Label-Free Biosensors for Laboratory-Based Diagnostics of Infections: Current Achievements and New Trends
Abstract
:1. Introduction
2. Lateral Flow Immunoassay (LFIA) as Simplified Formats of Modern Biosensors
3. Introduction to Biosensor Technologies
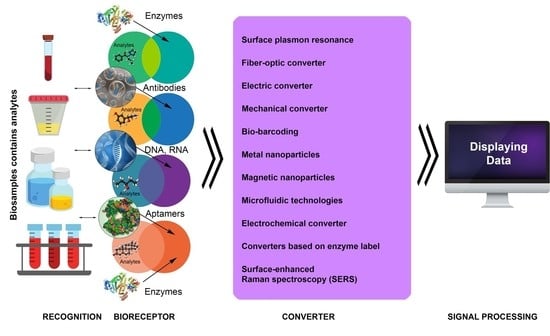
4. Main Types of Biosensors and Their Functions
4.1. Label-Free Biosensors
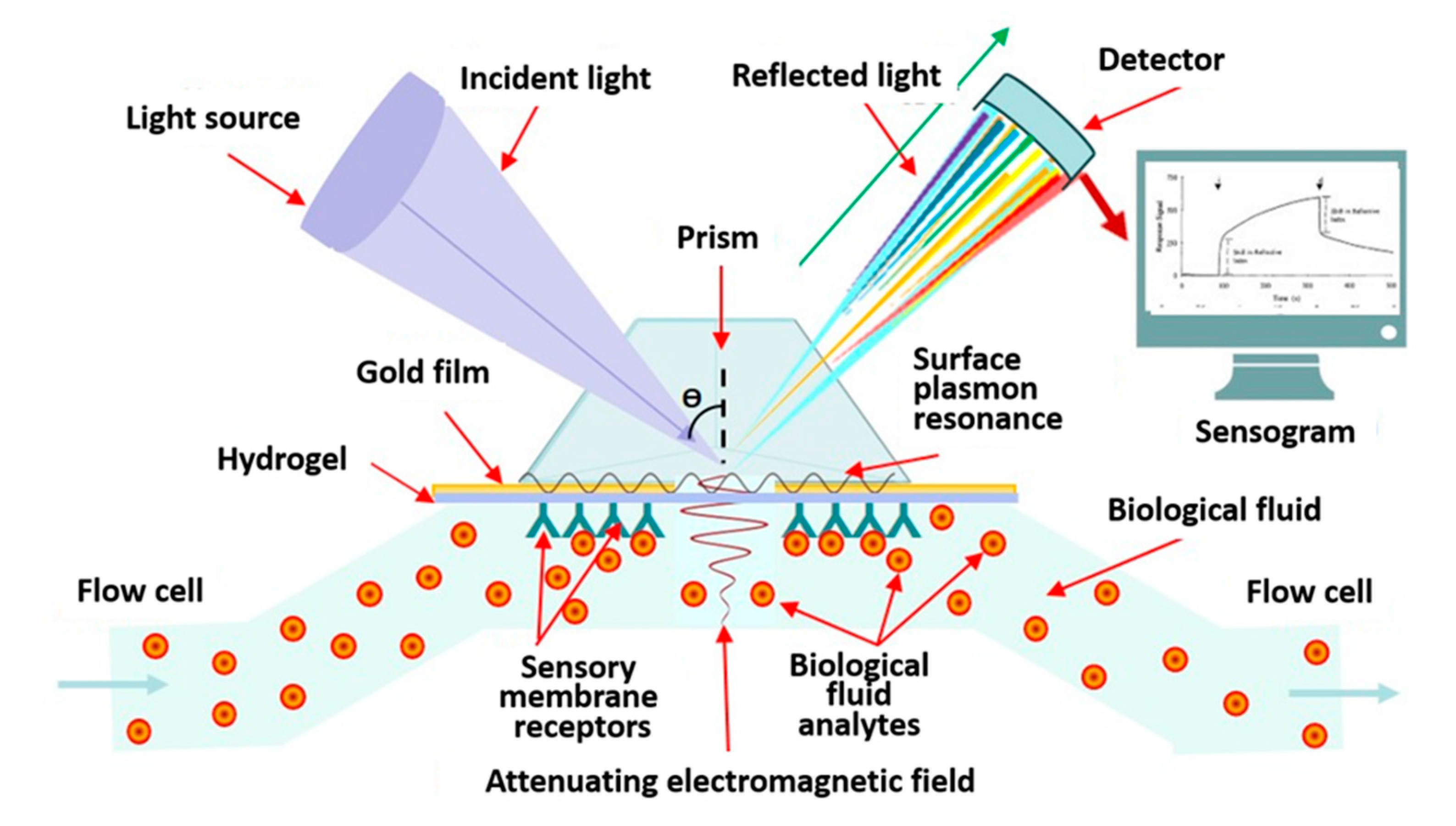
4.1.1. Label-Free Biosensors with Optical Converter
4.1.2. Electrochemical Label-Free Biosensors
4.1.3. Microwave Label-Free Biosensors
4.2. Mechanical Biosensors
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Havelaar, A.H.; Kirk, M.D.; Torgerson, P.R.; Gibb, H.J.; Hald, T.; Lake, R.J.; Praet, N.; Bellinger, D.C.; De Silva, N.R. World Health Organization Global Estimates and Regional Comparisons of the Burden of Foodborne Disease in 2010. PLOS Med. 2015, 12, e1001923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization (WHO) Infectious Disease Newsletter. Available online: https://www.who.int/topics/infectious_diseases/factsheets/ru/ (accessed on 9 December 2019).
