The Growing Interest in Development of Innovative Optical Aptasensors for the Detection of Antimicrobial Residues in Food Products
Abstract
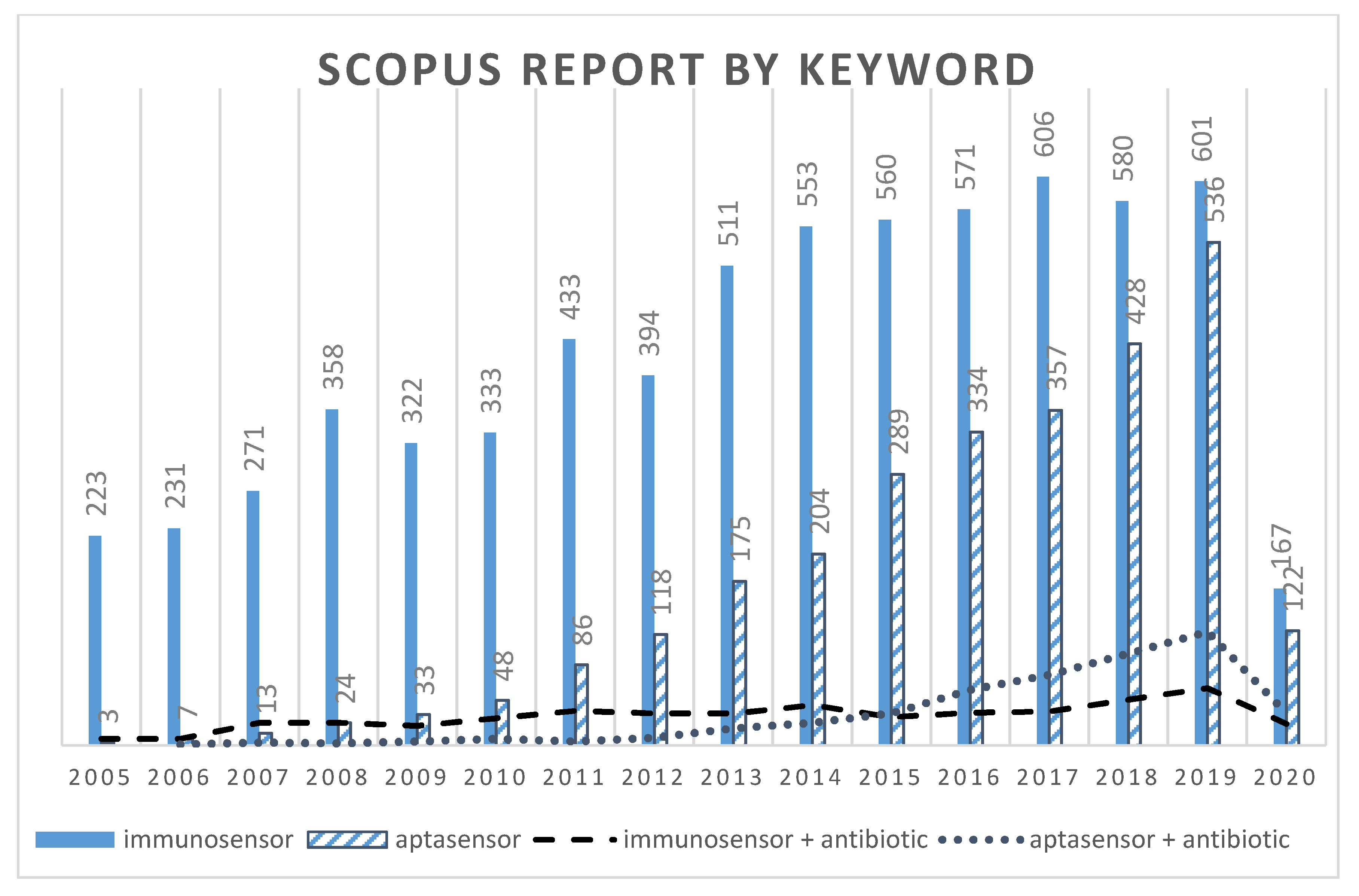
:1. Introduction
1.1. Antimicrobial Residues in Food Products
1.2. Screening Methods
1.3. Biosensors
2. Aptamers for Antimicrobial Residues
2.1. Aptamer Production
2.2. Aptamers’ Characteristics: Advantages and Drawbacks
- -
- A change of mass can be detected with mass-sensitive (e.g., quartz crystal microbalance (QCM), surface acoustic wave (SAW)) or optical biosensors (e.g., surface plasmon resonance (SPR));
- -
- A change of its structure and/or conformation (e.g., hairpin, pseudo-node), to fully interact with the target, could be detected by optical or electrochemical biosensors, when, for instance, the 3′ and 5′ extremities of the aptamer are labeled with specific tags (e.g., pairs fluorophores (e.g., carboxyfluorescein (FAM), cyanine)—quenchers (e.g., nanomaterials (e.g., quantum dots (QDs), gold nanoparticles (AuNPs)), or electrochemical tags (methylene blue (MB), ferrocene)). The conformation of a given aptamer is heavily affected by various factors, such as temperature, pH, type, and concentration of cations.
- -
- A change in electrochemical properties could be detected, even without labeling. Aptamers are polyanionic molecules. Therefore, their electrochemical properties could be used for the detection of a target analyte. A change in conductance could be observed and measured when the target analyte is bound to the aptamer compared to when the aptamer is free.
