Raman Imaging of Nanocarriers for Drug Delivery
Abstract
1. Introduction
2. Raman Spectroscopy
2.1. Spontaneous Raman Spectroscopy
2.2. Increasing Raman Sensitivity
2.2.1. Surface Enhanced Raman Scattering (SERS)
2.2.2. Resonance Raman Scattering
2.2.3. Coherent Raman
3. Confocal Raman Imaging
3.1. Analysis of Hyperspectral Images
3.2. Label-Free Imaging of Drug Delivery
3.3. Imaging with Bioorthogonal Labels
3.4. Imaging Intracellular Uptake of Nanocarriers
3.4.1. Uptake of Polymeric Nanocarriers
3.4.2. Imaging Lipid-Based Nanocarriers
3.5. Imaging Intracellular Degradation of Nanocarriers
4. Coherent Raman Imaging
4.1. In Vitro Imaging
4.2. Coherent Raman Imaging of Nanoparticle Interactions Ex Vivo
4.2.1. Drug Delivery to the Skin
4.2.2. Drug Delivery to Targeted Organs
4.2.3. Drug Delivery to the Brain
5. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Mudshinge, S.R.; Deore, A.B.; Patil, S.; Bhalgat, C.M. Nanoparticles: Emerging carriers for drug delivery. Saudi Pharm. J. 2011, 19, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, B.; Dey, N.S.; Maji, R.; Bhowmik, P.; Das, P.J.; Paul, P. Current Status and Future Scope for Nanomaterials in Drug Delivery. In Application of Nanotechnology in Drug Delivery; IntechOpen Limited: London, UK, 2014; pp. 525–544. ISBN 9789537619992. [Google Scholar]
- Mir, M.; Ahmed, N.; Rehman, A. Recent applications of PLGA based nanostructures in drug delivery. Colloids Surf. B Biointerfaces 2017, 159, 217–231. [Google Scholar] [CrossRef] [PubMed]
- Wicki, A.; Witzigmann, D.; Balasubramanian, V.; Huwyler, J. Nanomedicine in cancer therapy: Challenges, opportunities, and clinical applications. J. Control. Release 2015, 200, 138–157. [Google Scholar] [CrossRef] [PubMed]
- Allam, A.N.; El gamal, S.S.; Nagger, V.F. Bioavailability: A pharmaceutical review. J. Nov. Drug Deliv. Technol. 2011, 1, 77–93. [Google Scholar]
- Zhao, H.; Lin, Z.Y.; Yildirimer, L.; Dhinakar, A.; Zhao, X.; Wu, J. Polymer-based nanoparticles for protein delivery: Design, strategies and applications. J. Mater. Chem. B 2016, 4, 4060–4071. [Google Scholar] [CrossRef]
- Dechy-Cabaret, O.; Martin-Vaca, B.; Bourissou, D. Controlled ring-opening polymerization of lactide and glycolide. Chem. Rev. 2004, 104, 6147–6176. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Kulkarni, A.; Nabulsi, J.; Nagesha, D.K.; Cormack, R.; Makrigiorgos, M.G.; Sridhar, S. Facile synthesis of PEGylated PLGA nanoparticles encapsulating doxorubicin and its in vitro evaluation as potent drug delivery vehicle. Drug Deliv. Transl. Res. 2013, 3, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Friedman, A.; Claypool, S.; Liu, R. The Smart Targeting of Nanoparticles. Curr. Pharm. Des. 2013, 19, 6315–6329. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Herrero, E.; Fernández-Medarde, A. Advanced targeted therapies in cancer: Drug nanocarriers, the future of chemotherapy. Eur. J. Pharm. Biopharm. 2015, 93, 52–79. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Yang, F.; Xiong, F.; Gu, N. The smart drug delivery system and its clinical potential. Theranostics 2016, 6, 1306–1323. [Google Scholar] [CrossRef] [PubMed]