- Sheng, L.; Lu, Y.; Deng, S.; Liao, X.; Zhang, K.; Ding, T.; Gao, H.; Liu, D.; Deng, R.; Li, J. A transcription aptasensor: Amplified, label-free and culture-independent detection of foodborne pathogens via light-up RNA aptamers. Chem. Commun. (Camb.) 2019, 55, 10096–10099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, L.C., Jr.; Lyons, C. Electrode systems for continuous monitoring in cardiovascular surgery. Ann. N. Y. Acad. Sci. 1962, 102, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Jia, W.; Hou, C.; Lei, Y. Microbial biosensors: A review. Biosens. Bioelectron. 2011, 26, 1788–1799. [Google Scholar] [CrossRef] [PubMed]
- Sin, M.L.; Mach, K.E.; Wong, P.K.; Liao, J.C. Advances and challenges in biosensor-based diagnosis of infectious diseases. Expert Rev. Mol. Diagn. 2014, 14, 225–244. [Google Scholar] [CrossRef] [Green Version]
- Peltomaa, R.; Glahn-Martínez, B.; Benito-Peña, E.; Moreno-Bondi, M.C. Optical Biosensors for Label-Free Detection of Small Molecules. Sensors 2018, 18, 4126. [Google Scholar] [CrossRef] [Green Version]
- Zarei, M. Infectious pathogens meet point-of-care diagnostics. Biosens. Bioelectron. 2018, 106, 193–203. [Google Scholar] [CrossRef]
- Kozel, T.R.; Burnham-Marusich, A.R. Point-of-Care Testing for Infectious Diseases: Past, Present, and Future. J. Clin. Microbiol. 2017, 55, 2313–2320. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Chung, D.R.; Kang, M. A new point-of-care test for the diagnosis of infectious diseases based on multiplex lateral flow immunoassays. Analyst 2019, 144, 2460–2466. [Google Scholar] [CrossRef]
- Fabri-Faja, N.; Calvo-Lozano, O.; Dey, P.; Terborg, R.A.; Estevez, M.C.; Belushkin, A.; Yesilköy, F.; Duempelmann, L.; Altug, H.; Pruneri, V.; et al. Early sepsis diagnosis via protein and miRNA biomarkers using a novel point-of-care photonic biosensor. Anal. Chim. Acta. 2019, 1077, 232–242. [Google Scholar] [CrossRef] [Green Version]
- Min, J.; Nothing, M.; Coble, B.; Zheng, H.; Park, J.; Im, H.; Weber, G.F.; Castro, C.M.; Swirski, F.K.; Weissleder, R.; et al. Integrated Biosensor for Rapid and Point-of-Care Sepsis Diagnosis. ACS Nano 2018, 12, 3378–3384. [Google Scholar] [CrossRef] [PubMed]
- Safenkova, I.V.; Panferov, V.G.; Panferova, N.A.; Varitsev, Y.A.; Zherdev, A.V.; Dzantiev, B.B. Alarm lateral flow immunoassay for detection of the total infection caused by the five viruses. Talanta 2019, 195, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, C.S.; Uldum, S.A.; Sørensen, J.F.; Skovsted, I.C.; Otte, S.; Elverdal, P.L. Evaluation of a new lateral flow test for detection of Streptococcus pneumoniae and Legionella pneumophila urinary antigen. J. Microbiol. Methods 2015, 116, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, Q.; Meng, Q.; Wu, F.; Zhang, L.; Tang, Y.; Guan, Y.; An, L. Quantum dots-based lateral flow immunoassay combined with image analysis for semiquantitative detection of IgE antibody to mite. Int. J. Nanomed. 2017, 12, 4805–4812. [Google Scholar] [CrossRef] [Green Version]
- Boisen, M.L.; Oottamasathien, D.; Jones, A.B.; Millett, M.M.; Nelson, D.S.; Bornholdt, Z.A.; Fusco, M.L.; Abelson, D.M.; Oda, S.; Hartnett, J.N.; et al. Development of prototype filovirus recombinant antigen immunoassays. J. Infect. Dis. 2015, 212 (Suppl. 2), S359–S367. [Google Scholar] [CrossRef]
- Nielsen, K.; Yu, W.L.; Kelly, L.; Bermudez, R.; Renteria, T.; Dajer, A.; Fusco, M.L.; Abelson, D.M.; Oda, S.; Hartnett, J.N.; et al. Development of a lateral flow assay for rapid detection of bovine antibody to Anaplasma marginale. J. Immunoass. Immunochem. 2008, 29, 10–18. [Google Scholar] [CrossRef]
- Posthuma-Trumpie, G.A.; Korf, J.; van Amerongen, A. Lateral flow (immuno)assay: Its strengths, weaknesses, opportunities and threats. A literature survey. Anal. Bioanal. Chem. 2008, 393, 569–582. [Google Scholar] [CrossRef] [Green Version]
- Rohrman, B.A.; Leautaud, V.; Molyneux, E.; Richards–Kortum, R.R. A lateral flow assay for quantitative detection of amplified HIV-1 RNA. PLoS ONE 2012, 7, e45611. [Google Scholar] [CrossRef] [Green Version]
- Kamphee, H.; Chaiprasert, A.; Prammananan, T.; Wiriyachaiporn, N.; Kanchanatavee, A.; Dharakul, T. Rapid molecular detection of multidrug-resistant tuberculosis by PCR-nucleic acid lateral flow immunoassay. PLoS ONE 2015, 10, e0137791. [Google Scholar] [CrossRef] [Green Version]
- Pilavaki, E.; Demosthenous, A. Optimized Lateral Flow Immunoassay Reader for the Detection of Infectious Diseases in Developing Countries. Sensors (Basel) 2017, 17, 2673. [Google Scholar] [CrossRef] [Green Version]
- Zhaoa, S.; Wangb, S.; Zhanga, S.; Liua, J.; Dong, Y. State of the art: Lateral flow assay (LFA) biosensor for on-site rapid detection. Chin. Chem. Lett. 2018, 29, 1567–1577. [Google Scholar] [CrossRef]
- Espinosa, J.R.; Galván, M.; Quiñones, A.S.; Ayala, J.L.; Durón, S.M. DNA Biosensor Based on Double-Layer Discharge for the Detection of HPV Type 16. Sensors (Basel) 2019, 19, 3956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, M.; Slaughter, G. Label-Free MicroRNA Optical Biosensors. Nanomaterials 2019, 9, 1573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to biosensors. Essays Biochem. 2016, 60, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ragavan, K.V.; Kumar, S.; Swaraj, S.; Neethirajan, S. Advances in biosensors and optical assays for diagnosis and detection of malaria. Biosens. Bioelectron. 2018, 105, 188–210. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Nanda, R.; Sahoo, S.; Mohapatra, E. Biosensors in Health Care: The Milestones Achieved in Their Development towards Lab-On-Chip-Analysis. Biochem. Res. Int. 2016, 2016, 3130469. [Google Scholar] [CrossRef] [Green Version]