2.3. Aptamers for Antimicrobials
3. Aptasensors for the Detection of Antimicrobial Residues
3.1. Fluorescent Aptasensors
3.2. Colorimetric Aptasensors
3.3. Other Optical Aptasensors
3.3.1. Chemiluminescent (CL) Aptasensors
3.3.2. Surface Plasmon Resonance (SPR) Aptasensors
3.3.3. Surface-Enhanced Raman Scattering (SERS) Aptasensors
3.3.4. Resonance Rayleigh Scattering Spectra (RRSS) Aptasensors
4. Conclusions and Perspectives
Funding
Conflicts of Interest
References
- European Commission. Council Regulation (EU) (2010) No. 37/2010 of 22 December 2009 on pharmacologically active substances and their classification regarding maximum residue limits in foodstuffs of animal origin. Off. J. Eur. Union 2009, 15, 1–72. [Google Scholar]
- European Commission. Commission Regulation (EC) No. 470/2009 laying down Community procedures for the establishment of residue limits of pharmacologically active substances in foodstuffs of animal origin. Eur. Parliement Counc. Off. J. Eur. Union 2009, 152, 11–22. [Google Scholar]
- European Commission. Commission Regulation (EU) 2019/1871 of 7 November 2019 on reference points for action for non-allowed pharmacologically active substances present in food of animal origin and repealing Decision 2005/34/EC (Text with EEA relevance). Off. J. Eur. Union 2019, 289, 41–46. [Google Scholar]
- European Commission. Regulation (EU) 2017/625 of the European Parliament and of the Council of 15 March 2017 on official controls and other official activities performed to ensure the application of food and feed law, rules on animal health and welfare, plant health and plant protection products. Off. J. Eur. Union 2017, 95, 1–42. [Google Scholar]
- European Commission. Commission Decision (EC) N° 2002/657 of 12 August 2002 implementing Council Directive. 96/23/EC concerning the performance of analytical methods and interpretation of results. Off. J. Eur. Union 2002, 221, 8–36. [Google Scholar]
- Watkins, H.; Kožárová, I. Broad Spectrum Detection of Antibiotic Residues in Poultry Meat by a Multi-Plate Assay. Folia Vet. 2019, 63, 9–17. [Google Scholar] [CrossRef] [Green Version]
- Pikkemaat, M.G.; Rapallini, M.L.B.A.; Zuidema, T.; Elferink, J.W.A.; Oostra-van Dijk, S.; Driessen-van Lankveld, W.D.M. Screening methods for the detection of antibiotic residues in slaughter animals: Comparison of the European Union Four-Plate Test, the Nouws Antibiotic Test and the PremiTest (applied to muscle and kidney). Food Addit. Contam. 2011, 28, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Xing, C.; Jing, X.; Zhang, X.; Yuan, J. Ultrasensitive indirect competitive ELISA and strip sensor for detection of furazolidone metabolite in animal tissues. Food Agric. Immunol. 2017, 28, 1269–1282. [Google Scholar] [CrossRef] [Green Version]
- Gaudin, V.; Hedou, C.; Verdon, E. Validation of 2 ELISA kits for the screening of tylosin and streptomycin in honey according to the European decision EC/2002/657. Food Addit. Contam. Part A 2013, 30, 93–109. [Google Scholar] [CrossRef]
- Han, S.; Zhou, T.; Yin, B.; He, P. A sensitive and semi-quantitative method for determination of multi-drug residues in animal body fluids using multiplex dipstick immunoassay. Anal. Chim. Acta 2016, 927, 64–71. [Google Scholar] [CrossRef]
- Reybroeck, W.; Ooghe, S.; Brabander, H.D.; Daeseleire, E. Validation of the Tetrasensor Honey Test Kit for the Screening of Tetracyclines in Honey. J. Agric. Food Chem. 2007, 55, 8359–8366. [Google Scholar] [CrossRef] [PubMed]
- Dubreil, E.; Gautier, S.; Fourmond, M.P.; Bessiral, M.; Gaugain, M.; Verdon, E.; Pessel, D. Validation approach for a fast and simple targeted screening method for 75 antibiotics in meat and aquaculture products using LC-MS/MS. Food Addit. Contam. Part A 2017, 34, 453–468. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Park, S.J.; Park, H.C.; Hossain, M.A.; Kim, M.A.; Son, S.W.; Lim, C.M.; Kim, T.W.; Cho, B.H. Multiresidue Screening of Veterinary Drugs in Meat, Milk, Egg, and Fish Using Liquid Chromatography Coupled with Ion Trap Time-of-Flight Mass Spectrometry. Appl. Biochem. Biotechnol. 2016, 182, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Gooch, J.; Daniel, B.; Parkin, M.; Frascione, N. Developing aptasensors for forensic analysis. TrAC Trends Anal. Chem. 2017, 94, 150–160. [Google Scholar] [CrossRef] [Green Version]
- Yáñez-Sedeño, P.; Agüí, L.; Villalonga, R.; Pingarrón, J.M. Biosensors in forensic analysis. A review. Anal. Chim. Acta 2014, 823, 1–19. [Google Scholar] [CrossRef]
