Gold Nanoparticle-Based Colorimetric Strategies for Chemical and Biological Sensing Applications
Abstract
1. Introduction
2. Aggregation-Based Colorimetric Assays
2.1. Labeled Detection Methods
2.1.1. Cross-Linking Aggregation
2.1.2. Non-Cross-Linking Aggregation
2.1.3. Destabilization-Induced Aggregation
2.2. Label-Free Detection Methods
3. Etching-Based Colorimetric Detection
3.1. H2O2-Mediated Etching Reactions
3.2. Ion-Mediated Etching Reactions
4. Growth-Based Colorimetric Sensing Strategies
4.1. Non-Enzyme-Mediated Growth Reactions
4.2. Enzyme-Mediated Growth Reactions
5. Nanoenzyme-Based Colorimetric Sensing Strategies
5.1. Peroxidase-Like Activity
5.2. Other Enzyme-Like Activity
6. Summary and Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Rick, J.; Tsai, M.C.; Hwang, B. Biosensors Incorporating Bimetallic Nanoparticles. Nanomaterials 2016, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Jayanthi, V.S.A.; Das, A.B.; Saxena, U. Recent advances in biosensor development for the detection of cancer biomarkers. Biosens. Bioelectron. 2017, 91, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chinnasamy, T.; Lifson, M.A.; Inci, F.; Demirci, U. Flexible Substrate-Based Devices for Point-of-Care Diagnostics. Trends Biotechnol. 2016, 34, 909–921. [Google Scholar] [CrossRef] [PubMed]
- Choi, S. Powering point-of-care diagnostic devices. Biotechnol. Adv. 2016, 34, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Nilghaz, A.; Guan, L.; Tan, W.; Shen, W. Advances of Paper-Based Microfluidics for Diagnostics—The Original Motivation and Current Status. ACS Sens. 2016, 1, 1382–1393. [Google Scholar] [CrossRef]
- Chuang, T.L.; Chang, C.C.; Chu-Su, Y.; Wei, S.C.; Zhao, X.; Hsueh, P.R.; Lin, C.W. Disposable surface plasmon resonance aptasensor with membrane-based sample handling design for quantitative interferon-gamma detection. Lab Chip 2014, 14, 2968–2977. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.; Zakaria, S.; Deng, M.; Allen, N.; Tram, K.; Li, Y. Integrating Deoxyribozymes into Colorimetric Sensing Platforms. Sensors 2016, 16, 2061. [Google Scholar] [CrossRef]
- Shen, L.; Hagen, J.A.; Papautsky, I. Point-of-care colorimetric detection with a smartphone. Lab Chip 2012, 12, 4240–4243. [Google Scholar] [CrossRef]
- Polavarapu, L.; Perez-Juste, J.; Xu, Q.-H.; Liz-Marzan, L.M. Optical sensing of biological, chemical and ionic species through aggregation of plasmonic nanoparticles. J. Mater. Chem. C 2014, 2, 7460–7476. [Google Scholar] [CrossRef]
- Wang, Z.; Ma, L. Gold nanoparticle probes. Coord. Chem. Rev. 2009, 253, 1607–1618. [Google Scholar] [CrossRef]
- Reimers, J.R.; Ford, M.J.; Marcuccio, S.M.; Ulstrup, J.; Hush, N.S. Competition of van der Waals and chemical forces on gold–sulfur surfaces and nanoparticles. Nat. Rev. Chem. 2017, 1, 0017. [Google Scholar] [CrossRef]
- Chen, Y.; Xianyu, Y.; Jiang, X. Surface Modification of Gold Nanoparticles with Small Molecules for Biochemical Analysis. Acc. Chem. Res. 2017, 50, 310–319. [Google Scholar] [CrossRef] [PubMed]
- Couto, C.; Vitorino, R.; Daniel-da-Silva, A.L. Gold nanoparticles and bioconjugation: A pathway for proteomic applications. Crit. Rev. Biotechnol. 2017, 37, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Cao-Milán, R.; Liz-Marzán, L.M. Gold nanoparticle conjugates: Recent advances toward clinical applications. Expert Opin. Drug Deliv. 2014, 11, 741–752. [Google Scholar] [CrossRef] [PubMed]
- Yeh, S.I.; Fang, W.F.; Huang, C.J.; Wang, T.M.; Yang, J.T. The Visual Colorimetric Detection of Multi-nucleotide Polymorphisms on a Pneumatic Droplet Manipulation Platform. J. Vis. Exp. 2016, e54424. [Google Scholar] [CrossRef] [PubMed]
- Sattarahmady, N.; Tondro, G.H.; Gholchin, M.; Heli, H. Gold nanoparticles biosensor of Brucella spp. genomic DNA: Visual and spectrophotometric detections. Biochem. Eng. J. 2015, 97, 1–7. [Google Scholar] [CrossRef]
- Trantakis, I.A.; Sturla, S.J. Gold nanoprobes for detecting DNA adducts. Chem. Commun. 2014, 50, 15517–15520. [Google Scholar] [CrossRef]
- Chang, C.C.; Wei, S.C.; Wu, T.H.; Lee, C.H.; Lin, C.W. Aptamer-based colorimetric detection of platelet-derived growth factor using unmodified goldnanoparticles. Biosens. Bioelectron. 2013, 42, 119–123. [Google Scholar] [CrossRef]
- Chang, C.C.; Chen, C.Y.; Chen, C.P.; Lin, C.W. Facile colorimetric detection of human chorionic gonadotropin based on the peptide-induced aggregation of gold nanoparticles. Anal. Methods 2015, 7, 29–33. [Google Scholar] [CrossRef]
