphene oxide as an efficient signal-to-background enhancer for DNA detection with a long range resonance energy transfer strategy. Chem. Commun. 2011, 47, 11718–11720. [Google Scholar] [CrossRef]
- Shahrokhian, S.; Salimian, R. Ultrasensitive detection of cancer biomarkers using conducting polymer/electrochemically reduced graphene oxide-based biosensor: Application toward BRCA1 sensing. Sens. Actuators B Chem. 2018, 266, 160–169. [Google Scholar] [CrossRef]
- Lin, L.; Liu, Y.; Tang, L.; Li, J. Electrochemical DNA sensor by the assembly of graphene and DNA-conjugated gold nanoparticles with silver enhancement strategy. Analyst 2011, 136, 4732–4737. [Google Scholar] [CrossRef]
- Kim, J.; Cote, L.J.; Kim, F.; Huang, J. Visualizing graphene based sheets by fluorescence quenching microscopy. J. Am. Chem. Soc. 2009, 132, 260–267. [Google Scholar] [CrossRef]
- Zhang, P.; He, M.; Zeng, Y. Ultrasensitive microfluidic analysis of circulating exosomes using a nanostructured graphene oxide/polydopamine coating. Lab Chip 2016, 16, 3033–3042. [Google Scholar] [CrossRef] [Green Version]
- Chae, M.-S.; Kim, J.; Jeong, D.; Kim, Y.; Roh, J.H.; Lee, S.M.; Heo, Y.; Kang, J.Y.; Lee, J.H.; Yoon, D.S. Enhancing surface functionality of reduced graphene oxide biosensors by oxygen plasma treatment for Alzheimer‘s disease diagnosis. Biosens. Bioelectron. 2017, 92, 610–617. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, H.; Huang, Z.; Li, T.; Deng, A.; Kong, J. DNase I enzyme-aided fluorescence signal amplification based on graphene oxide-DNA aptamer interactions for colorectal cancer exosome detection. Talanta 2018, 184, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Im, H.; Yang, K.S.; Lee, H.; Castro, C.M. Nanotechnology Platforms for Cancer Exosome Analyses. In Diagnostic and Therapeutic Applications of Exosomes in Cancer; Elsevier: Amsterdam, The Netherlands, 2018; pp. 119–128. [Google Scholar]
- Arenaccio, C.; Chiozzini, C.; Ferrantelli, F.; Leone, P.; Olivetta, E.; Federico, M. Exosomes in therapy: Engineering, pharmacokinetics and future applications. Curr. Drug Targets 2019, 20, 87–95. [Google Scholar] [CrossRef]
- Wang, X.; Hu, X.; Kawazoe, N.; Yang, Y.; Chen, G. Manipulating cell nanomechanics using micropatterns. Adv. Funct. Mater. 2016, 26, 7634–7643. [Google Scholar] [CrossRef]
- Wu, K.-H.; Cheng, H.-H.; Mohammad, A.A.; Blakey, I.; Jack, K.; Gentle, I.R.; Wang, D.-W. Electron-beam writing of deoxygenated micro-patterns on graphene oxide film. Carbon 2015, 95, 738–745. [Google Scholar] [CrossRef]
- Zhu, B.; Niu, Z.; Wang, H.; Leow, W.R.; Wang, H.; Li, Y.; Zheng, L.; Wei, J.; Huo, F.; Chen, X.J.S. Microstructured graphene arrays for highly sensitive flexible tactile sensors. Carbon 2014, 10, 3625–3631. [Google Scholar] [CrossRef]
- Yan, C.; Sun, J.; Ding, J.J.B. Critical areas of cell adhesion on micropatterned surfaces. Biomaterial 2011, 32, 3931–3938. [Google Scholar] [CrossRef]
- Poellmann, M.J.; Barton, K.L.; Mishra, S.; Johnson, A.J. Patterned hydrogel substrates for cell culture with electrohydrodynamic jet printing. Macromol. Biosci. 2011, 11, 1164–1168. [Google Scholar] [CrossRef]