- IUPAC. Compendium of Chemical Terminology; McNaught, A.D., Wilkinson, A., Eds.; Blackwell Scientific Publications: Oxford, UK, 1997; Volume 2, ISBN 0-9678550-9-8. [Google Scholar] [CrossRef]
- Biomaterials Nanoarchitectonics; Ebara, M. (Ed.) Elsevier Inc.: Tsukuba, Japan, 2016; 362p. [Google Scholar] [CrossRef]
- Russell, C.; Ward, A.C.; Vezza, V.; Hoskisson, P.; Alcorn, D.; Steenson, D.P.; Corrigan, D.K. Development of a needle shaped microelectrode for electrochemical detection of the sepsis biomarker interleukin-6 (IL-6) in real time. Biosens. Bioelectron. 2019, 126, 806–814. [Google Scholar] [CrossRef]
- Kumar, S.; Tripathy, S.; Jyoti, A.; Singh, S.G. Recent advances in biosensors for diagnosis and detection of sepsis: A comprehensive review. Biosens. Bioelectron. 2019, 124–125, 205–215. [Google Scholar] [CrossRef]
- Mazzaracchio, V.; Neagu, D.; Porchetta, A.; Marcoccio, E.; Pomponi, A.; Faggioni, G.; D’Amore, N.; Notargiacomo, A.; Pea, M.; Moscone, D.; et al. A label-free impedimetric aptasensor for the detection of Bacillus anthracis spore simulant. Biosens. Bioelectron. 2019, 126, 640–646. [Google Scholar] [CrossRef]
- Waller, D.F.; Hew, B.E.; Holdaway, C.; Jen, M.; Peckham, G.D. Rapid Detection of Bacillus anthracis Spores Using Immunomagnetic Separation and Amperometry. Biosensors (Basel) 2016, 6, 61. [Google Scholar] [CrossRef] [Green Version]
- Wynn, D.; Deo, S.; Daunert, S. Engineering Rugged Field Assays to Detect Hazardous Chemicals Using Spore-Based Bacterial Biosensors. Methods Enzymol. 2017, 589, 51–85. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Son, K.; Liu, Y.; Revzin, A. Biosensors for Cell Analysis. Ann. Rev. Biomed. Eng. 2015, 17, 165–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biopolymer Composites in Electronics; Sadasivuni, K.K.; Cabibihan, J.-J.; Ponnamma, D.; AlMaadeed, M.A.; Kim, J. (Eds.) Elsevier Inc.: Amsterdam, The Netherlands, 2017; 544p. [Google Scholar] [CrossRef]
- Yagi, K. Applications of whole-cell bacterial sensors in biotechnology and environmental science. Appl. Microbiol. Biotechnol. 2007, 73, 1251–1258. [Google Scholar] [CrossRef] [PubMed]
- Mangwani, N.; Dash, H.R.; Chauhan, A.; Das, S. Bacterial quorum sensing: Functional features and potential applications in biotechnology. J. Mol. Microb. Biotechnol. 2012, 22, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; White, I.M.; Shopova, S.I.; Zhu, H.; Suter, J.D.; Sun, Y. Sensitive optical biosensors for unlabeled targets: A review. Anal. Chim. Acta. 2008, 620, 8–26. [Google Scholar] [CrossRef] [PubMed]
- Sang, S.; Wang, Y.; Feng, Q.; Wei, Y.; Ji, J.; Zhang, W. Progress of new label-free techniques for biosensors: A review. Crit. Rev. Biotechnol. 2016, 36, 465–481. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, H.; Hu, Z.; Yu, G.; Yang, D.; Zhao, J. Label and label-free based surface-enhanced Raman scattering for pathogen bacteria detection: A review. Biosens. Bioelectron. 2017, 94, 131–140. [Google Scholar] [CrossRef]
- McLinden, T.; Sargeant, J.M.; Thomas, M.K.; Papadopoulos, A.; Fazil, A. Component costs of foodborne illness: A scoping review. BMC Public Health 2014, 14, 509. [Google Scholar] [CrossRef] [Green Version]
- Novick, A.; Weiner, M. Enzyme induction as an all-or-none phenomenon. Proc. Natl. Acad. Sci. USA 1957, 43, 553–566. [Google Scholar] [CrossRef] [Green Version]
- Jacob, F.; Monod, J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961, 3, 318–356. [Google Scholar] [CrossRef]
- Wu, X.; Xu, C.; Tripp, R.A.; Huang, Y.W.; Zhao, Y. Detection and differentiation of foodborne pathogenic bacteria in mung bean sprouts using field deployable label-free SERS devices. Analyst 2013, 138, 3005–3012. [Google Scholar] [CrossRef] [PubMed]
- Van der Meer, J.R.; Belkin, S. Where microbiology meets microengineering: Design and applications of reporter bacteria. Nat. Rev. Microbiol. 2010, 8, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Marks, H.; Schechinger, M.; Garza, J.; Locke, A.; Coté, G. Surface enhanced Raman spectroscopy (SERS) for in vitro diagnostic testing at the point of care. Nanophotonics 2017, 6, 681–701. [Google Scholar] [CrossRef]
- Lee, T.; Park, S.Y.; Jang, H.; Kim, G.H.; Lee, Y.; Park, C.; Mohammadniaei, M.; Lee, M.H.; Min, J. Fabrication of electrochemical biosensor consisted of multi-functional DNA structure/porous au nanoparticle for avian influenza virus (H5N1) in chicken serum. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 99, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Faria, H.A.M.; Zucolotto, V. Label-free electrochemical DNA biosensor for zika virus identification. Biosens. Bioelectron. 2019, 131, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Teengam, P.; Siangproh, W.; Tuantranont, A.; Vilaivan, T.; Chailapakul, O.; Henry, C.S. Electrochemical impedance-based DNA sensor using pyrrolidinyl peptide nucleic acids for tuberculosis detection. Anal. Chim. Acta. 2018, 1044, 102–109. [Google Scholar] [CrossRef]