- Ahirwar, R.; Nahar, P. Development of a label-free gold nanoparticle-based colorimetric aptasensor for detection of human estrogen receptor alpha. Anal. Bioanal. Chem. 2016, 408, 327–332. [Google Scholar] [CrossRef]
- Hu, Q.; Fu, Y.; Xu, X.; Qiao, Z.; Wang, R.; Zhang, Y.; Li, Y. A colorimetric detection of acrylamide in potato chips based on nucleophile-initiated thiol-ene Michael addition. Analyst 2016, 141, 1136–1143. [Google Scholar] [CrossRef]
- Bai, W.; Zhu, C.; Liu, J.; Yan, M.; Yang, S.; Chen, A. Gold nanoparticle–based colorimetric aptasensor for rapid detection of six organophosphorous pesticides. Environ. Toxicol. Chem. 2015, 34, 2244–2249. [Google Scholar] [CrossRef]
- Du, J.; Zhu, B.; Leow, W.R.; Chen, S.; Sum, T.C.; Peng, X.; Chen, X. Colorimetric Detection of Creatinine Based on Plasmonic Nanoparticles via Synergistic Coordination Chemistry. Small 2015, 11, 4104–4110. [Google Scholar] [CrossRef]
- Yue, G.; Su, S.; Li, N.; Shuai, M.; Lai, X.; Astruc, D.; Zhao, P. Gold nanoparticles as sensors in the colorimetric and fluorescence detection of chemical warfare agents. Coord. Chem. Rev. 2016, 311, 75–84. [Google Scholar] [CrossRef]
- Wang, F.; Lu, Y.; Yang, J.; Chen, Y.; Jing, W.; He, L.; Liu, Y. A smartphone readable colorimetric sensing platform for rapid multiple protein detection. Analyst 2017, 142, 3177–3182. [Google Scholar] [CrossRef]
- Liu, G.; Zhang, R.; Li, L.; Huang, X.; Li, T.; Lu, M.; Xu, D.; Wang, J. Anti-Agglomeration Behavior and Sensing Assay of Chlorsulfuron Based on Acetamiprid-Gold Nanoparticles. Nanomaterials 2018, 8, 499. [Google Scholar] [CrossRef]
- Wang, F.; Sun, J.; Lu, Y.; Zhang, X.; Song, P.; Liu, Y. Dispersion-aggregation-dispersion colorimetric detection for mercury ions based on an assembly of gold nanoparticles and carbon nanodots. Analyst 2018, 143, 4741–4746. [Google Scholar] [CrossRef]
- Chang, C.C.; Lee, C.H.; Wu, T.H.; Chen, C.P.; Chen, C.Y.; Lin, C.W. Reversion of gold nanoparticle aggregates for the detection of Cu2+ and its application in immunoassays. Analyst 2017, 142, 4684–4690. [Google Scholar] [CrossRef]
- Wu, B.; Zou, F.; Wang, X.; Koh, K.; Wang, K.; Chen, H. The colorimetric assay of diamine oxidase activity with high sensitivity based on calixarene derivative-capped gold nanoparticles. Anal. Methods 2017, 9, 2153–2158. [Google Scholar] [CrossRef]
- Zou, L.; Shen, R.; Ling, L.; Li, G. Sensitive DNA detection by polymerase chain reaction with gold nanoparticles. Anal. Chim. Acta 2018, 1038, 105–111. [Google Scholar] [CrossRef]
- Liu, P.; Han, L.; Wang, F.; Petrenko, V.A.; Liu, A. Gold nanoprobe functionalized with specific fusion protein selection from phage display and its application in rapid, selective and sensitive colorimetric biosensing of Staphylococcus aureus. Biosens. Bioelectron. 2016, 82, 195–203. [Google Scholar] [CrossRef]
- Sajjanar, B.; Kakodia, B.; Bisht, D.; Saxena, S.; Singh, A.K.; Joshi, V.; Tiwari, A.K.; Kumar, S. Peptide-activated gold nanoparticles for selective visual sensing of virus. J. Nanopart. Res. 2015, 17, 234. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Y.; Chen, P.; Wang, Y.; Zhang, J.; Aili, D.; Liedberg, B. Biofunctionalized Gold Nanoparticles for Colorimetric Sensing of Botulinum Neurotoxin A Light Chain. Anal. Chem. 2014, 86, 2345–2352. [Google Scholar] [CrossRef]
- Parnsubsakul, A.; Oaew, S.; Surareungchai, W. Zwitterionic peptide-capped gold nanoparticles for colorimetric detection of Ni2+. Nanoscale 2018, 10, 5466–5473. [Google Scholar] [CrossRef]
- Shinde, S.; Kim, D.Y.; Saratale, R.G.; Syed, A.; Ameen, F.; Ghodake, G. A Spectral Probe for Detection of Aluminum (III) Ions Using Surface Functionalized Gold Nanoparticles. Nanomaterials 2017, 7, 287. [Google Scholar] [CrossRef]
- Chandrawati, R.; Stevens, M.M. Controlled assembly of peptide-functionalized gold nanoparticles for label-free detection of blood coagulation Factor XIII activity. Chem. Commun. 2014, 50, 5431–5434. [Google Scholar] [CrossRef]
- Retout, M.; Valkenier, H.; Triffaux, E.; Doneux, T.; Bartik, K.; Bruylants, G. Rapid and Selective Detection of Proteins by Dual Trapping Using Gold Nanoparticles Functionalized with Peptide Aptamers. ACS Sens. 2016, 1, 929–933. [Google Scholar] [CrossRef]
- Yang, H.; Tang, Z.; Wang, L.; Zhou, W.; Li, L.; Zhang, Y.; Chen, S. The reactivity study of peptide A3-capped gold and silver nanoparticles with heavy metal ions. Mater. Sci. Eng. B 2016, 210, 37–42. [Google Scholar] [CrossRef]
- Ding, X.; Ge, D.; Yang, K.L. Colorimetric protease assay by using gold nanoparticles and oligopeptides. Sens. Actuators B Chem. 2014, 201, 234–239. [Google Scholar] [CrossRef]