- Delekta, S.S.; Smith, A.D.; Li, J.; Östling, M.J.N. Inkjet printed highly transparent and flexible graphene micro-supercapacitors. Nanoscale 2017, 9, 6998–7005. [Google Scholar] [CrossRef]
- Cai, K.; Dong, H.; Chen, C.; Yang, L.; Jandt, K.D.; Deng, L.J.C.; Biointerfaces, S.B. Inkjet printing of laminin gradient to investigate endothelial cellular alignment. Colloids Surf. B Biointerfaces 2009, 72, 230–235. [Google Scholar] [CrossRef]
- Brinkmann, F.; Hirtz, M.; Haller, A.; Gorges, T.M.; Vellekoop, M.J.; Riethdorf, S.; Müller, V.; Pantel, K.; Fuchs, H. A versatile microarray platform for capturing rare cells. Sci. Rep. 2015, 5, 15342. [Google Scholar] [CrossRef] [Green Version]
- Yu, D.; Tang, L.; Dong, Z.; Loftis, K.A.; Ding, Z.; Cheng, J.; Qin, B.; Yan, J.; Li, W. Effective reduction of non-specific binding of blood cells in a microfluidic chip for isolation of rare cancer cells. Biomater. Sci. 2018, 6, 2871–2880. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-H.; Penmatsa, V.; Tajima, S.; Kawarada, H.; Wang, C. Direct amination on 3-dimensional pyrolyzed carbon micropattern surface for DNA detection. Mater. Lett. 2009, 63, 2680–2683. [Google Scholar] [CrossRef]
- Zhang, P.; Zhou, X.; He, M.; Shang, Y.; Tetlow, A.L.; Godwin, A.K.; Zeng, Y. Ultrasensitive detection of circulating exosomes with a 3D-nanopatterned microfluidic chip. Nat. Biomed. Eng. 2019, 3, 438. [Google Scholar] [CrossRef]
- Wunsch, B.H.; Smith, J.T.; Gifford, S.M.; Wang, C.; Brink, M.; Bruce, R.L.; Austin, R.H.; Stolovitzky, G.; Astier, Y. Nanoscale lateral displacement arrays for the separation of exosomes and colloids down to 20 nm. Nat. Nanotechnol. 2016, 11, 936–940. [Google Scholar] [CrossRef]
- Wu, L.; Ji, H.; Guan, Y.; Ran, X.; Ren, J.; Qu, X. A graphene-based chemical nose/tongue approach for the identification of normal, cancerous and circulating tumor cells. NPG Asia Mater. 2017, 9, e356. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Cao, B.; Sun, B.; Cao, Y.; Yang, K.; Lin, Y.-S. Highly-sensitive capture of circulating tumor cells using micro-ellipse filters. Sci. Rep. 2017, 7, 610. [Google Scholar] [CrossRef]
- Lee, D.-H.; Cho, H.-S.; Han, D.; Chand, R.; Yoon, T.-J.; Kim, Y.-S. Highly selective organic transistor biosensor with inkjet printed graphene oxide support system. J. Mater. Chem. B 2017, 5, 3580–3585. [Google Scholar] [CrossRef]
- Nobbenhuis, M.A.; Walboomers, J.M.; Helmerhorst, T.J.; Rozendaal, L.; Remmink, A.J.; Risse, E.K.; van der Linden, H.C.; Voorhorst, F.J.; Kenemans, P.; Meijer, C.J. Relation of human papilloma virus status to cervical lesions and consequences for cervical-cancer screening: A prospective study. Lancet 1999, 354, 20–25. [Google Scholar] [CrossRef]
- Teengam, P.; Siangproh, W.; Tuantranont, A.; Henry, C.S.; Vilaivan, T.; Chailapakul, O. Electrochemical paper-based peptide nucleic acid biosensor for detecting human papillomavirus. Anal. Chim. Acta 2017, 952, 32–40. [Google Scholar] [CrossRef]