- Aberl, F.; Kosslinger, C. Biosensor-based methods in clinical diagnosis. Mol. Diagn. Infect. Dis. 1998, 13, 503–517. [Google Scholar]
- Justino, C.I.; Rocha-Santos, T.A.; Duarte, A.C. Review of analytical figures of merit of sensors and biosensors in clinical applications. Trends Anal. Chem. 2010, 29, 1172–1183. [Google Scholar] [CrossRef]
- Velusamy, V.; Arshak, K.; Korostynska, O.; Oliwa, K.; Adley, C. An overview of foodborne pathogen detection: In the perspective of biosensors. Biotechnol. Adv. 2010, 28, 232–254. [Google Scholar] [CrossRef]
- Gaudin, V. Advances in biosensor development for the screening of antibiotic residues in food products of animal origin—A comprehensive review. Biosens. Bioelectron. 2017, 90, 363–377. [Google Scholar] [CrossRef]
- Miller, G.A.; Clark, R.C.; Jessee, E.J. Production of monoclonal antibodies to salinomycin. Hybridoma 1986, 5, 353–360. [Google Scholar] [CrossRef]
- van de Water, C.; Haagsma, N.; van Kooten, P.J.; van Eden, W. An enzyme linked immunosorbent assay for the determination of chloramphenicol using a monoclonal antibody. Z. Lebensm.-Unters. Forsch. 1987, 185, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Dixon-Holland, D.E. ELISA and its application for residue analysis of antibiotics and drugs in products of animal origin. In Analysis of Antibiotic/Drug Residues in Food Products of Animal Origin; Springer: Boston, MA, USA, 1992; pp. 57–74. [Google Scholar]
- Haagsma, N.; Van de Water, C. Immunochemical methods in the analysis of veterinary drug residues. In Analysis of Antibiotic/Drug Residues in Food Products of Animal Origin; Springer: Boston, MA, USA, 1992; pp. 81–97. [Google Scholar]
- Tombelli, S.; Minunni, M.; Mascini, M. Analytical applications of aptamers. Biosens. Bioelectron. 2005, 20, 2424–2434. [Google Scholar] [CrossRef] [PubMed]
- Jayasena, S.D. Aptamers: An Emerging Class of Molecules That Rival Antibodies in Diagnostics. Clin. Chem. 1999, 45, 1628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Bock, L.C.; Griffin, L.C.; Latham, J.A.; Vermaas, E.H.; Toole, J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef]
- Lorsch, J.R.; Szostak, J.W. In vitro selection of RNA aptamers specific for cyanocobalamin. Biochemistry 1994, 33, 973–982. [Google Scholar] [CrossRef]
- Yamamoto, R.; Toyoda, S.; Viljanen, P.; Machida, K.; Nishikawa, S.; Murakami, K.; Taira, K.; Kumar, P.K. In vitro selection of RNA aptamers that can bind specifically to Tat protein of HIV-1. Nucleic Acids Symp. Ser. 1995, 145–146. [Google Scholar]
- Leung, L.L.K. Application of combinatorial libraries and protein engineering to the discovery of novel anti-thrombotic drugs. Thromb. Haemost. 1995, 74, 373–376. [Google Scholar] [CrossRef]
- Kim, Y.S.; Gu, M.B. Advances in Aptamer Screening and Small Molecule Aptasensors. In Biosensors Based on Aptamers and Enzymes; Gu, M.B., Kim, H.-S., Eds.; Springer: Berlin, Germany, 2014; pp. 29–67. [Google Scholar]
- Conrad, R.; Ellington, A.D. Detecting Immobilized Protein Kinase C Isozymes with RNA Aptamers. Anal. Biochem. 1996, 242, 261–265. [Google Scholar] [CrossRef]
- Weiss, S.; Proske, D.; Neumann, M.; Groschup, M.H.; Kretzschmar, H.A.; Famulok, M.; Winnacker, E.L. RNA aptamers specifically interact with the prion protein PrP. J. Virol. 1997, 71, 8790–8797. [Google Scholar] [CrossRef] [Green Version]
- Davis, K.A.; Abrams, B.; Lin, Y.; Jayasena, S.D. Staining of cell surface human CD4 with 2′-F-pyrimidine-containing RNA aptamers for flow cytometry. Nucleic Acids Res. 1998, 26, 3915–3924. [Google Scholar] [CrossRef] [PubMed]
- Brody, E.N.; Gold, L. Aptamers as therapeutic and diagnostic agents. Rev. Mol. Biotechnol. 2000, 74, 5–13. [Google Scholar] [CrossRef]
- Muldoon, M.T.; Holtzapple, C.K.; Deshpande, S.S.; Beier, R.C.; Stanker, L.H. Development of a monoclonal antibody-based cELISA for the analysis of sulfadimethoxine. 1. Development and characterization of monoclonal antibodies and molecular modeling studies of antibody recognition. J. Agric. Food Chem. 2000, 48, 537–544. [Google Scholar] [CrossRef]
- Hasegawa, H.; Savory, N.; Abe, K.; Ikebukuro, K. Methods for Improving Aptamer Binding Affinity. Molecules 2016, 21, 421. [Google Scholar] [CrossRef]