- Ahn, J.; Choi, Y.; Lee, A.R.; Lee, J.H.; Jung, J.H. A duplex DNA-gold nanoparticle probe composed as a colorimetric biosensor for sequence-specific DNA-binding proteins. Analyst 2016, 141, 2040–2045. [Google Scholar] [CrossRef]
- Wang, J.; Zou, B.; Ma, Y.; Ma, X.; Sheng, N.; Rui, J.; Shao, Y.; Zhou, G. Closed-Tube PCR with Nested Serial Invasion Probe Visualization Using Gold Nanoparticles. Clin. Chem. 2017, 63, 852–860. [Google Scholar] [CrossRef]
- Singh, G.; Manohar, M.; Adegoke, A.A.; Stenström, T.A.; Shanker, R. Novel aptamer-linked nanoconjugate approach for detection of waterborne bacterial pathogens: An update. J. Nanopart. Res. 2016, 19, 4. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, B.; Liu, H.; Zhang, X.; Tan, W. Aptamer-conjugated gold nanoparticles for bioanalysis. Nanomedicine 2013, 8, 983–993. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, L.; Wei, W.; Zhao, H.; Zhou, Z.; Zhang, Y.; Liu, S. Colorimetric detection of influenza A virus using antibody-functionalized gold nanoparticles. Analyst 2015, 140, 3989–3995. [Google Scholar] [CrossRef]
- Basso, C.R.; Tozato, C.C.; Junior, J.P.A.; Pedrosa, V.A. A fast and highly sensitive method for the detection of canine distemper virus by the naked eye. Anal. Methods 2015, 7, 2264–2267. [Google Scholar] [CrossRef]
- Lesniewski, A.; Los, M.; Jonsson-Niedziółka, M.; Krajewska, A.; Szot, K.; Los, J.M.; Niedziolka-Jonsson, J. Antibody Modified Gold Nanoparticles for Fast and Selective, Colorimetric T7 Bacteriophage Detection. Bioconj. Chem. 2014, 25, 644–648. [Google Scholar] [CrossRef]
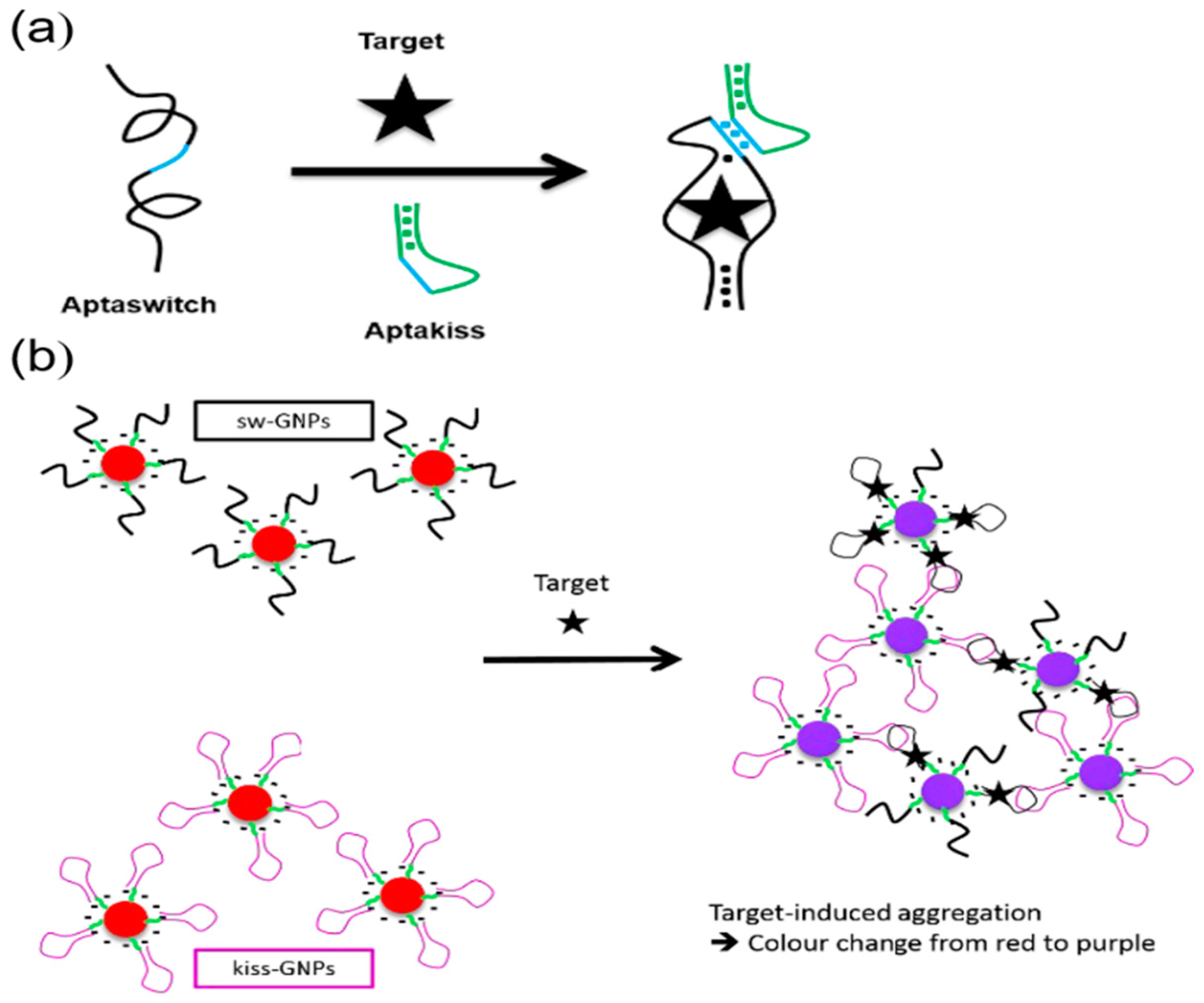
- Goux, E.; Dausse, E.; Guieu, V.; Azema, L.; Durand, G.; Henry, M.; Choisnard, L.; Toulme, J.J.; Ravelet, C.; Peyrin, E. A colorimetric nanosensor based on a selective target-responsive aptamer kissing complex. Nanoscale 2017, 9, 4048–4052. [Google Scholar] [CrossRef]
- Durand, G.; Lisi, S.; Ravelet, C.; Dausse, E.; Peyrin, E.; Toulmé, J.J. Riboswitches Based on Kissing Complexes for the Detection of Small Ligands. Angew. Chem. Int. Ed. 2014, 53, 6942–6945. [Google Scholar] [CrossRef]
- Guo, L.; Xu, Y.; Ferhan, A.R.; Chen, G.; Kim, D.-H. Oriented Gold Nanoparticle Aggregation for Colorimetric Sensors with Surprisingly High Analytical Figures of Merit. J. Am. Chem. Soc. 2013, 135, 12338–12345. [Google Scholar] [CrossRef]
- Chen, X.Y.; Ha, W.; Shi, Y.P. Sensitive colorimetric detection of melamine in processed raw milk using asymmetrically PEGylated gold nanoparticles. Talanta 2019, 194, 475–484. [Google Scholar] [CrossRef]
- Chen, X.Y.; Ma, R.T.; Ha, W.; Shi, Y.P. Direct colorimetric detection of aspartic acid in rat brain based on oriented aggregation of Janus gold nanoparticle. Sens. Actuators B Chem. 2018, 274, 668–675. [Google Scholar] [CrossRef]
- Wang, F.; Liu, S.; Lin, M.; Chen, X.; Lin, S.; Du, X.; Li, H.; Ye, H.; Qiu, B.; Lin, Z.; et al. Colorimetric detection of microcystin-LR based on disassembly of orient-aggregated gold nanoparticle dimers. Biosens. Bioelectron. 2015, 68, 475–480. [Google Scholar] [CrossRef]
- Xiong, Y.; Li, M.; Liu, H.; Xuan, Z.; Yang, J.; Liu, D. Janus PEGylated gold nanoparticles: A robust colorimetric probe for sensing nitrite ions in complex samples. Nanoscale 2017, 9, 1811–1815. [Google Scholar] [CrossRef]
- Shahdordizadeh, M.; Yazdian-Robati, R.; Ansari, N.; Ramezani, M.; Abnous, K.; Taghdisi, S.M. An aptamer-based colorimetric lead(II) assay based on the use of gold nanoparticles modified with dsDNA and exonuclease I. Microchim. Acta 2018, 185, 151. [Google Scholar] [CrossRef]
- Li, J.; Liu, R.; Zhang, Y.; Zhu, B.; Yao, X.; Lin, H.; Zhou, L.; Zhu, Z.; Yang, C. Application of DNA functionalized gold nanoparticles. Sci. Sin. Chim. 2015, 45, 1112–1144. [Google Scholar]
- Wang, Y.; Guo, J.; Guo, Y.; Zhang, X.; Ju, H. Enzymatically driven formation of palindromic DNA-Au nanoparticles for snowball assembly and colorimetric biosensing. Sens. Actuators B Chem. 2018, 267, 328–335. [Google Scholar] [CrossRef]
- Park, C.; Park, H.; Lee, H.J.; Lee, H.S.; Park, K.H.; Choi, C.H.; Na, S. Double amplified colorimetric detection of DNA using gold nanoparticles, enzymes and a catalytic hairpin assembly. Microchim. Acta 2018, 186, 34. [Google Scholar] [CrossRef]
- Osmani Bojd, M.; Kamaladini, H.; Haddadi, F.; Vaseghi, A. Thiolated AuNP probes and multiplex PCR for molecular detection of Staphylococcus epidermidis. Mol. Cell. Probes 2017, 34, 30–36. [Google Scholar] [CrossRef]