- Saraiva, C.; Praça, C.; Ferreira, R.; Santos, T.; Ferreira, L.; Bernardino, L. Nanoparticle-mediated brain drug delivery: Overcoming blood-brain barrier to treat neurodegenerative diseases. J. Control. Release 2016, 235, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Patel, T.; Zhou, J.; Piepmeier, J.M.; Saltzman, W.M. Polymeric nanoparticles for drug delivery to the central nervous system. Adv. Drug Deliv. Rev. 2012, 64, 701–705. [Google Scholar] [CrossRef] [PubMed]
- Rittchen, S.; Boyd, A.; Burns, A.; Park, J.; Fahmy, T.M.; Metcalfe, S.; Williams, A. Myelin repair invivo is increased by targeting oligodendrocyte precursor cells with nanoparticles encapsulating leukaemia inhibitory factor (LIF). Biomaterials 2015, 56, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Dong, X. Current strategies for brain drug delivery. Theranostics 2018, 8, 1481–1493. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, A.D.; Vanjari, Y.H.; Sancheti, K.H.; Belgamwar, V.S.; Surana, S.J.; Pardeshi, C.V. Nanotechnology-mediated nose to brain drug delivery for Parkinson’s disease: A mini review. J. Drug Target. 2015, 23, 775–788. [Google Scholar] [CrossRef] [PubMed]
- Bourganis, V.; Kammona, O.; Alexopoulos, A.; Kiparissides, C. Recent Advances in Carrier Mediated Nose-to-Brain Delivery of Pharmaceutics. Eur. J. Pharm. Biopharm. 2018, 128, 337–362. [Google Scholar] [CrossRef] [PubMed]
- Jana, S.; Gandhi, A.; Kk, S.; Sk, B. Natural Polymers and their Application in Drug Delivery and Biomedical Field. J. PharmaSciTech 2011, 1, 16–27. [Google Scholar]
- Makadia, H.K.; Siegel, S.J. Poly Lactic-co-Glycolic Acid (PLGA) as biodegradable controlled drug delivery carrier. Polymers 2011, 3, 1377–1397. [Google Scholar] [CrossRef] [PubMed]
- Allen, T.M.; Cullis, P.R. Liposomal drug delivery systems: From concept to clinical applications. Adv. Drug Deliv. Rev. 2013, 65, 36–48. [Google Scholar] [CrossRef] [PubMed]
- Hines, D.J.; Kaplan, D.L. Poly(lactic-co-glycolic) acid-controlled-release systems: Experimental and modeling insights. Crit. Rev. Ther. Drug Carrier Syst. 2013, 30, 257–276. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Rouaud, O.; Poncelet, D. Microencapsulation by solvent evaporation: State of the art for process engineering approaches. Int. J. Pharm. 2008, 363, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H. Thin-Film Hydration Followed by Extrusion Method for Liposome Preparation. In Liposomes: Methods and Protocols; Springer: New York, NY, USA, 2017; Volume 1522, pp. 17–22. ISBN 9781461491644. [Google Scholar]
- Xu, W.; Ling, P.; Zhang, T. Polymeric Micelles, a Promising Drug Delivery System to Enhance Bioavailability of Poorly Water-Soluble Drugs. J. Drug Deliv. 2013, 2013, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Gullotti, E.; Tong, L.; Highley, C.B.; Errabelli, D.R.; Hasan, T.; Cheng, J.X.; Kohane, D.S.; Yeo, Y. Intracellular drug delivery by poly(lactic-co-glycolic acid) nanoparticles, revisited. Mol. Pharm. 2009, 6, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Jonderian, A.; Maalouf, R. Formulation and in vitro interaction of rhodamine-B loaded PLGA nanoparticles with cardiac myocytes. Front. Pharmacol. 2016, 7, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Nair, K.L.; Jagadeeshan, S.; Nair, S.A.; Kumar, G.S.V. Biological evaluation of 5-fluorouracil nanoparticles for cancer chemotherapy and its dependence on the carrier, PLGA. Int. J. Nanomed. 2011, 6, 1685–1697. [Google Scholar]