- González-Pabón, M.J.; Figueredo, F.; Martínez-Casillas, D.C.; Cortón, E. Characterization of a new composite membrane for point of need paper-based micro-scale microbial fuel cell analytical devices. PLoS ONE 2019, 14, e0222538. [Google Scholar] [CrossRef]
- Lim, J.W.; Ha, D.; Lee, J.; Lee, S.K.; Kim, T. Review of Micro/Nanotechnologies for Microbial Biosensors. Front. Bioeng. Biotechnol. 2015, 3, 61. [Google Scholar] [CrossRef] [Green Version]
- Lei, Y.; Chen, W.; Mulchandani, A. Microbial biosensors. Anal. Chim. Acta 2006, 568, 200–210. [Google Scholar] [CrossRef]
- Urmann, K.; Reich, P.; Walter, J.G.; Beckmann, D.; Segal, E.; Scheper, T. Rapid and label-free detection of protein a by aptamer-tethered porous silicon nanostructures. J. Biotechnol. 2017, 257, 171–177. [Google Scholar] [CrossRef]
- Zhang, S.; Ma, L.; Ma, K.; Xu, B.; Liu, L.; Tian, W. Label-Free Aptamer-Based Biosensor for Specific Detection of Chloramphenicol Using AIE Probe and Graphene Oxide. ACS Omega 2018, 3, 12886–12892. [Google Scholar] [CrossRef] [PubMed]
- Gharatape, A.; Yari Khosroushahi, A. Optical Biomarker-based Biosensors for Cancer/Infectious Disease Medical Diagnoses. Appl. Immunohistochem. Mol. Morphol. 2019, 27, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Zhou, J. Application of biosensors to detection of epidemic diseases in animals. Res. Vet. Sci. 2018, 118, 444–448. [Google Scholar] [CrossRef] [PubMed]
- Kahraman, M.; Mullen, E.R.; Korkmaz, A.; Wachsmann-Hogiu, S. Fundamentals and applications of SERS-based bioanalytical sensing. Nanophotonics 2017, 6, 831–852. [Google Scholar] [CrossRef] [Green Version]
- Qiu, C.; Zhai, H.; Hou, J. Biosensors Design in Yeast and Applications in Metabolic Engineering. FEMS Yeast Res. 2019, 19, foz082. [Google Scholar] [CrossRef]
- Zhang, F.Z.; Keasling, J. Biosensors and their applications in microbial metabolic engineering. Trends Microbiol. 2011, 19, 323–329. [Google Scholar] [CrossRef]
- Dietrich, J.A.; McKee, A.E.; Keasling, J.D. High-throughput metabolic engineering: Advances in small-molecule screening and selection. Ann. Rev. Biochem. 2010, 79, 563–590. [Google Scholar] [CrossRef]
- Durrieu, C.; Lagarde, F.; Jaffrezic-Renault, N. Nanotechnology assets in biosensors design for environmental monitoring. In Nanomaterials: A Danger or a Promise; Brayner, R., Fiévet, F., Coradin, T., Eds.; Springer: London, UK, 2013; pp. 189–229. [Google Scholar]
- Chang, H.J.; Voyvodic, P.L.; Zúñiga, A.; Bonnet, J. Microbially derived biosensors for diagnosis, monitoring and epidemiology. Microb. Biotechnol. 2017, 10, 1031–1035. [Google Scholar] [CrossRef] [Green Version]
- Renella, G.; Giagnoni, L. Light dazzles from the black box: Whole-cell biosensors are ready to inform on fundamental soil biological processes. Chem. Biol. Technol. Agric. 2016, 3, 8. [Google Scholar] [CrossRef] [Green Version]
- Park, M.; Tsai, S.L.; Chen, W. Microbial biosensors: Engineered microorganisms as the sensing machinery. Sensors (Basel) 2013, 13, 5777–5795. [Google Scholar] [CrossRef] [Green Version]
- Roy, V.; Adams, B.L.; Bentley, W.E. Developing next generation antimicrobials by intercepting AI-2 mediated quorum sensing. Enzym. Microb. Technol. 2011, 49, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Amine, A.; Arduini, F.; Moscone, D.; Palleschi, G. Recent advances in biosensors based on enzyme inhibition. Biosens. Bioelectron. 2016, 76, 180–194. [Google Scholar] [CrossRef] [PubMed]
- Barreiros dos Santos, M.; Agusil, J.P.; Prieto-Simón, B.; Sporer, C.; Teixeira, V.; Samitier, J. Highly sensitive detection of pathogen Escherichia coli O157:H7 by electrochemical impedance spectroscopy. Biosens. Bioelectron. 2013, 45, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Rocchitta, G.; Spanu, A.; Babudieri, S.; Latte, G.; Madeddu, G.; Galleri, G.; Nuvoli, S.; Bagella, P.; Demartis, M.I.; Fiore, V.; et al. Enzyme Biosensors for Biomedical Applications: Strategies for Safeguarding Analytical Performances in Biological Fluids. Sensors (Basel) 2016, 16, 780. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Marrakchi, M.; Xu, D.; He, D.; Andreescu, S. Biosensors based on modularly designed synthetic peptides for recognition, detection and live/dead differentiation of pathogenic bacteria. Biosens. Bioelectron. 2016, 80, 9–16. [Google Scholar] [CrossRef]
- Golichenari, B.; Velonia, K.; Nosrati, R.; Nezami, A.; Farokhi-Fard, A.; Abnous, K.; Behravan, J.; Tsatsakis, A.M. Label-free nano-biosensing on the road to tuberculosis detection. Biosens. Bioelectron. 2018, 113, 124–135. [Google Scholar] [CrossRef]
- Golichenari, B.; Nosrati, R.; Farokhi-Fard, A.; Faal Maleki, M.; Gheibi Hayat, S.M.; Ghazvini, K.; Vaziri, F.; Behravan, J. Electrochemical-based biosensors for detection of Mycobacterium tuberculosis and tuberculosis biomarkers. Crit. Rev. Biotechnol. 2019, 39, 1056–1077. [Google Scholar] [CrossRef]
- Mowbray, S.E.; Amiri, A.M. A Brief Overview of Medical Fiber Optic Biosensors and Techniques in the Modification for Enhanced Sensing Ability. Diagnostics (Basel) 2019, 9, 23. [Google Scholar] [CrossRef] [Green Version]