- Miao, X.; Ning, X.; Li, Z.; Cheng, Z. Sensitive detection of miRNA by using hybridization chain reaction coupled with positively charged gold nanoparticles. Sci. Rep. 2016, 6, 32358. [Google Scholar] [CrossRef]
- Xu, C.; Lan, L.; Yao, Y.; Ping, J.; Li, Y.; Ying, Y. An unmodified gold nanorods-based DNA colorimetric biosensor with enzyme-free hybridization chain reaction amplification. Sens. Actuators B Chem. 2018, 273, 642–648. [Google Scholar] [CrossRef]
- He, H.; Dai, J.; Meng, Y.; Duan, Z.; Zhou, C.; Zheng, B.; Du, J.; Guo, Y.; Xiao, D. Self-assembly of DNA nanoparticles through multiple catalyzed hairpin assembly for enzyme-free nucleic acid amplified detection. Talanta 2018, 179, 641–645. [Google Scholar] [CrossRef]
- Chang, C.C.; Chen, C.P.; Chen, C.Y.; Lin, C.W. DNA base-stacking assay utilizing catalytic hairpin assembly-induced gold nanoparticle aggregation for colorimetric protein sensing. Chem. Commun. 2016, 52, 4167–4170. [Google Scholar] [CrossRef]
- Sato, K.; Hosokawa, K.; Maeda, M. Rapid Aggregation of Gold Nanoparticles Induced by Non-Cross-Linking DNA Hybridization. J. Am. Chem. Soc. 2003, 125, 8102–8103. [Google Scholar] [CrossRef]
- Kanayama, N.; Sekine, T.; Ozasa, K.; Kishi, S.; Nyu, T.; Hayashi, T.; Maeda, M. Terminal-Specific Interaction between Double-Stranded DNA Layers: Colloidal Dispersion Behavior and Surface Force. Langmuir 2016, 32, 13296–13304. [Google Scholar] [CrossRef]
- Sekine, T.; Kanayama, N.; Ozasa, K.; Nyu, T.; Hayashi, T.; Maeda, M. Stochastic Binding Process of Blunt-End Stacking of DNA Molecules Observed by Atomic Force Microscopy. Langmuir 2018, 34, 15078–15083. [Google Scholar] [CrossRef]
- Akiyama, Y.; Wang, G.; Shiraishi, S.; Kanayama, N.; Takarada, T.; Maeda, M. Rapid Naked-Eye Discrimination of Cytochrome P450 Genetic Polymorphism through Non-Crosslinking Aggregation of DNA-Functionalized Gold Nanoparticles. ChemistryOpen 2016, 5, 508–512. [Google Scholar] [CrossRef]
- Kanayama, N.; Takarada, T.; Maeda, M. Rapid naked-eye detection of mercury ions based on non-crosslinking aggregation of double-stranded DNA-carrying gold nanoparticles. Chem. Commun. 2011, 47, 2077–2079. [Google Scholar] [CrossRef]
- Wang, G.; Akiyama, Y.; Takarada, T.; Maeda, M. Rapid Non-Crosslinking Aggregation of DNA-Functionalized Gold Nanorods and Nanotriangles for Colorimetric Single-Nucleotide Discrimination. Chem. Eur. J. 2016, 22, 258–263. [Google Scholar] [CrossRef]
- Fujita, M.; Katafuchi, Y.; Ito, K.; Kanayama, N.; Takarada, T.; Maeda, M. Structural study on gold nanoparticle functionalized with DNA and its non-cross-linking aggregation. J. Colloid Interface Sci. 2012, 368, 629–635. [Google Scholar] [CrossRef]
- Wang, G.; Akiyama, Y.; Shiraishi, S.; Kanayama, N.; Takarada, T.; Maeda, M. Cross-Linking versus Non-Cross-Linking Aggregation of Gold Nanoparticles Induced by DNA Hybridization: A Comparison of the Rapidity of Solution Color Change. Bioconj. Chem. 2017, 28, 270–277. [Google Scholar] [CrossRef]
- Chang, C.C.; Wang, G.; Takarada, T.; Maeda, M. Target-Recycling-Amplified Colorimetric Detection of Pollen Allergen Using Non-Cross-Linking Aggregation of DNA-Modified Gold Nanoparticles. ACS Sens. 2019, 4, 362–369. [Google Scholar] [CrossRef]
- McVey, C.; Huang, F.; Elliott, C.; Cao, C. Endonuclease controlled aggregation of gold nanoparticles for the ultrasensitive detection of pathogenic bacterial DNA. Biosens. Bioelectron. 2017, 92, 502–508. [Google Scholar] [CrossRef]
- Aldewachi, H.S.; Woodroofe, N.; Turega, S.; Gardiner, P.H.E. Optimization of gold nanoparticle-based real-time colorimetric assay of dipeptidyl peptidase IV activity. Talanta 2017, 169, 13–19. [Google Scholar] [CrossRef]
- Aldewachi, H.; Woodroofe, N.; Gardiner, P. Study of the Stability of Functionalized Gold Nanoparticles for the Colorimetric Detection of Dipeptidyl Peptidase IV. Appl. Sci. 2018, 8, 2589. [Google Scholar] [CrossRef]
- Zhang, W.; Tang, Y.; Liu, J.; Jiang, L.; Huang, W.; Huo, F.W.; Tian, D. Colorimetric Assay for Heterogeneous-Catalyzed Lipase Activity: Enzyme-Regulated Gold Nanoparticle Aggregation. J. Agric. Food Chem. 2015, 63, 39–42. [Google Scholar] [CrossRef]
- Ma, X.; Kou, X.; Xu, Y.; Yang, D.; Miao, P. Colorimetric sensing strategy for heparin assay based on PDDA-induced aggregation of gold nanoparticles. Nanoscale Adv. 2019, 1, 486–489. [Google Scholar] [CrossRef]
- Zhu, X.; Zhang, N.; Zhang, Y.; Liu, B.; Chang, Z.; Zhou, Y.; Hao, Y.; Ye, B.; Xu, M. A sensitive gold nanoparticle-based aptasensor for colorimetric detection of Aβ1–40 oligomers. Anal. Methods 2018, 10, 641–645. [Google Scholar] [CrossRef]