- Raman, C.V.; Kariamanikkam, K.S. A new type of secondary radiation. Nature 1928, 121, 501–502. [Google Scholar] [CrossRef]
- Yamakoshi, H.; Dodo, K.; Palonpon, A.; Ando, J.; Fujita, K.; Kawata, S.; Sodeoka, M. Alkyne-tag Raman imaging for visualization of mobile small molecules in live cells. J. Am. Chem. Soc. 2012, 134, 20681–20689. [Google Scholar] [CrossRef] [PubMed]
- Mil’shtein, S. Infrared scanning for biomedical applications. Scanning 2006, 28, 274–277. [Google Scholar] [CrossRef] [PubMed]
- Ember, K.J.I.; Hoeve, M.A.; McAughtrie, S.L.; Bergholt, M.S.; Dwyer, B.J.; Stevens, M.M.; Faulds, K.; Forbes, S.J.; Campbell, C.J. Raman spectroscopy and regenerative medicine: A review. NPJ Regen. Med. 2017, 2, 12. [Google Scholar] [CrossRef] [PubMed]
- Garrett, N.L.; Godfrey, L.; Lalatsa, A.; Serrano, D.R.; Uchegbu, I.F.; Schatzlein, A.; Moger, J. Detecting polymeric nanoparticles with coherent anti-stokes Raman scattering microscopy in tissues exhibiting fixative-induced autofluorescence. Proc. SPIE 2015, 9329, 932922. [Google Scholar]
- Müller, M.; Schins, J.M. Imaging the thermodynamic state of lipid membranes with multiplex CARS microscopy. J. Phys. Chem. B 2002, 106, 3715–3723. [Google Scholar] [CrossRef]
- Cialla-May, D.; Zheng, X.S.; Weber, K.; Popp, J. Recent progress in surface-enhanced Raman spectroscopy for biological and biomedical applications: From cells to clinics. Chem. Soc. Rev. 2017, 46, 3945–3961. [Google Scholar] [CrossRef] [PubMed]
- Bruzas, I.; Lum, W.; Gorunmez, Z.; Sagle, L. Advances in surface-enhanced Raman spectroscopy (SERS) substrates for lipid and protein characterization: Sensing and beyond. Analyst 2018, 143, 3990–4008. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Auchinvole, C.; Fisher, K.; Campbell, C.J. Quantitative measurement of redox potential in hypoxic cells using SERS nanosensors. Nanoscale 2014, 6, 12104–12110. [Google Scholar] [CrossRef] [PubMed]
- Granger, J.H.; Schlotter, N.E.; Crawford, A.C.; Porter, M.D. Prospects for point-of-care pathogen diagnostics using surface-enhanced Raman scattering (SERS). Chem. Soc. Rev. 2016, 45, 3865–3882. [Google Scholar] [CrossRef] [PubMed]
- Robert, B. Resonance Raman spectroscopy. Photosynth. Res. 2009, 101, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.; Dent, G. Modern Raman Spectroscopy—A Practical Approach; John Wiley & Sons Ltd.: Chichester, UK, 2005; ISBN 0471496685. [Google Scholar]
- Sloan-Dennison, S.; Shand, N.C.; Graham, D.; Faulds, K. Resonance Raman detection of antioxidants using an iron oxide nanoparticle catalysed decolourisation assay. Analyst 2017, 142, 4715–4720. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Heo, J.; Lim, C.; Pliss, A.; Kachynski, A.V.; Kuzmin, A.N.; Kim, S.; Prasad, P.N. Organelle specific imaging in live cells and immuno-labeling using resonance Raman probe. Biomaterials 2015, 53, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, D.; Cheng, J.-X. Coherent Raman Scattering Microscopy in Biology and Medicine. Annu. Rev. Biomed. Eng. 2015, 17, 415–445. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.-X.; Xie, X.S. Vibrational spectroscopic imaging of living systems: An emerging platform for biology and medicine. Science 2015, 350, aaa870–aaa871. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.-X. Coherent Anti-Stokes Raman Scattering Microscopy. Appl. Spectrosc. 2007, 61, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Maker, P.D.; Terhune, R.W. Study of optical effects due to an induced polarization third order in the electric field strength. Phys. Rev. 1965, 137, 801–818. [Google Scholar] [CrossRef]