- Abisado, R.G.; Benomar, S.; Klaus, J.R.; Dandekar, A.A.; Chandler, J.R. Bacterial Quorum Sensing and Microbial Community Interactions. mBio 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Bhardwaj, N.; Bhardwaj, S.K.; Mehta, J.; Kim, K.H.; Deep, A. MOF-Bacteriophage Biosensor for Highly Sensitive and Specific Detection of Staphylococcus aureus. ACS Appl. Mater. Interfaces 2017, 9, 33589–33598. [Google Scholar] [CrossRef]
- Nasrin, F.; Chowdhury, A.D.; Takemura, K.; Lee, J.; Adegoke, O.; Deo, V.K.; Abe, F.; Suzuki, T.; Park, E.Y. Single-step detection of norovirus tuning localized surface plasmon resonance-induced optical signal between gold nanoparticles and quantum dots. Biosens. Bioelectron. 2018, 122, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Damborský, P.; Švitel, J.; Katrlík, J. Optical biosensors. Essays Biochem. 2016, 60, 91–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nquyet, N.T.; Yen, L.T.H.; Doan, V.Y.; Hoang, N.L.; Van Thu, V.; Lan, H.; Trung, T.; Pham, V.H.; Tam, P.D. A label-free and highly sensitive DNA biosensor based on the core-shell structured CeO2-NR@Ppy nanocomposite for Salmonella detection. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 96, 790–797. [Google Scholar] [CrossRef]
- Campuzano, S.; Yáñez-Sedeño, P.; Pingarrón, J.M. Molecular Biosensors for Electrochemical Detection of Infectious Pathogens in Liquid Biopsies: Current Trends and Challenges. Sensors (Basel) 2017, 17, 2533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Oueslati, R.; Cheng, C.; Zhao, L.; Chen, J.; Almeida, R.; Wu, J. Rapid, highly sensitive detection of Gram-negative bacteria with lipopolysaccharide based disposable aptasensor. Biosens. Bioelectron. 2018, 112, 48–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Xu, J.; Liu, J.; Wang, X.; Chen, B. Disease-Related Detection with Electrochemical Biosensors: A Review. Sensors (Basel) 2017, 17, 2375. [Google Scholar] [CrossRef]
- Zanchetta, G.; Lanfranco, R.; Giavazzi, F.; Bellini, T.; Buscaglia, M. Emerging applications of label-free optical biosensors. Nanophotonics 2017, 6, 158. [Google Scholar] [CrossRef]
- Dudak, F.C.; Boyaci, I.H. Rapid and label-free bacteria detection by surface plasmon resonance (SPR) biosensors. Biotechnol. J. 2009, 4, 1003–1011. [Google Scholar] [CrossRef]
- Zhang, Y.; Tian, J.; Li, K.; Tian, H.; Xu, W. Label-free visual biosensor based on cascade amplification for the detection of Salmonella. Anal. Chim. Acta 2019, 1075, 144–151. [Google Scholar] [CrossRef]
- Yang, F.; Chang, T.L.; Liu, T.; Wu, D.; Du, H.; Liang, J.; Tian, F. Label-free detection of Staphylococcus aureus bacteria using long-period fiber gratings with functional polyelectrolyte coatings. Biosens. Bioelectron. 2019, 133, 147–153. [Google Scholar] [CrossRef]
- Wang, Z.; Yao, X.; Wang, R.; Ji, Y.; Yue, T.; Sun, J.; Li, T.; Wang, J.; Zhang, D. Label-free strip sensor based on surface positively charged nitrogen-rich carbon nanoparticles for rapid detection of Salmonella enteritidis. Biosens. Bioelectron. 2019, 132, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.H.; Huang, S.C.; Chen, K.P.; Li, B.R.; Li, Y.K. Effective Construction of a High-Capacity Boronic Acid Layer on a Quartz Crystal Microbalance Chip for High-Density Antibody Immobilization. Sensors (Basel) 2019, 19, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, S.; Soylu, M.C.; Kirimli, C.E.; Wu, W.; Sen, B.; Joshi, S.G.; Emery, C.L.; Au, G.; Niu, X.; Hamilton, R.; et al. Rapid, label-free genetic detection of enteropathogens in stool without genetic isolation or amplification. Biosens. Bioelectron. 2019, 130, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Silva, N.F.D.; Magalhães, J.M.C.S.; Barroso, M.F.; Oliva-Teles, T.; Freire, C.; Delerue-Matos, C. In situ formation of gold nanoparticles in polymer inclusion membrane: Application as platform in a label-free potentiometric immunosensor for Salmonella typhimurium detection. Talanta 2019, 194, 134–142. [Google Scholar] [CrossRef]
- Srinivasan, S.; Ranganathan, V.; DeRosa, M.C.; Murari, B.M. Label-free aptasensors based on fluorescent screening assays for the detection of Salmonella typhimurium. Anal. Biochem. 2018, 559, 17–23. [Google Scholar] [CrossRef]
- Rubab, M.; Shahbaz, H.M.; Olaimat, A.N.; Oh, D.H. Biosensors for rapid and sensitive detection of Staphylococcus aureus in food. Biosens. Bioelectron. 2018, 105, 49–57. [Google Scholar] [CrossRef]
- Wu, R.; Ma, Y.; Pan, J.; Lee, S.H.; Liu, J.; Zhu, H.; Gu, R.; Shea, K.J.; Pan, G. Efficient. Capture, Rapid Killing and Ultrasensitive Detection of Bacteria by a Nano-Decorated Multi-Functional Electrode Sensor. Biosens. Bioelectron. 2018, 101, 52–59. [Google Scholar] [CrossRef]
- Kimuda, S.G.; Biraro, I.A.; Bagaya, B.S.; Raynes, J.G.; Cose, S. Characterising antibody avidity in individuals of varied Mycobacterium tuberculosis infection status using surface plasmon resonance. PLoS ONE 2018, 13, e0205102. [Google Scholar] [CrossRef]
- Rebelo, R.; Barbosa, A.I.; Caballero, D.; Kwon, I.K.; Oliveira, J.M.; Kundu, S.C.; Reis, R.L.; Correlo, V.M. 3D biosensors in advanced medical diagnostics of high mortality diseases. Biosens. Bioelectron. 2019, 130, 20–39. [Google Scholar] [CrossRef]