- Jiang, Y.; Shi, M.; Liu, Y.; Wan, S.; Cui, C.; Zhang, L.; Tan, W. Aptamer/AuNP Biosensor for Colorimetric Profiling of Exosomal Proteins. Angew. Chem. Int. Ed. 2017, 56, 11916–11920. [Google Scholar] [CrossRef]
- Luo, C.; Wen, W.; Lin, F.; Zhang, X.; Gu, H.; Wang, S. Simplified aptamer-based colorimetric method using unmodified gold nanoparticles for the detection of carcinoma embryonic antigen. RSC Adv. 2015, 5, 10994–10999. [Google Scholar] [CrossRef]
- Chang, C.C.; Chen, C.Y.; Zhao, X.; Wu, T.H.; Wei, S.C.; Lin, C.-W. Label-free colorimetric aptasensor for IgE using DNA pseudoknot probe. Analyst 2014, 139, 3347–3351. [Google Scholar] [CrossRef]
- Qi, M.; Tu, C.; Dai, Y.; Wang, W.; Wang, A.; Chen, J. A simple colorimetric analytical assay using gold nanoparticles for specific detection of tetracycline in environmental water samples. Anal. Methods 2018, 10, 3402–3407. [Google Scholar] [CrossRef]
- Yin, X.; Wang, S.; Liu, X.; He, C.; Tang, Y.; Li, Q.; Liu, J.; Su, H.; Tan, T.; Dong, Y. Aptamer-based Colorimetric Biosensing of Ochratoxin A in Fortified White Grape Wine Sample Using Unmodified Gold Nanoparticles. Anal. Sci. 2017, 33, 659–664. [Google Scholar] [CrossRef]
- Martínez-Aquino, C.; Costero, A.M.; Gil, S.; Gaviña, P. Resorcinol Functionalized Gold Nanoparticles for Formaldehyde Colorimetric Detection. Nanomaterials 2019, 9, 302. [Google Scholar] [CrossRef]
- Zhou, M.; Lin, T.; Gan, X. Colorimetric aggregation assay for silver(I) based on the use of aptamer modified gold nanoparticles and C-Ag(I)-C interaction. Microchim. Acta 2017, 184, 4671–4677. [Google Scholar] [CrossRef]
- Wu, Y.; Zhan, S.; Wang, L.; Zhou, P. Selection of a DNA aptamer for cadmium detection based on cationic polymer mediated aggregation of gold nanoparticles. Analyst 2014, 139, 1550–1561. [Google Scholar] [CrossRef]
- Chen, Z.; Guo, J.; Ma, H.; Zhou, T.; Li, X. A simple colorimetric sensor for potassium ion based on DNA G-quadruplex conformation and salt-induced gold nanoparticles aggregation. Anal. Methods 2014, 6, 8018–8021. [Google Scholar] [CrossRef]
- Chang, C.C.; Chen, C.Y.; Chuang, T.L.; Wu, T.H.; Wei, S.C.; Liao, H.; Lin, C.W. Aptamer-based colorimetric detection of proteins using a branched DNA cascade amplification strategy and unmodified gold nanoparticles. Biosens. Bioelectron. 2016, 78, 200–205. [Google Scholar] [CrossRef]
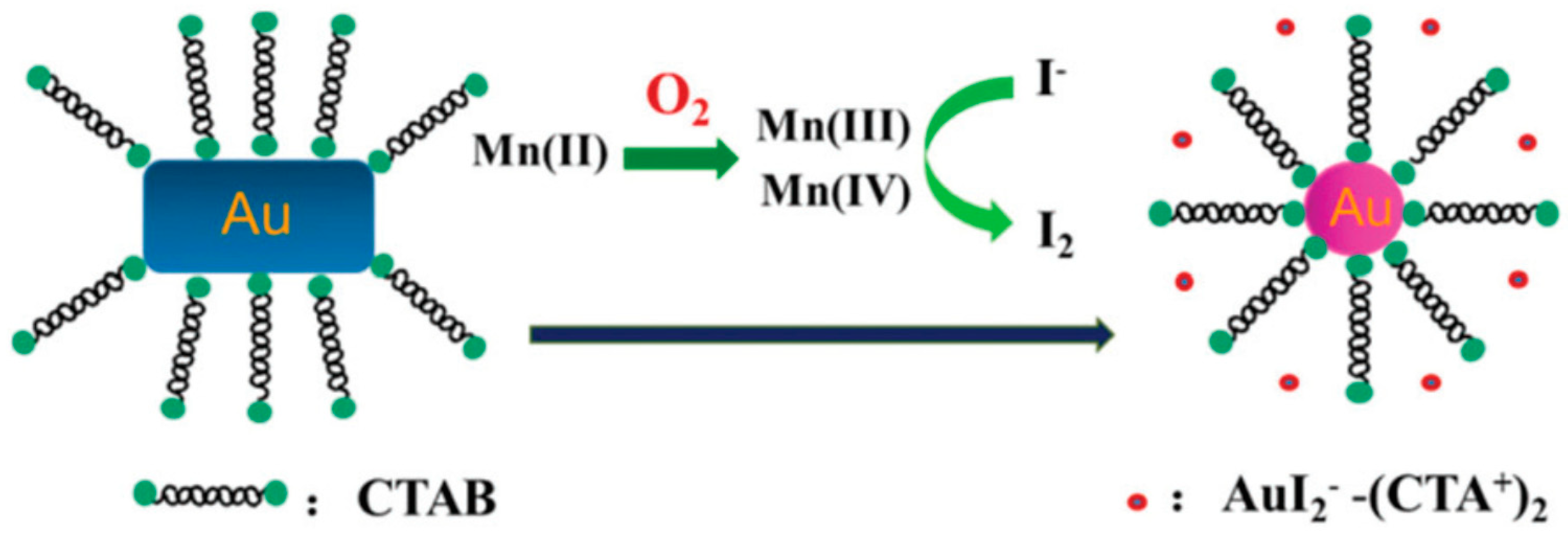
- Weng, G.; Dong, X.; Zhao, J.; Li, J.; Zhu, J.; Zhao, J. Selective oxidative etching of CTAC-stabilized multi-branched gold nanoparticles: Application in spectral sensing of iodide ions. J. Nanopart. Res. 2018, 20, 256. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, H.Z.; Liu, J.J.; Yuan, D.; Li, R.S.; Huang, C.Z. Time-resolved visual detection of heparin by accelerated etching of gold nanorods. Analyst 2018, 143, 824–828. [Google Scholar] [CrossRef]
- Murphy, C.J.; Thompson, L.B.; Alkilany, A.M.; Sisco, P.N.; Boulos, S.P.; Sivapalan, S.T.; Yang, J.A.; Chernak, D.J.; Huang, J. The Many Faces of Gold Nanorods. J. Phys. Chem. Lett. 2010, 1, 2867–2875. [Google Scholar] [CrossRef]
- Chandrasekar, G.; Mougin, K.; Haidara, H.; Vidal, L.; Gnecco, E. Shape and size transformation of gold nanorods (GNRs) via oxidation process: A reverse growth mechanism. Appl. Surface Sci. 2011, 257, 4175–4179. [Google Scholar] [CrossRef]
- Yang, H.; Liu, A.; Wei, M.; Liu, Y.; Lv, B.; Wei, W.; Zhang, Y.; Liu, S. Visual, Label-Free Telomerase Activity Monitor via Enzymatic Etching of Gold Nanorods. Anal. Chem. 2017, 89, 12094–12100. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, Z.; Pan, D.; Chen, L. Fenton-like Reaction-Mediated Etching of Gold Nanorods for Visual Detection of Co2+. Langmuir 2015, 31, 643–650. [Google Scholar] [CrossRef]
- Weng, G.; Li, J.; Zhu, J.; Zhao, J. Plasmonic sensing of CTAB in gold nanorods solution based on Cu(II) ions-mediated H2O2 etching effect. J. Nanopart. Res. 2014, 16, 2728. [Google Scholar] [CrossRef]
- Saa, L.; Coronado-Puchau, M.; Pavlov, V.; Liz-Marzan, L.M. Enzymatic etching of gold nanorods by horseradish peroxidase and application to blood glucose detection. Nanoscale 2014, 6, 7405–7409. [Google Scholar] [CrossRef]
- Saa, L.; Grinyte, R.; Sánchez-Iglesias, A.; Liz-Marzán, L.M.; Pavlov, V. Blocked Enzymatic Etching of Gold Nanorods: Application to Colorimetric Detection of Acetylcholinesterase Activity and Its Inhibitors. ACS Appl. Mater. Interfaces 2016, 8, 11139–11146. [Google Scholar] [CrossRef]
- Lu, S.; Chen, L.; Yang, P.; Matras-Postolek, K. Highly sensitive visual detection of catalase based on the accelerating decomposition of H2O2 using Au nanorods as a sensor. RSC Adv. 2016, 6, 19620–19625. [Google Scholar] [CrossRef]