- Zumbusch, A.; Holtom, G.R.; Xie, X.S. Three-Dimensional Vibrational Imaging by Coherent Anti-Stokes Raman Scattering. Phys. Rev. Lett. 1999, 82, 4142–4145. [Google Scholar] [CrossRef]
- Evans, C.L.; Xie, X.S. Coherent Anti-Stokes Raman Scattering Microscopy: Chemical Imaging for Biology and Medicine. Annu. Rev. Anal. Chem. 2008, 1, 883–909. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.L.; Potma, E.O.; Puoris’haag, M.; Cote, D.; Lin, C.P.; Xie, X.S. Chemical imaging of tissue in vivo with video-rate coherent anti-Stokes Raman scattering microscopy. Proc. Natl. Acad. Sci. USA 2005, 102, 16807–16812. [Google Scholar] [CrossRef] [PubMed]
- Hiramatsu, K.; Ideguchi, T.; Yonamine, Y.; Lee, S.; Luo, Y.; Hashimoto, K.; Ito, T.; Hase, M.; Park, J.; Kasai, Y.; et al. High-throughput label-free molecular fingerprinting flow cytometry. Sci. Adv. 2019, 5, eaau0241. [Google Scholar] [CrossRef] [PubMed]
- Woodbury, E.J.; Ng, W.K. Ruby Laser Operation in the Near IR. Proc. IRE 1962, 50, 2367. [Google Scholar]
- Yakovlev, V.V.; Petrov, G.I.; Zhang, H.F.; Noojin, G.D.; Denton, M.L.; Thomas, R.J.; Scully, M.O. Stimulated Raman scattering: Old physics, new applications. J. Mod. Opt. 2009, 56, 1970–1973. [Google Scholar] [CrossRef] [PubMed]
- Tipping, W.J.; Lee, M.; Serrels, A.; Brunton, V.G.; Hulme, A.N. Stimulated Raman scattering microscopy: An emerging tool for drug discovery. Chem. Soc. Rev. 2016, 45, 2075–2089. [Google Scholar] [CrossRef] [PubMed]
- Min, W.; Freudiger, C.W.; Lu, S.; Xie, X.S. Coherent Nonlinear Optical Imaging: Beyond Fluorescence Microscopy. Annu. Rev. Phys. Chem. 2011, 62, 507–530. [Google Scholar] [CrossRef] [PubMed]
- Krafft, C.; Schie, I.W.; Meyer, T.; Schmitt, M.; Popp, J. Developments in spontaneous and coherent Raman scattering microscopic imaging for biomedical applications. Chem. Soc. Rev. 2016, 45, 1819–1849. [Google Scholar] [CrossRef] [PubMed]
- Gomes da Costa, S.; Richter, A.; Schmidt, U.; Breuninger, S.; Hollricher, O. Confocal Raman microscopy in life sciences. Morphologie 2019, 103, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Michael, R.; Lenferink, A.; Vrensen, G.F.J.M.; Gelpi, E.; Barraquer, R.I.; Otto, C. Hyperspectral Raman imaging of neuritic plaques and neurofibrillary tangles in brain tissue from Alzheimer’s disease patients. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kong, K.; Kendall, C.; Stone, N.; Notingher, I. Raman spectroscopy for medical diagnostics—From in-vitro biofluid assays to in-vivo cancer detection. Adv. Drug Deliv. Rev. 2015, 89, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Surmacki, J.; Musial, J.; Kordek, R.; Abramczyk, H. Raman imaging at biological interfaces: Applications in breast cancer diagnosis. Mol. Cancer 2013, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kong, K.; Rowlands, C.J.; Varma, S.; Perkins, W.; Leach, I.H.; Koloydenko, A.A.; Williams, H.C.; Notingher, I. Diagnosis of tumors during tissue-conserving surgery with integrated autofluorescence and Raman scattering microscopy. Proc. Natl. Acad. Sci. USA 2013, 110, 15189–15194. [Google Scholar] [CrossRef] [PubMed]