- Kwon, Y.S.; Ahmad Raston, N.H.; Gu, M.B. An ultra-sensitive colorimetric detection of tetracyclines using the shortest aptamer with highly enhanced affinity. Chem. Commun. 2014, 50, 40–42. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.E.; Wu, H.; Niu, Y.; Cai, J. Improving the stability of aptamers by chemical modification. Curr. Med. Chem. 2011, 18, 4126–4138. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.; Duan, N.; Wu, S.; Xu, B.; Wang, Z. Chemiluminescent aptasensor for chloramphenicol based on N-(4-aminobutyl)-N-ethylisoluminol-functionalized flower-like gold nanostructures and magnetic nanoparticles. Anal. Bioanal. Chem. 2015, 407, 7907–7915. [Google Scholar] [CrossRef]
- González-Fernández, E.; de-los-Santos-Álvarez, N.; Lobo-Castañón, M.J.; Miranda-Ordieres, A.J.; Tuñón-Blanco, P. Aptamer-Based Inhibition Assay for the Electrochemical Detection of Tobramycin Using Magnetic Microparticles. Electroanalysis 2011, 23, 43–49. [Google Scholar] [CrossRef]
- Ha, N.R.; Jung, I.P.; La, I.J.; Jung, H.S.; Yoon, M.Y. Ultra-sensitive detection of kanamycin for food safety using a reduced graphene oxide-based fluorescent aptasensor. Sci. Rep. 2017, 7, 40305. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Gu, C.; Wang, C.; Song, B.; Zhou, X.; Lou, X.; He, M. Evanescent wave aptasensor for continuous and online aminoglycoside antibiotics detection based on target binding facilitated fluorescence quenching. Biosens. Bioelectron. 2018, 102, 646–651. [Google Scholar] [CrossRef]
- Yan, C.; Zhang, J.; Yao, L.; Xue, F.; Lu, J.; Li, B.; Chen, W. Aptamer-mediated colorimetric method for rapid and sensitive detection of chloramphenicol in food. Food Chem. 2018, 260, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Guo, Y.; Wang, X.; Sun, X. Multiplexed aptasensor based on metal ions labels for simultaneous detection of multiple antibiotic residues in milk. Biosens. Bioelectron. 2018, 115, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Gan, N.; Shen, Z.; Cao, J.; Hu, F.; Li, T. Microchip electrophoresis based aptasensor for multiplexed detection of antibiotics in foods via a stir-bar assisted multi-arm junctions recycling for signal amplification. Biosens. Bioelectron. 2019, 130, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Youn, H.; Lee, K.; Her, J.; Jeon, J.; Mok, J.; So, J.I.; Shin, S.; Ban, C. Aptasensor for multiplex detection of antibiotics based on FRET strategy combined with aptamer/graphene oxide complex. Sci. Rep. 2019, 9, 7659. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Gan, N.; Zhou, Y.; Li, T.; Hu, F.; Cao, Y.; Chen, Y. Novel label-free and high-throughput microchip electrophoresis platform for multiplex antibiotic residues detection based on aptamer probes and target catalyzed hairpin assembly for signal amplification. Biosens. Bioelectron. 2017, 97, 100–106. [Google Scholar] [CrossRef]
- Hao, L.; Gu, H.; Duan, N.; Wu, S.; Wang, Z. A chemiluminescent aptasensor for simultaneous detection of three antibiotics in milk. Anal. Methods 2016, 8, 7929–7936. [Google Scholar] [CrossRef]
- Wu, Y.-Y.; Huang, P.; Wu, F.-Y. A label-free colorimetric aptasensor based on controllable aggregation of AuNPs for the detection of multiplex antibiotics. Food Chem. 2020, 304, 125377. [Google Scholar] [CrossRef]
- Nguyen, T.; Hilton, J.P.; Lin, Q. Emerging applications of aptamers to micro- and nanoscale biosensing. Microfluid. Nanofluidics 2009, 6, 347. [Google Scholar] [CrossRef]
- Wang, T.; Chen, C.; Larcher, L.M.; Barrero, R.A.; Veedu, R.N. Three decades of nucleic acid aptamer technologies: Lessons learned, progress and opportunities on aptamer development. Biotechnol. Adv. 2019, 37, 28–50. [Google Scholar] [CrossRef]
- Pfeiffer, F.; Mayer, G. Selection and Biosensor Application of Aptamers for Small Molecules. Front. Chem. 2016, 4, 25. [Google Scholar] [CrossRef] [Green Version]
- Kaur, H.; Bruno, J.G.; Kumar, A.; Sharma, T.K. Aptamers in the Therapeutics and Diagnostics Pipelines. Theranostics 2018, 8, 4016–4032. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature 1992, 355, 850–852. [Google Scholar] [CrossRef] [PubMed]
- Mehta, J.; Van Dorst, B.; Rouah-Martin, E.; Herrebout, W.; Scippo, M.L.; Blust, R.; Robbens, J. In vitro selection and characterization of DNA aptamers recognizing chloramphenicol. J. Biotechnol. 2011, 155, 361–369. [Google Scholar] [CrossRef]
- Burke, D.H.; Hoffman, D.C.; Brown, A.; Hansen, M.; Pardi, A.; Gold, L. RNA aptamers to the peptidyl transferase inhibitor chloramphenicol. Chem. Biol. 1997, 4, 833–843. [Google Scholar] [CrossRef] [Green Version]
- Sadeghi, A.S.; Mohsenzadeh, M.; Abnous, K.; Taghdisi, S.M.; Ramezani, M. Development and characterization of DNA aptamers against florfenicol: Fabrication of a sensitive fluorescent aptasensor for specific detection of florfenicol in milk. Talanta 2018, 182, 193–201. [Google Scholar] [CrossRef]