- Erdem, Ö.; Saylan, Y.; Cihangir, N.; Denizli, A. Molecularly imprinted nanoparticles based plasmonic sensors for real-time Enterococcus faecalis detection. Biosens. Bioelectron. 2019, 126, 608–614. [Google Scholar] [CrossRef]
- Hoyos-Nogués, M.; Gil, F.J.; Mas-Moruno, C. Antimicrobial Peptides: Powerful Biorecognition Elements to Detect Bacteria in Biosensing Technologies. Molecules 2018, 23, 1683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liebana, S.; Brandao, D.; Alegret, S.; Pividori, M.I. Electrocehmical immunosensors, genosensors and phagosensors for Salmonela detection. Anal. Methods 2014, 6, 8858–8873. [Google Scholar] [CrossRef]
- Cui, F.; Xu, Y.; Wang, R.; Liu, H.; Chen, L.; Zhang, Q.; Mu, X. Label-free impedimetric glycan biosensor for quantitative evaluation interactions between pathogenic bacteria and mannose. Biosens. Bioelectron. 2018, 103, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Bu, T.; Huang, Q.; Yan, L.; Zhang, W.; Dou, L.; Huang, L.; Yang, Q.; Zhao, B.; Yang, B.; Li, T.; et al. Applicability of biological dye tracer in strip biosensor for ultrasensitive detection of pathogenic bacteria. Food Chem. 2019, 274, 816–821. [Google Scholar] [CrossRef]
- Zarei, S.S.; Soleimanian-Zad, S.; Ensafi, A.A. An impedimetric aptasensor for Shigella dysenteriae using a gold nanoparticle-modified glassy carbon electrode. Mikrochim. Acta. 2018, 185, 538. [Google Scholar] [CrossRef]
- Chuensirikulchai, K.; Laopajon, W.; Phunpae, P.; Apiratmateekul, N.; Surinkaew, S.; Tayapiwatana, C.; Pata, S.; Kasinrerk, W. Sandwich antibody-based biosensor system for identification of Mycobacterium tuberculosis complex and nontuberculous mycobacteria. J Immunoass. Immunochem. 2019, 29, 590–604. [Google Scholar] [CrossRef]
- Kravets, V.G.; Kabashin, A.V.; Barnes, W.L.; Grigorenko, A.N. Plasmonic Surface Lattice Resonances: A Review of Properties and Applications. Chem. Rev. 2018, 118, 12–5912. [Google Scholar] [CrossRef]
- Ermolaeva, T.N.; Kalmykova, E.N. Piezoelectric immunosensors: Analytical potentials and outlooks. Russ. Chem. Rev. 2006, 75, 397. [Google Scholar] [CrossRef]
- Muratsugu, M.; Ohta, F.; Miya, Y.; Hosokawa, T.; Kurosawa, S.; Kamo, N.; Ikeda, H. Quartz crystal microbalance for the detection of microgram quantities of human serum albumin: Relationship between the frequency change and the mass of protein adsorbed. Anal. Chem. 1993, 65, 2933–2937. [Google Scholar] [CrossRef]
- Savas, S.; Altintas, Z. Graphene Quantum Dots as Nanozymes for Electrochemical Sensing of Yersinia enterocolitica in Milk and Human Serum. Materials (Basel) 2019, 12, 2189. [Google Scholar] [CrossRef] [Green Version]
- Russel, M.; Sophocleous, M.; JiaJia, S.; Xu, W.; Xiao, L.; Maskow, T.; Alam, M.; Georgiou, J. High-frequency, dielectric spectroscopy for the detection of electrophysiological/biophysical differences in different bacteria types and concentrations. Anal. Chim. Acta 2018, 1028, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Piekarz, I.; Górska, S.; Odrobina, S.; Drab, M.; Wincza, K.; Gamian, A.; Gruszczynski, S. A microwave matrix sensor for multipoint label-free Escherichia coli detection. Biosens. Bioelectron. 2019, 147, 111784. [Google Scholar] [CrossRef] [PubMed]
- Biagi, M.C.; Fabregas, R.; Gramse, G.; van der Hofstadt, M.; Juárez, A.; Kienberger, F.; Fumagalli, L.; Gomila, G. Nanoscale Electric Permittivity of Single Bacterial Cells at Gigahertz Frequencies by Scanning Microwave Microscopy. ACS Nano 2016, 10, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Esteban-Ferrer, D.; Edwards, M.A.; Fumagalli, L.; Juárez, A.; Gomila, G. Electric Polarization Properties of Single Bacteria Measured with Electrostatic Force Microscopy. ACS Nano 2014, 8, 9843–9849. [Google Scholar] [CrossRef] [PubMed]
- Checa, M.; Millan-Solsona, R.; Blanco, N.; Torrents, E.; Fabregas, R.; Gomila, G. Mapping the dielectric constant of a single bacterial cell at the nanoscale with scanning dielectric force volume microscopy. Nanoscale. 2019, 11, 20809–20819. [Google Scholar] [CrossRef]
- Flores-Cosío, G.; Herrera-López, E.J.; Arellano-Plaza, M.; Gschaedler-Mathis, A.; Sanchez, A.; Amaya-Delgado, L. Dielectric property measurements as a method to determine the physiological state of Kluyveromyces marxianus and Saccharomyces cerevisiae stressed with furan aldehydes. Appl. Microbiol. Biotechnol. 2019, 103, 9633–9642. [Google Scholar] [CrossRef]
- Mehrotra, P.; Chatterjee, B.; Sen, S. EM-Wave Biosensors: A Review of RF, Microwave, mm-Wave and Optical Sensing. Sensors (Basel) 2019, 19, 1013. [Google Scholar] [CrossRef] [Green Version]
- Oberoi, K.S.; Daya, K.S.; Tirumalai, P.S. Microwave Sensor for Detection of E. Coli in Water. In Proceedings of the Sixth International Conference on Sensing Technology (ICST), Kolkata, India, 18–21 December 2012. [Google Scholar] [CrossRef]
- Narang, R.; Mohammadi, S.; Ashani, M.M.; Narang, R.; Mohammadi, S.; Ashani, M.M.; Sadabadi, H.; Hejazi, H.; Zarifi, M.H.; Sanati-Nezhad, A. Sensitive, Real-time and Non-Intrusive Detection of Concentration and Growth of Pathogenic Bacteria using Microfluidic-Microwave Ring Resonator Biosensor. Sci. Rep. 2018, 8, 15807. [Google Scholar] [CrossRef]