- Ashton, L.; Hollywood, K.A.; Goodacre, R. Making colourful sense of Raman images of single cells. Analyst 2015, 140, 1852–1858. [Google Scholar] [CrossRef] [PubMed]
- Shinzawa, H.; Awa, K.; Kanematsu, W.; Ozaki, Y. Multivariate data analysis for Raman spectroscopic imaging. J. Raman Spectrosc. 2009, 40, 1720–1725. [Google Scholar] [CrossRef]
- Nascimento, J.M.P.; Dias, J.M.B. Vertex Component Analysis: A Fast Algorith to Unmix Hyperspectral Data. IEEE Trans. Geosci. Remote Sens. 2005, 43, 898–910. [Google Scholar] [CrossRef]
- Klein, K.; Gigler, A.M.; Aschenbrenner, T.; Monetti, R.; Bunk, W.; Jamitzky, F.; Morfill, G.; Stark, R.W.; Schlegel, J. Label-free live-cell imaging with confocal Raman microscopy. Biophys. J. 2012, 102, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Kang, C.; Liu, F.; Zhou, Y.; Luo, L.; Qiao, H. RGD Peptide-Based Target Drug Delivery of Doxorubicin Nanomedicine. Drug Dev. Res. 2017, 78, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Romero, G.; Qiu, Y.; Murray, R.A.; Moya, S.E. Study of intracellular delivery of doxorubicin from poly(lactide-co-glycolide) nanoparticles by means of fluorescence lifetime imaging and confocal Raman microscopy. Macromol. Biosci. 2013, 13, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Romero, G.; Ochoteco, O.; Sanz, D.J.; Estrela-Lopis, I.; Donath, E.; Moya, S.E. Poly(Lactide-co-Glycolide) Nanoparticles, Layer by Layer Engineered for the Sustainable Delivery of AntiTNF-α. Macromol. Biosci. 2013, 13, 903–912. [Google Scholar] [CrossRef] [PubMed]
- Romero, G.; Rojas, E.; Estrela-Lopis, I.; Donath, E.; Moya, S.E. Spontaneous confocal Raman microscopy-a tool to study the uptake of nanoparticles and carbon nanotubes into cells. Nanoscale Res. Lett. 2011, 6, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Shen, Y.; Hu, F.; Min, W. Applications of vibrational tags in biological imaging by Raman microscopy. Analyst 2017, 142, 4018–4029. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Hu, F.; Chen, Z.; Shen, Y.; Zhang, L.; Min, W. Live-Cell Bioorthogonal Chemical Imaging: Stimulated Raman Scattering Microscopy of Vibrational Probes. Acc. Chem. Res. 2016, 49, 1494–1502. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Hu, F.; Shen, Y.; Chen, Z.; Yu, Y.; Lin, C.C.; Wang, M.C.; Min, W. Live-cell imaging of alkyne-tagged small biomolecules by stimulated Raman scattering. Nat. Methods 2014, 11, 410–412. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Chen, T.; Zhu, Y.; Li, A.; Huang, Y.; Chen, X. Live-cell stimulated Raman scattering imaging of alkyne-tagged biomolecules. Angew. Chem. Int. Ed. 2014, 53, 5827–5831. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Paley, D.W.; Wei, L.; Weisman, A.L.; Friesner, R.A.; Nuckolls, C.; Min, W. Multicolor Live-Cell Chemical Imaging by Isotopically Edited Alkyne Vibrational Palette. J. Am. Chem. Soc. 2014, 136, 8027–8033. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Lamprecht, M.R.; Wei, L.; Morrison, B.; Min, W. Bioorthogonal chemical imaging of metabolic activities in live mammalian hippocampal tissues with stimulated Raman scattering. Sci. Rep. 2016, 6, 39660. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Chen, Z.; Zhang, L.; Shen, Y.; Wei, L.; Min, W. Vibrational Imaging of Glucose Uptake Activity in Live Cells and Tissues by Stimulated Raman Scattering. Angew. Chem. Int. Ed. 2015, 54, 9821–9825. [Google Scholar] [CrossRef] [PubMed]