- Baugh, C.; Grate, D.; Wilson, C. 2.8 Å crystal structure of the malachite green. J. Mol. Biol. 2000, 301, 117–128. [Google Scholar] [CrossRef]
- Niazi, J.H.; Lee, S.J.; Kim, Y.S.; Gu, M.B. ssDNA aptamers that selectively bind oxytetracycline. Bioorg. Med. Chem. 2008, 16, 1254–1261. [Google Scholar] [CrossRef]
- Berens, C.; Thain, A.; Schroeder, R. A tetracycline-binding RNA aptamer. Bioorg. Med. Chem. 2001, 9, 2549–2556. [Google Scholar] [CrossRef]
- Niazi, J.H.; Lee, S.J.; Gu, M.B. Single-stranded DNA aptamers specific for antibiotics tetracyclines. Bioorg. Med. Chem. 2008, 16, 7245–7253. [Google Scholar] [CrossRef]
- Schürer, H.; Stembera, K.; Knoll, D.; Mayer, G.; Blind, M.; Förster, H.H.; Famulok, M.; Welzel, P.; Hahn, U. Aptamers that bind to the antibiotic moenomycin A. Bioorg. Med. Chem. 2001, 9, 2557–2563. [Google Scholar] [CrossRef]
- Paniel, N.; Istamboulié, G.; Triki, A.; Lozano, C.; Barthelmebs, L.; Noguer, T. Selection of DNA aptamers against penicillin G using Capture-SELEX for the development of an impedimetric sensor. Talanta 2017, 162, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, C.S.R. Aptamers to Antibiotics. In The Aptamer Handbook; Klussmann, D.S., Ed.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2006; pp. 116–130. [Google Scholar]
- Mehlhorn, A.; Rahimi, P.; Joseph, Y. Aptamer-Based Biosensors for Antibiotic Detection: A Review. Biosensors 2018, 8, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Yin, S.; Li, Y.; Lu, D.; Zhang, J.; Sun, C. Application of aptamers in detection and chromatographic purification of antibiotics in different matrices. TrAC Trends Anal. Chem. 2017, 95, 1–22. [Google Scholar] [CrossRef]
- Rowe, A.A.; Miller, E.A.; Plaxco, K.W. Reagentless Measurement of Aminoglycoside Antibiotics in Blood Serum via an Electrochemical, Ribonucleic Acid Aptamer-Based Biosensor. Anal. Chem. 2010, 82, 7090–7095. [Google Scholar] [CrossRef] [Green Version]
- Piro, B.; Shi, S.; Reisberg, S.; Noël, V.; Anquetin, G. Comparison of Electrochemical Immunosensors and Aptasensors for Detection of Small Organic Molecules in Environment, Food Safety, Clinical and Public Security. Biosensors 2016, 6, 7. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.M.; Hu, Y.; Yang, Y.K.; Liu, H.; Fang, G.Z.; Lu, X.; Wang, S. Emerging functional nanomaterials for the detection of food contaminants. Trends Food Sci. Technol. 2018, 71, 94–106. [Google Scholar] [CrossRef]
- Zhou, Y.; Mahapatra, C.; Chen, H.; Peng, X.; Ramakrishna, S.; Nanda, H.S. Recent developments in fluorescent aptasensors for detection of antibiotics. Curr. Opin. Biomed. Eng. 2020, 13, 16–24. [Google Scholar] [CrossRef]
- Wang, J.; Lu, T.; Hu, Y.; Wang, X.; Wu, Y. A label-free and carbon dots based fluorescent aptasensor for the detection of kanamycin in milk. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 226, 117651. [Google Scholar] [CrossRef]
- Wang, Y.; Ma, T.; Ma, S.; Liu, Y.; Tian, Y.; Wang, R.; Jiang, Y.; Hou, D.; Wang, J. Fluorometric determination of the antibiotic kanamycin by aptamer-induced FRET quenching and recovery between MoS2 nanosheets and carbon dots. Microchim. Acta 2017, 184, 203–210. [Google Scholar] [CrossRef]
- Ramezani, M.; Danesh, N.M.; Lavaee, P.; Abnous, K.; Taghdisi, S.M. A selective and sensitive fluorescent aptasensor for detection of kanamycin based on catalytic recycling activity of exonuclease III and gold nanoparticles. Sens. Actuators B Chem. 2016, 222, 1–7. [Google Scholar] [CrossRef]
- He, Y.; Wen, X.; Zhang, B.; Fan, Z. Novel aptasensor for the ultrasensitive detection of kanamycin based on grapheneoxide quantum-dot-linked single-stranded DNA-binding protein. Sens. Actuators B Chem. 2018, 265, 20–26. [Google Scholar] [CrossRef]
- Chen, J.; Li, Z.; Ge, J.; Yang, R.; Zhang, L.; Qu, L.B.; Wang, H.Q.; Zhang, L. An aptamer-based signal-on bio-assay for sensitive and selective detection of Kanamycin A by using gold nanoparticles. Talanta 2015, 139, 226–232. [Google Scholar] [CrossRef]
- Ling, K.; Jiang, H.; Zhang, L.; Li, Y.; Yang, L.; Qiu, C.; Li, F.R. A self-assembling RNA aptamer-based nanoparticle sensor for fluorometric detection of Neomycin B in milk. Anal. Bioanal. Chem. 2016, 408, 3593–3600. [Google Scholar] [CrossRef]
- Lee, A.Y.; Ha, N.R.; Jung, I.P.; Kim, S.H.; Kim, A.R.; Yoon, M.Y. Development of a ssDNA aptamer for detection of residual benzylpenicillin. Anal. Biochem. 2017, 531, 1–7. [Google Scholar] [CrossRef]