- Zhu, Q.; Wu, J.; Zhao, J.; Ni, W. Role of Bromide in Hydrogen Peroxide Oxidation of CTAB-Stabilized Gold Nanorods in Aqueous Solutions. Langmuir 2015, 31, 4072–4077. [Google Scholar] [CrossRef]
- DuChene, J.S.; Niu, W.; Abendroth, J.M.; Sun, Q.; Zhao, W.; Huo, F.; Wei, W.D. Halide Anions as Shape-Directing Agents for Obtaining High-Quality Anisotropic Gold Nanostructures. Chem. Mater. 2013, 25, 1392–1399. [Google Scholar] [CrossRef]
- Weng, G.; Dong, X.; Li, J.; Zhao, J. Halide ions can trigger the oxidative etching of gold nanorods with the iodide ions being the most efficient. J. Mater. Sci. 2016, 51, 7678–7690. [Google Scholar] [CrossRef]
- Sun, S.; Gao, M.; Lei, G.; Zou, H.; Ma, J.; Huang, C. Visually monitoring the etching process of gold nanoparticles by KI/I2 at single-nanoparticle level using scattered-light dark-field microscopic imaging. Nano Res. 2016, 9, 1125–1134. [Google Scholar] [CrossRef]
- Cheng, X.; Huang, Y.; Yuan, C.; Dai, K.; Jiang, H.; Ma, J. Colorimetric detection of α-glucosidase activity based on the etching of gold nanorods and its application to screen anti-diabetic drugs. Sens. Actuators B Chem. 2019, 282, 838–843. [Google Scholar] [CrossRef]
- Wu, S.; Li, D.; Gao, Z.; Wang, J. Controlled etching of gold nanorods by the Au(III)-CTAB complex, and its application to semi-quantitative visual determination of organophosphorus pesticides. Microchim. Acta 2017, 184, 4383–4391. [Google Scholar] [CrossRef]
- Chang, C.C.; Wang, G.; Takarada, T.; Maeda, M. Iodine-Mediated Etching of Triangular Gold Nanoplates for Colorimetric Sensing of Copper Ion and Aptasensing of Chloramphenicol. ACS Appl. Mater. Interfaces 2017, 9, 34518–34525. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, Z.; Chen, L. Ultrasensitive Visual Sensing of Molybdate Based on Enzymatic-like Etching of Gold Nanorods. Langmuir 2015, 31, 9253–9259. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, Z.; Cheng, F.; Zhang, Y.; Chen, L. Highly sensitive on-site detection of glucose in human urine with naked eye based on enzymatic-like reaction mediated etching of gold nanorods. Biosens. Bioelectron. 2017, 89, 932–936. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, Z.; Cheng, F.; Zhang, Y.; Chen, L. Iodine-mediated etching of gold nanorods for plasmonic sensing of dissolved oxygen and salt iodine. Analyst 2016, 141, 2955–2961. [Google Scholar] [CrossRef]
- Wen, T.; Zhang, H.; Tang, X.; Chu, W.; Liu, W.; Ji, Y.; Hu, Z.; Hou, S.; Hu, X.; Wu, X. Copper Ion Assisted Reshaping and Etching of Gold Nanorods: Mechanism Studies and Applications. J. Phys. Chem. C 2013, 117, 25769–25777. [Google Scholar] [CrossRef]
- Alex, S.A.; Satija, J.; Khan, M.A.; Bhalerao, G.M.; Chakravarty, S.; Kasilingam, B.; Sivakumar, A.; Chandrasekaran, N.; Mukherjee, A. Etching-based transformation of dumbbell-shaped gold nanorods facilitated by hexavalent chromium and their possible application as a plasmonic sensor. Anal. Methods 2015, 7, 5583–5592. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, Z.; Qu, C.; Chen, L. Highly Sensitive Visual Detection of Copper Ions Based on the Shape-Dependent LSPR Spectroscopy of Gold Nanorods. Langmuir 2014, 30, 3625–3630. [Google Scholar] [CrossRef]
- Zhang, Y.; Leng, Y.; Miao, L.; Xin, J.; Wu, A. The colorimetric detection of Pb2+ by using sodium thiosulfate and hexadecyl trimethyl ammonium bromide modified gold nanoparticles. Dalton Trans. 2013, 42, 5485–5490. [Google Scholar] [CrossRef]
- Lee, Y.F.; Huang, C.C. Colorimetric Assay of Lead Ions in Biological Samples Using a Nanogold-Based Membrane. ACS Appl. Mater. Interfaces 2011, 3, 2747–2754. [Google Scholar] [CrossRef]
- Chen, Y.-Y.; Chang, H.T.; Shiang, Y.C.; Hung, Y.L.; Chiang, C.K.; Huang, C.C. Colorimetric Assay for Lead Ions Based on the Leaching of Gold Nanoparticles. Anal. Chem. 2009, 81, 9433–9439. [Google Scholar] [CrossRef]
- Lan, Y.J.; Lin, Y.-W. A non-aggregation colorimetric method for trace lead(ii) ions based on the leaching of gold nanorods. Anal. Methods 2014, 6, 7234–7242. [Google Scholar] [CrossRef]
- de la Rica, R.; Stevens, M.M. Plasmonic ELISA for the ultrasensitive detection of disease biomarkers with the naked eye. Nat. Nanotechnol. 2012, 7, 821–824. [Google Scholar] [CrossRef]
- Zayats, M.; Baron, R.; Popov, I.; Willner, I. Biocatalytic Growth of Au Nanoparticles: From Mechanistic Aspects to Biosensors Design. Nano Lett. 2005, 5, 21–25. [Google Scholar] [CrossRef]