- Long, R.; Zhang, L.; Shi, L.; Shen, Y.; Hu, F.; Zeng, C.; Min, W. Two-color vibrational imaging of glucose metabolism using stimulated Raman scattering. Chem. Commun. 2018, 54, 152–155. [Google Scholar] [CrossRef] [PubMed]
- Gaschler, M.M.; Hu, F.; Feng, H.; Linkermann, A.; Min, W.; Stockwell, B.R. Determination of the Subcellular Localization and Mechanism of Action of Ferrostatins in Suppressing Ferroptosis. ACS Chem. Biol. 2018, 13, 1013–1020. [Google Scholar] [CrossRef] [PubMed]
- Tipping, W.J.; Lee, M.; Serrels, A.; Brunton, V.G.; Hulme, A.N. Imaging drug uptake by bioorthogonal stimulated Raman scattering microscopy. Chem. Sci. 2017, 8, 5606–5615. [Google Scholar] [CrossRef] [PubMed]
- Behzadi, S.; Serpooshan, V.; Tao, W.; Hamaly, M.A.; Alkawareek, M.Y.; Dreaden, E.C.; Brown, D.; Alkilany, A.M.; Farokhzad, O.C.; Mahmoudi, M. Cellular uptake of nanoparticles: Journey inside the cell. Chem. Soc. Rev. 2017, 46, 4218–4244. [Google Scholar] [CrossRef] [PubMed]
- Gratton, S.E.; Ropp, P.A.; Pohlhaus, P.D.; Luft, J.C.; Madden, V.J.; Napier, M.E.; DeSimone, J.M. The effect of particle design on cellular internalization pathways. Proc. Natl. Acad. Sci. USA 2008, 105, 11613–11618. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, T.; Wang, Y.; Liu, L.; Lv, F.; Li, Z.; Huang, Y.; Schanze, K.S.; Wang, S. Conjugated Polymer with Intrinsic Alkyne Units for Synergistically Enhanced Raman Imaging in Living Cells. Angew. Chem. Int. Ed. 2017, 56, 13455–13458. [Google Scholar] [CrossRef] [PubMed]
- Frankel, A.D.; Pabo, C.O. Cellular uptake of the tat protein from human immunodeficiency virus. Cell 1988, 55, 1189–1193. [Google Scholar] [CrossRef]
- Chernenko, T.; Buyukozturk, F.; Miljkovic, M.; Carrier, R.; Diem, M.; Amiji, M. Label-free Raman microspectral analysis for comparison of cellular uptake and distribution between nontargeted and EGFR-targeted biodegradable polymeric nanoparticles. Drug Deliv. Transl. Res. 2013, 3, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Matthäus, C.; Schubert, S.; Schmitt, M.; Krafft, C.; Dietzek, B.; Schubert, U.S.; Popp, J. Resonance Raman spectral imaging of intracellular uptake of β-carotene loaded poly(d,l-lactide-co-glycolide) nanoparticles. ChemPhysChem 2013, 14, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Karami, N.; Moghimipour, E.; Salimi, A. Liposomes as a Novel Drug Delivery System: Fundamental and Pharmaceutical Application. Asian J. Pharm. 2018, 12, 31–41. [Google Scholar]
- Matthäus, C.; Kale, A.; Chernenko, T.; Torchilin, V.; Diem, M. New ways of imaging uptake and intracellular fate of liposomal drug carrier systems inside individual cells, based on Raman microscopy. Mol. Pharm. 2008, 5, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Chernenko, T.; Sawant, R.R.; Milijkovic, M.; Quintero, L.; Diem, M.; Torchilin, V. Raman Microscopy for Non-Invasive Imaging of Pharmaceutical Nanocarriers: Intracellular Distribution of Cationic Liposomes of Different Compositions. Mol. Pharm. 2012, 9, 930–936. [Google Scholar] [CrossRef] [PubMed]