- Tu, C.; Dai, Y.; Zhang, Y.; Wang, W.; Wu, L. A simple fluorescent strategy based on triple-helix molecular switch for sensitive detection of chloramphenicol. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 224, 117415. [Google Scholar] [CrossRef]
- Ma, X.; Li, H.; Qiao, S.; Huang, C.; Liu, Q.; Shen, X.; Geng, Y.; Xu, W.; Sun, C. A simple and rapid sensing strategy based on structure-switching signaling aptamers for the sensitive detection of chloramphenicol. Food Chem. 2020, 302, 125359. [Google Scholar] [CrossRef]
- Sharma, R.; Akshath, U.S.; Bhatt, P.; Raghavarao, K.S. Fluorescent aptaswitch for chloramphenicol detection—Quantification enabled by immobilization of aptamer. Sens. Actuators B Chem. 2019, 290, 110–117. [Google Scholar] [CrossRef]
- Zhang, S.; Ma, L.; Ma, K.; Xu, B.; Liu, L.; Tian, W. Label-Free Aptamer-Based Biosensor for Specific Detection of Chloramphenicol Using AIE Probe and Graphene Oxide. ACS Omega 2018, 3, 12886–12892. [Google Scholar] [CrossRef]
- Alibolandi, M.; Hadizadeh, F.; Vajhedin, F.; Abnous, K.; Ramezani, M. Design and fabrication of an aptasensor for chloramphenicol based on energy transfer of CdTe quantum dots to graphene oxide sheet. Mater. Sci. Eng. C 2015, 48, 611–619. [Google Scholar] [CrossRef]
- Liu, S.; Bai, J.; Huo, Y.; Ning, B.; Peng, Y.; Li, S.; Han, D.; Kang, W.; Gao, Z. A zirconium-porphyrin MOF-based ratiometric fluorescent biosensor for rapid and ultrasensitive detection of chloramphenicol. Biosens. Bioelectron. 2020, 149, 111801. [Google Scholar] [CrossRef]
- Yang, Q.; Zhou, L.; Wu, Y.X.; Zhang, K.; Cao, Y.; Zhou, Y.; Wu, D.; Hu, F.; Gan, N. A two dimensional metal–organic framework nanosheets-based fluorescence resonance energy transfer aptasensor with circular strand-replacement DNA polymerization target-triggered amplification strategy for homogenous detection of antibiotics. Anal. Chim. Acta 2018, 1020, 1–8. [Google Scholar] [CrossRef]
- Wang, Y.; Gan, N.; Zhou, Y.; Li, T.; Cao, Y.; Chen, Y. Novel single-stranded DNA binding protein-assisted fluorescence aptamer switch based on FRET for homogeneous detection of antibiotics. Biosens. Bioelectron. 2017, 87, 508–513. [Google Scholar] [CrossRef]
- Miao, Y.B.; Ren, H.X.; Gan, N.; Zhou, Y.; Cao, Y.; Li, T.; Chen, Y. A homogeneous and “off–on” fluorescence aptamer-based assay for chloramphenicol using vesicle quantum dot-gold colloid composite probes. Anal. Chim. Acta 2016, 929, 49–55. [Google Scholar] [CrossRef]
- Yan, Z.; Yi, H.; Wang, L.; Zhou, X.; Yan, R.; Zhang, D.; Wang, S.; Su, L.; Zhou, S. Fluorescent aptasensor for ofloxacin detection based on the aggregation of gold nanoparticles and its effect on quenching the fluorescence of Rhodamine B. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 221, 117203. [Google Scholar] [CrossRef]
- Dolati, S.; Ramezani, M.; Nabavinia, M.S.; Soheili, V.; Abnous, K.; Taghdisi, S.M. Selection of specific aptamer against enrofloxacin and fabrication of graphene oxide based label-free fluorescent assay. Anal. Biochem. 2018, 549, 124–129. [Google Scholar] [CrossRef]
- Babaei, M.; Jalalian, S.H.; Bakhtiari, H.; Ramezani, M.; Abnous, K.; Taghdisi, S.M. Aptamer-Based Fluorescent Switch for Sensitive Detection of Oxytetracycline. Aust. J. Chem. 2017, 70, 718–723. [Google Scholar] [CrossRef]
- Song, K.M.; Jeong, E.; Jeon, W.; Jo, H.; Ban, C. A coordination polymer nanobelt (CPNB)-based aptasensor for sulfadimethoxine. Biosens. Bioelectron. 2012, 33, 113–119. [Google Scholar] [CrossRef]
- Xing, Y.P.; Liu, C.; Zhou, X.H.; Shi, H.C. Label-free detection of kanamycin based on a G-quadruplex DNA aptamer-based fluorescent intercalator displacement assay. Sci. Rep. 2015, 5, 8125. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, W.; Tan, S.; Chen, T. Label-Free and Simple G-quadruplex-based Turn-Off Fluorescence Assay for the Detection of Kanamycin. Anal. Lett. 2018, 51, 1718–1729. [Google Scholar] [CrossRef]
- Taghdisi, S.M.; Danesh, N.M.; Nameghi, M.A.; Ramezani, M.; Abnous, K. A label-free fluorescent aptasensor for selective and sensitive detection of streptomycin in milk and blood serum. Food Chem. 2016, 203, 145–149. [Google Scholar] [CrossRef]
- Gan, N.; Ou, C.; Tang, H.; Zhou, Y.; Cao, J. A homogenous “signal-on” aptasensor for antibiotics based on a single stranded DNA binding protein-quantum dot aptamer probe coupling exonuclease-assisted target recycling for signal amplification. RSC Adv. 2017, 7, 8381–8387. [Google Scholar]