- Zhou, Y.; Ma, Z. Colorimetric detection of Hg2+ by Au nanoparticles formed by H2O2 reduction of HAuCl4 using Au nanoclusters as the catalyst. Sens. Actuators B Chem. 2017, 241, 1063–1068. [Google Scholar] [CrossRef]
- Zhao, Y.; Gui, L.; Chen, Z. Colorimetric detection of Hg2+ based on target-mediated growth of gold nanoparticles. Sens. Actuators B Chem. 2017, 241, 262–267. [Google Scholar] [CrossRef]
- Shen, L.; Chen, J.; Li, N.; He, P.; Li, Z. Rapid colorimetric sensing of tetracycline antibiotics with in situ growth of gold nanoparticles. Anal. Chim. Acta 2014, 839, 83–90. [Google Scholar] [CrossRef]
- Wang, S.; Hu, X.; Tan, L.; Liao, Q.; Chen, Z. Colorimetric detection of lysozyme based on its effect on the growth of gold nanoparticles induced by the reaction of chloroauric acid and hydroxylamine. Microchim. Acta 2016, 183, 3135–3141. [Google Scholar] [CrossRef]
- Wang, Y.; Satyavolu, N.S.R.; Lu, Y. Sequence-specific control of inorganic nanomaterials morphologies by biomolecules. Curr. Opin. Colloid Interface Sci. 2018, 38, 158–169. [Google Scholar] [CrossRef]
- Tan, L.H.; Yue, Y.; Satyavolu, N.S.R.; Ali, A.S.; Wang, Z.; Wu, Y.; Lu, Y. Mechanistic Insight into DNA-Guided Control of Nanoparticle Morphologies. J. Am. Chem. Soc. 2015, 137, 14456–14464. [Google Scholar] [CrossRef]
- Soh, J.H.; Lin, Y.; Rana, S.; Ying, J.Y.; Stevens, M.M. Colorimetric Detection of Small Molecules in Complex Matrixes via Target-Mediated Growth of Aptamer-Functionalized Gold Nanoparticles. Anal. Chem. 2015, 87, 7644–7652. [Google Scholar] [CrossRef]
- Zhu, Q.; Li, T.; Ma, Y.; Wang, Z.; Huang, J.; Liu, R.; Gu, Y. Colorimetric detection of cholic acid based on an aptamer adsorbed gold nanoprobe. RSC Adv. 2017, 7, 19250–19256. [Google Scholar] [CrossRef]
- Liu, D.; Yang, J.; Wang, H.F.; Wang, Z.; Huang, X.; Wang, Z.; Niu, G.; Hight Walker, A.R.; Chen, X. Glucose Oxidase-Catalyzed Growth of Gold Nanoparticles Enables Quantitative Detection of Attomolar Cancer Biomarkers. Anal. Chem. 2014, 86, 5800–5806. [Google Scholar] [CrossRef]
- Gao, Z.; Deng, K.; Wang, X.D.; Miró, M.; Tang, D. High-Resolution Colorimetric Assay for Rapid Visual Readout of Phosphatase Activity Based on Gold/Silver Core/Shell Nanorod. ACS Appl. Mater. Interfaces 2014, 6, 18243–18250. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, Z.L.; Zheng, S.Y. Highly sensitive DNA detection using cascade amplification strategy based on hybridization chain reaction and enzyme-induced metallization. Biosens. Bioelectron. 2015, 66, 520–526. [Google Scholar] [CrossRef]
- Zhou, C.H.; Wu, Z.; Chen, J.J.; Xiong, C.; Chen, Z.; Pang, D.W.; Zhang, Z.L. Biometallization-Based Electrochemical Magnetoimmunosensing Strategy for Avian Influenza A (H7N9) Virus Particle Detection. Chem. Asian J. 2015, 10, 1387–1393. [Google Scholar] [CrossRef]
- Zhou, C.H.; Zhao, J.Y.; Pang, D.W.; Zhang, Z.L. Enzyme-Induced Metallization as a Signal Amplification Strategy for Highly Sensitive Colorimetric Detection of Avian Influenza Virus Particles. Anal. Chem. 2014, 86, 2752–2759. [Google Scholar] [CrossRef]
- Chen, J.; Jackson, A.A.; Rotello, V.M.; Nugen, S.R. Colorimetric Detection of Escherichia coli Based on the Enzyme-Induced Metallization of Gold Nanorods. Small 2016, 12, 2469–2475. [Google Scholar] [CrossRef]
- Guo, Y.; Wu, J.; Li, J.; Ju, H. A plasmonic colorimetric strategy for biosensing through enzyme guided growth of silver nanoparticles on gold nanostars. Biosens. Bioelectron. 2016, 78, 267–273. [Google Scholar] [CrossRef]
- Xu, S.; Ouyang, W.; Xie, P.; Lin, Y.; Qiu, B.; Lin, Z.; Chen, G.; Guo, L. Highly Uniform Gold Nanobipyramids for Ultrasensitive Colorimetric Detection of Influenza Virus. Anal. Chem. 2017, 89, 1617–1623. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, C.; Cai, N.; Long, S.; Yu, F. Negatively charged gold nanoparticles as an intrinsic peroxidase mimic and their applications in the oxidation of dopamine. J. Mater. Sci. 2014, 49, 7143–7150. [Google Scholar] [CrossRef]
- Jv, Y.; Li, B.; Cao, R. Positively-charged gold nanoparticles as peroxidiase mimic and their application in hydrogen peroxide and glucose detection. Chem. Commun. 2010, 46, 8017–8019. [Google Scholar] [CrossRef]
- Jiang, C.; Zhu, J.; Li, Z.; Luo, J.; Wang, J.; Sun, Y. Chitosan–gold nanoparticles as peroxidase mimic and their application in glucose detection in serum. RSC Adv. 2017, 7, 44463–44469. [Google Scholar] [CrossRef]
- Jiang, C.; Li, Z.; Wu, Y.; Guo, W.; Wang, J.; Jiang, Q. Colorimetric Detection of Hg2+ Based on Enhancement of Peroxidase-like Activity of Chitosan-Gold Nanoparticles. Bull. Korean Chem. Soc. 2018, 39, 625–630. [Google Scholar] [CrossRef]