- Cazzolli, G.; Caponi, S.; Defant, A.; Gambi, C.M.C.; Marchetti, S.; Mattarelli, M.; Montagna, M.; Rossi, B.; Rossi, F.; Viliani, G. Aggregation processes in micellar solutions: A Raman study. J. Raman Spectrosc. 2012, 43, 1877–1883. [Google Scholar] [CrossRef]
- Van Apeldoorn, A.A.; Van Manen, H.J.; Bezemer, J.M.; De Bruijn, J.D.; Van Blitterswijk, C.A.; Otto, C. Raman imaging of PLGA microsphere degradation inside macrophages. J. Am. Chem. Soc. 2004, 126, 13226–13227. [Google Scholar] [CrossRef] [PubMed]
- Chernenko, T.; Matthäus, C.; Milane, L.; Quintero, L.; Amiji, M.; Diem, M. Label-free raman spectral imaging of intracellular delivery and degradation of polymeric nanoparticle systems. ACS Nano 2009, 3, 3552–3559. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Brucks, S.D.; Lambert, T.H.; Campos, L.M.; Min, W. Stimulated Raman scattering of polymer nanoparticles for multiplexed live-cell imaging. Chem. Commun. 2017, 53, 6187–6190. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Yan, S.; Xiao, L.; Ji, R.; Yang, L.; Miao, A.J.; Wang, P. Label-Free Imaging of Nanoparticle Uptake Competition in Single Cells by Hyperspectral Stimulated Raman Scattering. Small 2018, 14, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Prausnitz, M.R.; Langer, R. Transdermal drug delivery. Nat. Biotechnol. 2008, 26, 1261–1268. [Google Scholar] [CrossRef] [PubMed]
- Prausnitz, M.R.; Mitragotri, S.; Langer, R. Current status and future potential of transdermal drug delivery. Nat. Rev. Drug Discov. 2004, 3, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Drutis, D.M.; Hancewicz, T.M.; Pashkovski, E.; Feng, L.; Mihalov, D.; Holtom, G.; Ananthapadmanabhan, K.P.; Xie, X.S.; Misra, M. Three-dimensional chemical imaging of skin using stimulated Raman scattering microscopy. J. Biomed. Opt. 2014, 19, 111604. [Google Scholar] [CrossRef] [PubMed]
- Saar, B.G.; Contreras-Rojas, L.R.; Xie, X.S.; Guy, R.H. Imaging drug delivery to skin with stimulated Raman scattering microscopy. Mol. Pharm. 2011, 8, 969–975. [Google Scholar] [CrossRef] [PubMed]
- Saar, B.G.; Freudiger, C.W.; Reichman, J.; Stanley, C.M.; Holtom, G.R.; Xie, X.S. Video-Rate Molecular Imaging in Vivo with Stimulated Raman Scattering. Science 2010, 330, 1368–1370. [Google Scholar] [CrossRef] [PubMed]
- Chiu, W.S.; Belsey, N.A.; Garrett, N.L.; Moger, J.; Delgado-Charro, M.B.; Guy, R.H. Molecular diffusion in the human nail measured by stimulated Raman scattering microscopy. Proc. Natl. Acad. Sci. USA 2015, 112, 7725–7730. [Google Scholar] [CrossRef] [PubMed]
- Belsey, N.A.; Garrett, N.L.; Contreras-Rojas, L.R.; Pickup-Gerlaugh, A.J.; Price, G.J.; Moger, J.; Guy, R.H. Evaluation of drug delivery to intact and porated skin by coherent Raman scattering and fluorescence microscopies. J. Control. Release 2014, 174, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Herkenne, C.; Alberti, I.; Naik, A.; Kalia, Y.N.; Mathy, F.X.; Préat, V.; Guy, R.H. In vivo methods for the assessment of topical drug bioavailability. Pharm. Res. 2008, 25, 87–103. [Google Scholar] [CrossRef] [PubMed]
- Francis, A.T.; Nguyen, T.T.; Lamm, M.S.; Teller, R.; Forster, S.P.; Xu, W.; Rhodes, T.; Smith, R.L.; Kuiper, J.; Su, Y.; et al. In Situ Stimulated Raman Scattering (SRS) Microscopy Study of the Dissolution of Sustained-Release Implant Formulation. Mol. Pharm. 2018, 15, 5793–5801. [Google Scholar] [CrossRef] [PubMed]