- Zeng, J.; Gan, N.; Zhang, K.; He, L.; Lin, J.; Hu, F.; Cao, Y. Zero background and triple-signal amplified fluorescence aptasensor for antibiotics detection in foods. Talanta 2019, 199, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.B.; Ren, H.X.; Gan, N.; Cao, Y.; Li, T.; Chen, Y. Fluorescent aptasensor for chloramphenicol detection using DIL-encapsulated liposome as nanotracer. Biosens. Bioelectron. 2016, 81, 454–459. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Xu, H.; Li, G.; Yang, X.; Choi, M.M. Fluorescence quenching for chloramphenicol detection in milk based on protein-stabilized Au nanoclusters. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 149, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhang, H.; Shi, Z.; Duan, N.; Fang, C.; Dai, S.; Wang, Z. Aptamer-based fluorescence biosensor for chloramphenicol determination using upconversion nanoparticles. Food Control 2015, 50, 597–604. [Google Scholar] [CrossRef]
- Fang, C.; Wu, S.; Duan, N.; Dai, S.; Wang, Z. Highly sensitive aptasensor for oxytetracycline based on upconversion and magnetic nanoparticles. Anal. Methods 2015, 7, 2585–2593. [Google Scholar] [CrossRef]
- Sun, C.; Su, R.; Bie, J.; Sun, H.; Qiao, S.; Ma, X.; Sun, R.; Zhang, T. Label-free fluorescent sensor based on aptamer and thiazole orange for the detection of tetracycline. Dyes Pigments 2018, 149, 867–875. [Google Scholar] [CrossRef]
- Qin, O.; Liu, Y.; Chen, Q.; Guo, Z.; Zhao, J.; Li, H.; Hu, W. Rapid and specific sensing of tetracycline in food using a novel upconversion aptasensor. Food Control 2017, 81, 156–163. [Google Scholar]
- Zhou, C.; Zou, H.; Sun, C.; Ren, D.; Xiong, W.; Li, Y. Fluorescent aptasensor for detection of four tetracycline veterinary drugs in milk based on catalytic hairpin assembly reaction and displacement of G-quadruplex. Anal. Bioanal. Chem. 2018, 410, 2981–2989. [Google Scholar] [CrossRef]
- Liu, X.; Gao, T.; Gao, X.; Ma, T.; Tang, Y.; Zhu, L.; Li, J. An aptamer based sulfadimethoxine assay that uses magnetized upconversion nanoparticles. Microchim. Acta 2017, 184, 3557–3563. [Google Scholar] [CrossRef]
- Liu, C.; Lu, C.; Tang, Z.; Chen, X.; Wang, G.; Sun, F. Aptamer-functionalized magnetic nanoparticles for simultaneous fluorometric determination of oxytetracycline and kanamycin. Microchim. Acta 2015, 182, 2567–2575. [Google Scholar] [CrossRef]
- Ma, P.; Ye, H.; Deng, J.; Khan, I.M.; Yue, L.; Wang, Z. A fluorescence polarization aptasensor coupled with polymerase chain reaction and streptavidin for chloramphenicol detection. Talanta 2019, 205, 120119. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Su, J.; Lin, H.; Sun, X.; Liu, B.; Kankala, R.K.; Zhou, S.F. Luminescent carbon nanodots based aptasensors for rapid detection of kanamycin residue. Talanta 2019, 202, 452–459. [Google Scholar] [CrossRef]
- Ye, T.; Peng, Y.; Yuan, M.; Cao, H.; Yu, J.; Li, Y.; Xu, F. A “turn-on” fluorometric assay for kanamycin detection by using silver nanoclusters and surface plasmon enhanced energy transfer. Microchim. Acta 2019, 186, 40. [Google Scholar] [CrossRef]
- Feng, C.; Dai, S.; Wang, L. Optical aptasensors for quantitative detection of small biomolecules: A review. Biosens. Bioelectron. 2014, 59, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Alyamani, B.J.; Alsager, O.A.; Zourob, M. Label-Free Fluorescent Aptasensor for Small Targets via Displacement of Groove Bound Curcumin Molecules. Sensors 2019, 19, 4181. [Google Scholar] [CrossRef] [Green Version]
- Bagheri, E.; Abnous, K.; Alibolandi, M.; Ramezani, M.; Taghdisi, S.M. Triple-helix molecular switch-based aptasensors and DNA sensors. Biosens. Bioelectron. 2018, 111, 1–9. [Google Scholar] [CrossRef]
- Sharifi, S.; Vahed, S.Z.; Ahmadian, E.; Dizaj, S.M.; Eftekhari, A.; Khalilov, R.; Ahmadi, M.; Hamidi-Asl, E.; Labib, M. Detection of pathogenic bacteria via nanomaterials-modified aptasensors. Biosens. Bioelectron. 2019, 111933. [Google Scholar] [CrossRef]
- Wang, L.; Wu, A.; Wei, G. Graphene-based aptasensors: From molecule–interface interactions to sensor design and biomedical diagnostics. Analyst 2018, 143, 1526–1543. [Google Scholar] [CrossRef]
- Karimzadeh, A.; Hasanzadeh, M.; Shadjou, N.; de la Guardia, M. Optical bio(sensing) using nitrogen doped graphene quantum dots: Recent advances and future challenges. TrAC Trends Anal. Chem. 2018, 108, 110–121. [Google Scholar] [CrossRef]
- Bose, A.; Wong, T.W. Chapter 11—Nanotechnology-Enabled Drug Delivery for Cancer Therapy. In Nanotechnology Applications for Tissue Engineering; Thomas, S., Grohens, Y., Ninan, N., Eds.; William Andrew Publishing: Oxford, UK, 2015; pp. 173–193. [Google Scholar]