- Wu, Y.; Chen, Y.; Li, Y.; Huang, J.; Yu, H.; Wang, Z. Accelerating peroxidase-like activity of gold nanozymes using purine derivatives and its application for monitoring of occult blood in urine. Sens. Actuators B Chem. 2018, 270, 443–451. [Google Scholar] [CrossRef]
- Shah, J.; Purohit, R.; Singh, R.; Karakoti, A.S.; Singh, S. ATP-enhanced peroxidase-like activity of gold nanoparticles. J. Colloid Interface Sci. 2015, 456, 100–107. [Google Scholar] [CrossRef]
- Zhao, J.; Wu, Y.; Tao, H.; Chen, H.; Yang, W.; Qiu, S. Colorimetric detection of streptomycin in milk based on peroxidase-mimicking catalytic activity of gold nanoparticles. RSC Adv. 2017, 7, 38471–38478. [Google Scholar] [CrossRef]
- Sun, S.; Zhao, R.; Feng, S.; Xie, Y. Colorimetric zearalenone assay based on the use of an aptamer and of gold nanoparticles with peroxidase-like activity. Microchim. Acta 2018, 185, 535. [Google Scholar] [CrossRef]
- Li, R.S.; Liu, H.; Chen, B.B.; Zhang, H.Z.; Huang, C.Z.; Wang, J. Stable gold nanoparticles as a novel peroxidase mimic for colorimetric detection of cysteine. Anal. Methods 2016, 8, 2494–2501. [Google Scholar] [CrossRef]
- Zhao, Y.; Huang, Y.; Zhu, H.; Zhu, Q.; Xia, Y. Three-in-One: Sensing, Self-Assembly, and Cascade Catalysis of Cyclodextrin Modified Gold Nanoparticles. J. Am. Chem. Soc. 2016, 138, 16645–16654. [Google Scholar] [CrossRef]
- Chen, Z.; Tan, L.; Hu, L.; Zhang, Y.; Wang, S.; Lv, F. Real Colorimetric Thrombin Aptasensor by Masking Surfaces of Catalytically Active Gold Nanoparticles. ACS Appl. Mater. Interfaces 2016, 8, 102–108. [Google Scholar] [CrossRef]
- Wei, X.; Chen, Z.; Tan, L.; Lou, T.; Zhao, Y. DNA-Catalytically Active Gold Nanoparticle Conjugates-Based Colorimetric Multidimensional Sensor Array for Protein Discrimination. Anal. Chem. 2017, 89, 556–559. [Google Scholar] [CrossRef]
- Lin, J.H.; Huang, K.H.; Zhan, S.W.; Yu, C.J.; Tseng, W.L.; Hsieh, M.M. Inhibition of catalytic activity of fibrinogen-stabilized gold nanoparticles via thrombin-induced inclusion of nanoparticle into fibrin: Application for thrombin sensing with more than 104-fold selectivity. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 210, 59–65. [Google Scholar] [CrossRef]
- Chang, C.C.; Chen, C.P.; Lee, C.H.; Chen, C.Y.; Lin, C.W. Colorimetric detection of human chorionic gonadotropin using catalytic gold nanoparticles and a peptide aptamer. Chem. Commun. 2014, 50, 14443–14446. [Google Scholar] [CrossRef]
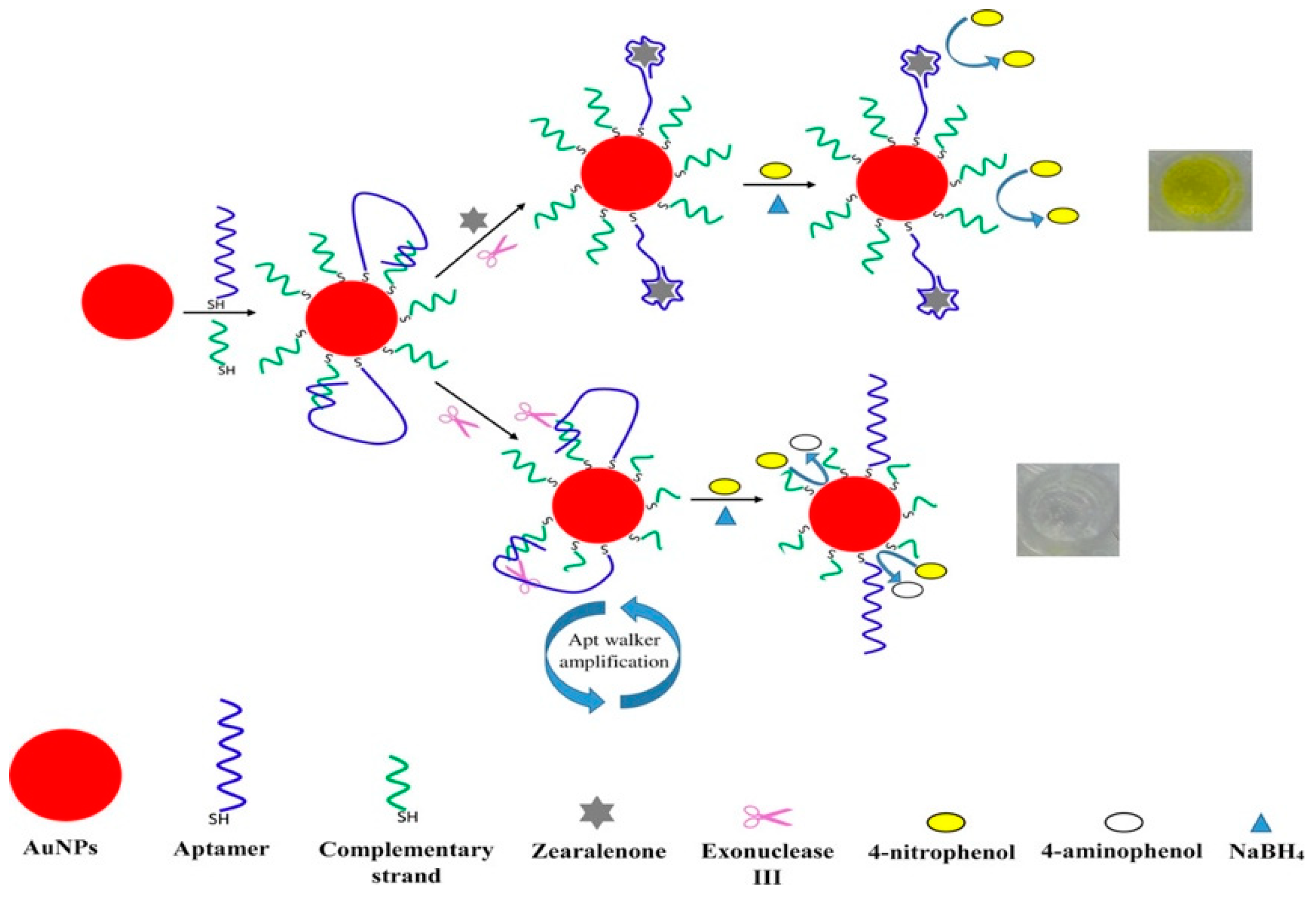
- Taghdisi, S.M.; Danesh, N.M.; Ramezani, M.; Emrani, A.S.; Abnous, K. Novel Colorimetric Aptasensor for Zearalenone Detection Based on Nontarget-Induced Aptamer Walker, Gold Nanoparticles, and Exonuclease-Assisted Recycling Amplification. ACS Appl. Mater. Interfaces 2018, 10, 12504–12509. [Google Scholar] [CrossRef]
- Wu, T.H.; Chang, C.C.; Vaillant, J.; Bruyant, A.; Lin, C.W. DNA biosensor combining single-wavelength colorimetry and a digital lock-in amplifier within a smartphone. Lab Chip 2016, 16, 4527–4533. [Google Scholar] [CrossRef]
- Smith, J.E.; Chávez, J.L.; Hagen, J.A.; Kelley-Loughnane, N. Design and Development of Aptamer-Gold Nanoparticle Based Colorimetric Assays for In-the-field Applications. JoVE 2016, e54063. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Y.; Yu, H.; Tian, L.; Wang, Z. Portable and smart devices for monitoring heavy metal ions integrated with nanomaterials. TrAC Trends Anal. Chem. 2018, 98, 190–200. [Google Scholar] [CrossRef]











© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, C.-C.; Chen, C.-P.; Wu, T.-H.; Yang, C.-H.; Lin, C.-W.; Chen, C.-Y. Gold Nanoparticle-Based Colorimetric Strategies for Chemical and Biological Sensing Applications. Nanomaterials 2019, 9, 861. https://doi.org/10.3390/nano9060861
Chang C-C, Chen C-P, Wu T-H, Yang C-H, Lin C-W, Chen C-Y. Gold Nanoparticle-Based Colorimetric Strategies for Chemical and Biological Sensing Applications. Nanomaterials. 2019; 9(6):861. https://doi.org/10.3390/nano9060861
Chicago/Turabian StyleChang, Chia-Chen, Chie-Pein Chen, Tzu-Heng Wu, Ching-Hsu Yang, Chii-Wann Lin, and Chen-Yu Chen. 2019. "Gold Nanoparticle-Based Colorimetric Strategies for Chemical and Biological Sensing Applications" Nanomaterials 9, no. 6: 861. https://doi.org/10.3390/nano9060861
APA StyleChang, C.-C., Chen, C.-P., Wu, T.-H., Yang, C.-H., Lin, C.-W., & Chen, C.-Y. (2019). Gold Nanoparticle-Based Colorimetric Strategies for Chemical and Biological Sensing Applications. Nanomaterials, 9(6), 861. https://doi.org/10.3390/nano9060861