- Teixeira, S.R.; Lloyd, C.; Yao, S.; Gazze, A.S.; Whitaker, I.S.; Francis, L.; Conlan, R.S.; Azzopardi, E. Polyaniline-graphene based α-amylase biosensor with a linear dynamic range in excess of 6 orders of magnitude. Biosens. Bioelectron. 2016, 85, 395–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivastava, R.; Fraser, C.; Gentleman, D.; Jamieson, L.A.; Murphy, M.J. Hyperamylasaemia: Not the usual suspects. BMJ 2005, 331, 890–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Zhang, L.; Pan, S.; Gu, B.; Zhen, Y.; Yan, J.; Zhou, Y. Amylase: Sensitive tumor marker for amylase-producing lung adenocarcinoma. J. Thorac. Dis. 2013, 5, E167–E169. [Google Scholar] [PubMed]
- Haque, M.H.; Gopalan, V.; Yadav, S.; Islam, M.N.; Eftekhari, E.; Li, Q.; Carrascosa, L.G.; Nguyen, N.-T.; Lam, A.K.; Shiddiky, M.J. Detection of regional DNA methylation using DNA-graphene affinity interactions. Biosens. Bioelectron. 2017, 87, 615–621. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.W.; Tseng, M.L.; Fu, Y.M.; Kang, C.H.; Cheng, Y.T.; Kuo, P.H.; Tzeng, C.K.; Chiou, S.H.; Wu, C.Y.; Chen, G.Y. Printable Graphene Oxide Micropatterns for a Bio-Subretinal Chip. Adv. Healthc. Mater. 2018, 7, 1800365. [Google Scholar] [CrossRef]



| Type of GO-Based DNA-Based Sensors | Description of Method | Sensitivity | References |
|---|---|---|---|
| Optical | Multicolor fluorescent DNA nanoprobe | 100 pM | [84] |
| Fluorescein amidites (FAM) labeled DNA probe | 1 nM | [91] | |
| Molecular beacon | 2 nM | [96] | |
| DNA probe and DNA-intercalating dyes (SYBR Green I) | 0.31 nM | [97] | |
| Electrochemical | MoS2/graphene nanosheet-modified electrodes | 0.01 fM | [93] |
| Gold nanoparticle labeled reporter DNA (DNA-r.AuNP) and DNA-c modified glassy carbon electrode (GCE)/Gr | 1 fM | [95] | |
| PP3CA/ERGO/GCE | 3 fM | [98] | |
| cDNA2 modified AuNPs with catalyzed silver staining and GCE-GR/cDNA1 | 72 pM | [99] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.-L.; Chen, C.-Y.; Hsieh, J.C.-H.; Yu, Z.-Y.; Cheng, S.-J.; Hsieh, K.Y.; Yang, J.-W.; Kumar, P.V.; Lin, S.-F.; Chen, G.-Y. Graphene Oxide-Based Biosensors for Liquid Biopsies in Cancer Diagnosis. Nanomaterials 2019, 9, 1725. https://doi.org/10.3390/nano9121725
Chen S-L, Chen C-Y, Hsieh JC-H, Yu Z-Y, Cheng S-J, Hsieh KY, Yang J-W, Kumar PV, Lin S-F, Chen G-Y. Graphene Oxide-Based Biosensors for Liquid Biopsies in Cancer Diagnosis. Nanomaterials. 2019; 9(12):1725. https://doi.org/10.3390/nano9121725
Chicago/Turabian StyleChen, Shiue-Luen, Chong-You Chen, Jason Chia-Hsun Hsieh, Zih-Yu Yu, Sheng-Jen Cheng, Kuan Yu Hsieh, Jia-Wei Yang, Priyank V Kumar, Shien-Fong Lin, and Guan-Yu Chen. 2019. "Graphene Oxide-Based Biosensors for Liquid Biopsies in Cancer Diagnosis" Nanomaterials 9, no. 12: 1725. https://doi.org/10.3390/nano9121725
APA StyleChen, S.-L., Chen, C.-Y., Hsieh, J. C.-H., Yu, Z.-Y., Cheng, S.-J., Hsieh, K. Y., Yang, J.-W., Kumar, P. V., Lin, S.-F., & Chen, G.-Y. (2019). Graphene Oxide-Based Biosensors for Liquid Biopsies in Cancer Diagnosis. Nanomaterials, 9(12), 1725. https://doi.org/10.3390/nano9121725