- Tigli, O.; Zaghloul, M. A Novel Circular SAW (Surface Acoustic Wave) Device in CMOS. Proc. IEEE Sens. 2007, 474–477. [Google Scholar] [CrossRef]
- Alvarez, M.; Lechuga, L.M. Microcantilever-based platforms as biosensing tools. Analyst 2010, 135, 827–836. [Google Scholar] [CrossRef]
- Wu, G.; Datar, R.H.; Hansen, K.M.; Thundat, T.; Cote, R.J.; Majumdar, A. Bioassay of prostate-specific antigen (PSA) using microcantilevers. Nat. Biotechnol. 2001, 19, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Sang, S.; Zhao, Y.; Zhang, W.; Li, P.; Hu, J.; Li, G. Surface stress-based biosensors. Biosens. Bioelectron. 2014, 51, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Dahlin, A.V. Plasmonic Biosensors: An Integrated View of Refractometric Detection; De Monfort University: Leicester, UK, 2012; Volume 4, 316p. [Google Scholar] [CrossRef]
- Ilica, B.; Yang, Y.; Craighead, H.G. Virus detection using nanoelectromechanical devices. Appl. Phys. Lett. 2004, 85, 2604. [Google Scholar] [CrossRef] [Green Version]
- Arlett, J.L.; Myers, E.B.; Roukes, M.L. Comparative advantages of mechanical biosensors. Nat. Nanotechnol. 2011, 6, 203–215. [Google Scholar] [CrossRef] [Green Version]
- Barton, R.A.; Ilic, B.; Verbridge, S.S.; Cipriany, B.R.; Parpia, J.M.; Craighead, H.G. Fabrication of a nanomechanical mass sensor containing a nanofluidic channel. Nano Lett. 2010, 10, 2058–2063. [Google Scholar] [CrossRef]
- Lee, J.; Moon, S.U.; Lee, Y.S.; Ali, B.A.; Al-Khedhairy, A.A.; Ali, D.; Ahmed, J.; Al Salem, A.M.; Kim, S. Quantum dot-based molecular beacon to monitor intracellular microRNAs. Sensors (Basel) 2015, 15, 12872–12883. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.; Kim, J.P.; Sim, S.J.; Lee, J. A multisized piezoelectric microcantilever biosensor array for the quantitative analysis of mass and surface stress. Appl. Phys. Lett. 2008, 93, 102902. [Google Scholar] [CrossRef]
- Kuss, S.; Amin, H.M.A.; Compton, R.G. Electrochemical Detection of Pathogenic Bacteria-Recent Strategies, Advances and Challenges. Chem. Asian J. 2018, 13, 2758–2769. [Google Scholar] [CrossRef]
- Muniandy, S.; Teh, S.J.; Appaturi, J.N.; Thong, K.L.; Lai, C.W.; Ibrahim, F.; Leo, B.F. A reduced graphene oxide-titanium dioxide nanocomposite based electrochemical aptasensor for rapid and sensitive detection of Salmonella enterica. Bioelectrochemistry 2019, 127, 136–144. [Google Scholar] [CrossRef]
- Singh, R.; Mukherjee, M.D.; Sumana, G.; Gupta, R.K.; Sood, S.; Malhotra, B.D. Biosensors for pathogen detection: A smart approach towards clinical diagnosis. Sens. Actuators B Chem. 2014, 197, 385–404. [Google Scholar] [CrossRef]
- Gonzalez-Sapienza, G.; Rossotti, M.A.; Tabares-da Rosa, S. Single-Domain Antibodies as Versatile Affinity Reagents for Analytical and Diagnostic Applications. Front. Immunol. 2017, 8, 977. [Google Scholar] [CrossRef] [PubMed]
- Bever, C.S.; Dong, J.X.; Vasylieva, N.; Barnych, B.; Cui, Y.; Xu, Z.L.; Hammock, B.D.; Gee, S.J. VHH antibodies: Emerging reagents for the analysis of environmental chemicals. Anal. Bioanal. Chem. 2016, 408, 5985–6002. [Google Scholar] [CrossRef]
- Shriver-Lake, L.C.; Liu, J.L.; Zabetakis, D.; Sugiharto, V.A.; Lee, C.; Defang, G.N.; Wu, S.L.; Anderson, G.P.; Goldman, E.R. Selection and Characterization of Anti-Dengue NS1 Single Domain Antibodies. Sci. Rep. 2018, 8, 18086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goode, J.; Dillon, G.; Liu, P.A. The development and optimisation of nanobody based electrochemical immunosensors for IgG. Sens. Actuators B Chem. 2016, 234, 478–484. [Google Scholar] [CrossRef] [Green Version]
- Della Pia, E.A.; Martinez, K.L. Single domain antibodies as a powerful tool for high quality surface plasmon resonance studies. PLoS ONE 2015, 10, e0124303. [Google Scholar] [CrossRef] [PubMed]
- Manjavacas, A.; Zundel, L.; Sanders, S. Analysis of the Limits of the Near-Field Produced by Nanoparticle Arrays. ACS Nano 2019, 13, 10682–10693. [Google Scholar] [CrossRef]




| Advantages | Disadvantages | References |
|---|---|---|
| ◾ Cheap, rapid, inexpensive, and easy to apply tests. ◾ Long shelf-life of test systems. ◾ Test systems do not require special temperature conditions for storage. ◾ No additional special equipment is required. ◾ They do not need qualified personnel. ◾ They can be used by general practice physicians or patients at home. ◾ Visual result is clear and easily distinguishable. ◾ Tests are usually sold as kits with a set of all the items needed to perform the test. ◾ Possible increase in sensitivity of test systems by the use of plasmon resonance, surface-enhanced Raman scattering (SERS), chemiluminescent or fluorescent labels. ◾ Possibility of multiplexed formats of test systems | ◾ Suitable only for primary screening and require confirmation of positive results by independent methods. ◾ Special equipment (scanners, reflectometers, CCD cameras) and software are required to obtain quantitative results. ◾ Technological improvement of the method increases cost and duration of the analysis. ◾ In a competitive format, response negatively correlates with concentration. ◾ Possible technical errors in application of specimen may affect the accuracy and reproducibility of result. ◾ Increase in sensitivity of tests is based on the use of gold, silver, or enzyme nanoparticles, which limits shelf-life, increases cost of analysis, and breaks the one-step rule of application. ◾ Tested specimen must be in the form of a solution. Preliminary dissolution of dry specimens is mandatory. ◾ When the analyte content in the solution is low, the specimen needs to be concentrated. | [14] [17,21] [15,19] [16,18,20] [18,19,20] [16,18,20] [13] [14,16,17] [18] [13,15,16] [18,19] |