- Garrett, N.L.; Lalatsa, A.; Begley, D.; Mihoreanu, L.; Uchegbu, I.F.; Schätzlein, A.G.; Moger, J. Label-free imaging of polymeric nanomedicines using coherent anti-stokes Raman scattering microscopy. J. Raman Spectrosc. 2012, 43, 681–688. [Google Scholar] [CrossRef]
- Garrett, N.L.; Lalatsa, A.; Uchegbu, I.; Schätzlein, A.; Moger, J. Exploring uptake mechanisms of oral nanomedicines using multimodal nonlinear optical microscopy. J. Biophotonics 2012, 5, 458–468. [Google Scholar] [CrossRef] [PubMed]
- Lalatsa, A.; Garrett, N.L.; Ferrarelli, T.; Moger, J.; Schätzlein, A.G.; Uchegbu, I.F. Delivery of peptides to the blood and brain after oral uptake of quaternary ammonium palmitoyl glycol chitosan nanoparticles. Mol. Pharm. 2012, 9, 1764–1774. [Google Scholar] [CrossRef] [PubMed]
- Lalatsa, A.; Schatzlein, A.G.; Uchegbu, I.F. Strategies to deliver peptide drugs to the brain. Mol. Pharm. 2014, 11, 1081–1093. [Google Scholar] [CrossRef] [PubMed]
- Godfrey, L.; Iannitelli, A.; Garrett, N.L.; Moger, J.; Imbert, I.; King, T.; Porreca, F.; Soundararajan, R.; Lalatsa, A.; Schätzlein, A.G.; et al. Nanoparticulate peptide delivery exclusively to the brain produces tolerance free analgesia. J. Control. Release 2018, 270, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Mazza, M.; Notman, R.; Anwar, J.; Rodger, A.; Hicks, M.; Parkinson, G.; McCarthy, D.; Daviter, T.; Moger, J.; Garrett, N.; et al. Nanofiber-based delivery of therapeutic peptides to the brain. ACS Nano 2013, 7, 1016–1026. [Google Scholar] [CrossRef] [PubMed]
- Fisusi, F.A.; Siew, A.; Chooi, K.W.; Okubanjo, O.; Garrett, N.; Lalatsa, K.; Serrano, D.; Summers, I.; Moger, J.; Stapleton, P.; et al. Lomustine Nanoparticles Enable Both Bone Marrow Sparing and High Brain Drug Levels - A Strategy for Brain Cancer Treatments. Pharm. Res. 2016, 33, 1289–1303. [Google Scholar] [CrossRef] [PubMed]
- Ventola, C.L. Progress in Nanomedicine: Approved and Investigational Nanodrugs. Pharm. Ther. 2017, 42, 742–755. [Google Scholar]
- Hossain, S.; Chowdhury, E.H.; Akaike, T. Nanoparticles and toxicity in therapeutic delivery: The ongoing debate. Ther. Deliv. 2011, 2, 125–132. [Google Scholar] [CrossRef] [PubMed]

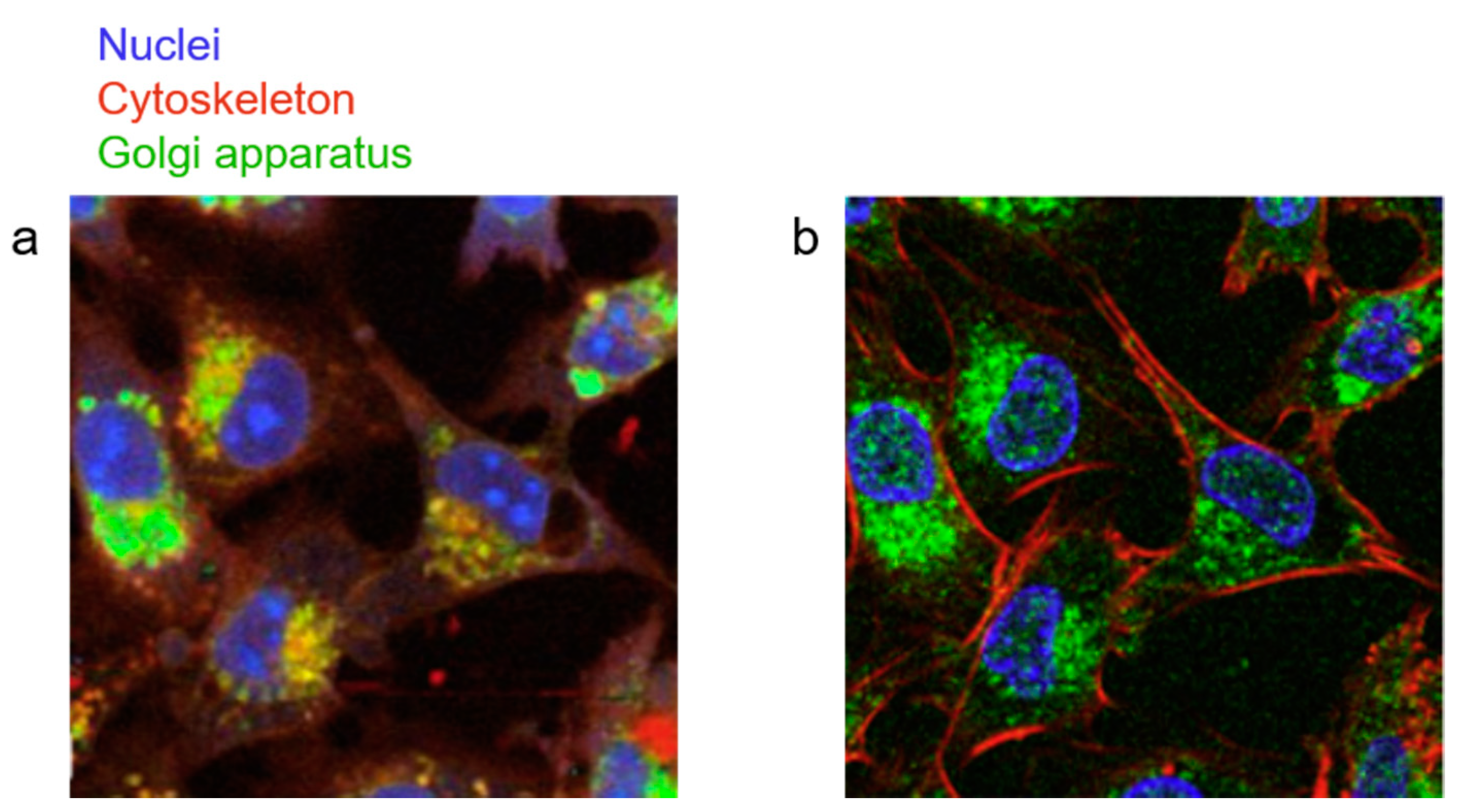





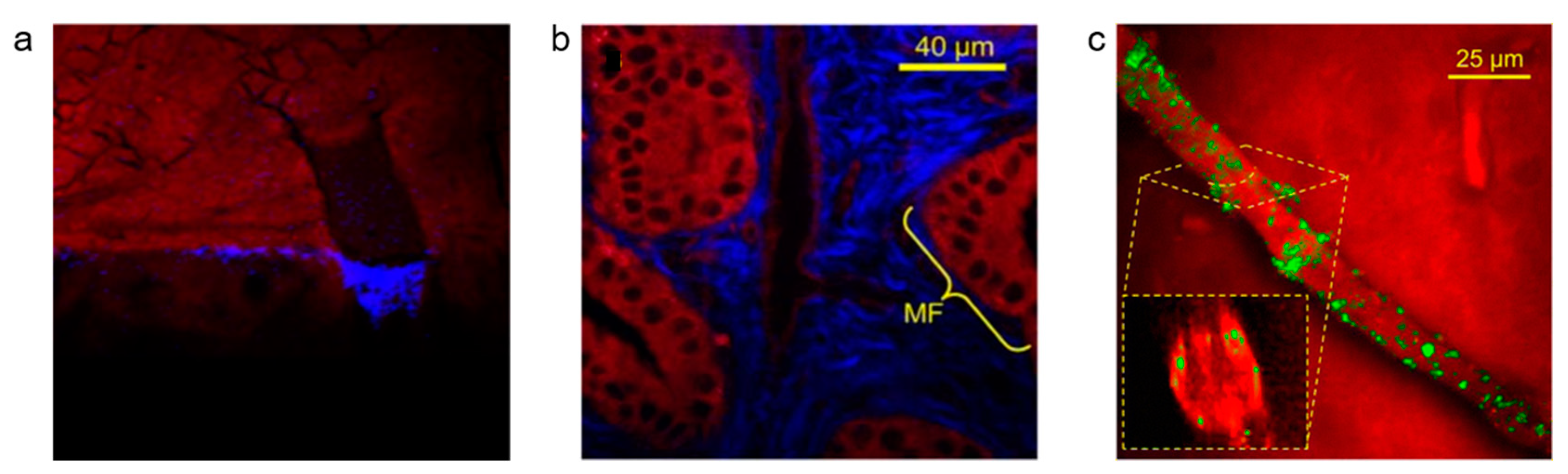
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vanden-Hehir, S.; Tipping, W.J.; Lee, M.; Brunton, V.G.; Williams, A.; Hulme, A.N. Raman Imaging of Nanocarriers for Drug Delivery. Nanomaterials 2019, 9, 341. https://doi.org/10.3390/nano9030341
Vanden-Hehir S, Tipping WJ, Lee M, Brunton VG, Williams A, Hulme AN. Raman Imaging of Nanocarriers for Drug Delivery. Nanomaterials. 2019; 9(3):341. https://doi.org/10.3390/nano9030341
Chicago/Turabian StyleVanden-Hehir, Sally, William J. Tipping, Martin Lee, Valerie G. Brunton, Anna Williams, and Alison N. Hulme. 2019. "Raman Imaging of Nanocarriers for Drug Delivery" Nanomaterials 9, no. 3: 341. https://doi.org/10.3390/nano9030341
APA StyleVanden-Hehir, S., Tipping, W. J., Lee, M., Brunton, V. G., Williams, A., & Hulme, A. N. (2019). Raman Imaging of Nanocarriers for Drug Delivery. Nanomaterials, 9(3), 341. https://doi.org/10.3390/nano9030341