| Advantages | References |
|---|---|
| ● A simplified pattern of analysis. | [3,29,49,51,81] |
| ● Reduced analysis time (rapid response time). | [7,29,82] |
| ● Lower cost of analysis. | [7,28,80] |
| ● Reduced consumption of organic solvents. | [33,64,78,83] |
| ● Portability and small dimensions. | [33,43,73] |
| ● No need in qualified medical personnel. | [3,7,39,64,83] |
| ● Opportunity to quantify biomolecules in real-time mode. | [25,26,78,84,85] |
| ● Target analytes are detected in natural forms, without. modifications and labels. | [22,33,73,80,82] |
| ● High sensitivity. | [22,25,26,43,64,85,86] |
| ● Direct measurement of analytes. | [43,51,64,80] |
| ● Opportunity to detect small molecules. | [3,7,25,26,43,79] |
| ● Opportunity of multiplexing. | [28,29,64,83] |
| ● Access to kinetic and thermodynamic parameters. | [22,26,39,80,86] |
| Recognizing Bioreceptor | Conversion Method | Test Models of Pathogens, Sensitivity | References |
|---|---|---|---|
| Bacteriophage | Photoluminescence | S. aureus 4 × 108 ufc/mL | [70] |
| Antimicrobial peptides | Impedancemetry | E. coli, S. aureus, P. aeruginosa, S. epidermidis, 102 ufc/mL | [75] |
| Antibacterial nanoparticles Zn-CuO and graphene oxide Man/MUA-MH/Au * | Impedancemetry and electrochemical impedance spectroscopy | E. coli, S. aureus 50 ufc/mL and antibacterial effect 100% (30 min) | [93] |
| Thiolated protein G on: - gold electrodes - gold nanoparticles | Cyclic voltammetry and electrochemical impedance spectroscopy | S. typhimurium, 2.16 × 106 ufc/mL E. coli, 50–103 ufc/mL | [102] |
| Enzymes | Electrochemical | E. coli O157:H7 150 ufc/mL | [68] |
| Nucleic acids (DNA, RNA) | Electrochemical | S. aureus, 140 ufc/mL S. typhimurium, 48 ufc/mL | [72] |
| Nucleic acids (DNA, RNA) | Electrochemical | S. aureus, M. tuberculosis | [73] |
| Aptamer on AuNP | Autofluorescence quenching | S. typhimurium, 48 ufc/mL | [92] |
| Monoclonal antibodies | Optical | S. enteritidis, 80 ufc/mL Listeria monocytogenes | [103] |
| Thiolated aptamer | Impedancemetry | Shigella dysenteriae | [104] |
| Nucleic acids (DNA, RNA) | Electrochemical impedance spectroscopy | M. tuberculosis | [50] |
| Monoclonal antibodies | Surface plasmon resonance | Enterococcus faecalis, 104–108 ufc/mL | [99] |
| Aptamer | Impedancemetry | Bacillus cereus, 104–106 ufc/mL Bacillus anthracis (spores) | [32] |
| Nucleic acids (DNA) | Cyclic voltammetry and electrochemical impedance spectroscopy | Salmonella spp. | [81] |
| Enzyme Simulator (Graphene Quantum Dots, GQD) | Electrochemical | Yersinia enterocolitica, 5 (milk)–30 (serum) ufc/mL | [105] |
| Monoclonal antibodies | Surface plasmon resonance | S. aureus, 224 ufc/mL, 30 min | [71] |
| Monoclonal antibodies | Visualization | Salmonella enteritidis, 102–108 ufc/mL | [88] |
| DNA, aptamer | Electrochemical | Bird flu virus H5N1 (AIV) | [48] |
| Nucleic acids (DNA) | Electrochemical impedance | Zika virus, 25.0 ± 1.7 hМ. | [49] |
| Aptamer (rGO-TiO2) | Electrochemical | S. enterica Typhimurium, 101–108 ufc/mL | [98] |
| Nucleic acids (DNA) | Piezoelectric | Clostridium difficile, sensitivity 95% and specificity 95% | [90] |
| Monoclonal antibodies | Surface plasmon resonance | M. tuberculosis, 102–106 ufc/mL | [93,101] |
| Aptamer | Fluorescent | S. enterica Typhimurium, 6–10 ufc/mL | [92] |
| Monoclonal antibodies | Potentiometry | S. enterica Typhimurium, 6 ufc/mL | [91] |
| Nucleic acids (DNA) | Electrochemical impedance | M. tuberculosis, 102–106 ufc/mL | [50] |
| Aptamer (RNA) | Fluorescent | S. aureus, 102–106 ufc/mL | [3] |
| Nicolson-Ross-Weir method | Dielectric spectroscopy | Bacillus Subtilis, 2.10–1.30 × 109 ufc/mL E. coli 1.60–1.00 × 109 ufc/mL | [106] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andryukov, B.G.; Besednova, N.N.; Romashko, R.V.; Zaporozhets, T.S.; Efimov, T.A. Label-Free Biosensors for Laboratory-Based Diagnostics of Infections: Current Achievements and New Trends. Biosensors 2020, 10, 11. https://doi.org/10.3390/bios10020011
Andryukov BG, Besednova NN, Romashko RV, Zaporozhets TS, Efimov TA. Label-Free Biosensors for Laboratory-Based Diagnostics of Infections: Current Achievements and New Trends. Biosensors. 2020; 10(2):11. https://doi.org/10.3390/bios10020011
Chicago/Turabian StyleAndryukov, Boris G., Natalya N. Besednova, Roman V. Romashko, Tatyana S. Zaporozhets, and Timofey A. Efimov. 2020. "Label-Free Biosensors for Laboratory-Based Diagnostics of Infections: Current Achievements and New Trends" Biosensors 10, no. 2: 11. https://doi.org/10.3390/bios10020011
APA StyleAndryukov, B. G., Besednova, N. N., Romashko, R. V., Zaporozhets, T. S., & Efimov, T. A. (2020). Label-Free Biosensors for Laboratory-Based Diagnostics of Infections: Current Achievements and New Trends. Biosensors, 10(2), 11. https://doi.org/10.3390/bios10020011