- Stanisavljevic, M.; Krizkova, S.; Vaculovicova, M.; Kizek, R.; Adam, V. Quantum dots-fluorescence resonance energy transfer-based nanosensors and their application. Biosens. Bioelectron. 2015, 74, 562–574. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.W.; Zou, X.M.; Song, S.H.; Chen, G.H. Quantum Dots Applied to Methodology on Detection of Pesticide and Veterinary Drug Residues. J. Agric. Food Chem. 2018, 66, 1307–1319. [Google Scholar] [CrossRef] [PubMed]
- Fenzl, C.; Hirsch, T.; Baeumner, A.J. Nanomaterials as versatile tools for signal amplification in (bio)analytical applications. TrAC Trends Anal. Chem. 2016, 79, 306–316. [Google Scholar] [CrossRef]
- Xu, Y.; Lu, C.; Sun, Y.; Shao, Y.; Cai, Y.; Zhang, Y.; Miao, J.; Miao, P. A colorimetric aptasensor for the antibiotics oxytetracycline and kanamycin based on the use of magnetic beads and gold nanoparticles. Microchim. Acta 2018, 185, 548. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Gan, N.; Li, T.; Zhang, H.; Cao, Y.; Jiang, Q. A colorimetric aptasensor for chloramphenicol in fish based on double-stranded DNA antibody labeled enzyme-linked polymer nanotracers for signal amplification. Sens. Actuators B Chem. 2015, 220, 679–687. [Google Scholar] [CrossRef]
- Luan, Q.; Miao, Y.; Gan, N.; Cao, Y.; Li, T.; Chen, Y. A POCT colorimetric aptasensor for streptomycin detection using porous silica beads- enzyme linked polymer aptamer probes and exonuclease-assisted target recycling for signal amplification. Sens. Actuators B Chem. 2017, 251, 349–358. [Google Scholar] [CrossRef]
- Luan, Q.; Gan, N.; Cao, Y.; Li, T. Mimicking an Enzyme-Based Colorimetric Aptasensor for Antibiotic Residue Detection in Milk Combining Magnetic Loop-DNA Probes and CHA-Assisted Target Recycling Amplification. J. Agric. Food Chem. 2017, 65, 5731–5740. [Google Scholar] [CrossRef]
- Cui, X.; Li, R.; Liu, X.; Wang, J.; Leng, X.; Song, X.; Pei, Q.; Wang, Y.; Liu, S.; Huang, J. Low-background and visual detection of antibiotic based on target-activated colorimetric split peroxidase DNAzyme coupled with dual nicking enzyme signal amplification. Anal. Chim. Acta 2018, 997, 1–8. [Google Scholar] [CrossRef]
- Wang, C.; Chen, D.; Wang, Q.; Tan, R. Kanamycin detection based on the catalytic ability enhancement of gold nanoparticles. Biosens. Bioelectron. 2017, 91, 262–267. [Google Scholar] [CrossRef]
- Zhao, J.; Wu, Y.; Tao, H.; Chen, H.; Yang, W.; Qiu, S. Colorimetric detection of streptomycin in milk based on peroxidase-mimicking catalytic activity of gold nanoparticles. RSC Adv. 2017, 7, 38471–38478. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Zhang, Y.; Xie, Y.; Sun, Y.; Bi, K.; Cui, Z.; Zhao, L.; Fan, W. An aptamer-based colorimetric sensor for streptomycin and its application in food inspection. Chem. Res. Chin. Univ. 2017, 33, 714–720. [Google Scholar] [CrossRef]
- Ma, Q.; Wang, Y.; Jia, J.; Xiang, Y. Colorimetric aptasensors for determination of tobramycin in milk and chicken eggs based on DNA and gold nanoparticles. Food Chem. 2018, 249, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Yan, S.; Lai, X.; Xu, Y.; Liu, T.; Xiang, Y. Colorimetric Aptasensor for Detection of Malachite Green in Fish Sample Based on RNA and Gold Nanoparticles. Food Anal. Methods 2018, 11, 1668–1676. [Google Scholar] [CrossRef]
- Luan, Q.; Xi, Y.; Gan, N.; Cao, Y.; Li, T.; Chen, Y. A facile colorimetric aptamer assay for small molecule detection in food based on a magnetic single-stranded DNA binding protein-linked composite probe. Sens. Actuators B Chem. 2017, 239, 979–987. [Google Scholar] [CrossRef]
- Luan, Q.; Xiong, X.; Gan, N.; Cao, Y.; Li, T.; Wu, D.; Dong, Y.; Hu, F. A multiple signal amplified colorimetric aptasensor for antibiotics measurement using DNAzyme labeled Fe-MIL-88-Pt as novel peroxidase mimic tags and CSDP target-triggered cycles. Talanta 2018, 187, 27–34. [Google Scholar] [CrossRef]
- Huang, W.; Zhang, H.; Lai, G.; Liu, S.; Li, B.; Yu, A. Sensitive and rapid aptasensing of chloramphenicol by colorimetric signal transduction with a DNAzyme-functionalized gold nanoprobe. Food Chem. 2019, 270, 287–292. [Google Scholar] [CrossRef]
- Javidi, M.; Housaindokht, M.R.; Verdian, A.; Razavizadeh, B.M. Detection of chloramphenicol using a novel apta-sensing platform based on aptamer terminal-lock in milk samples. Anal. Chim. Acta 2018, 1039, 116–123. [Google Scholar] [CrossRef]
- Du, J.; Jiang, Q.; Lu, X.; Chen, L.; Zhang, Y.; Xiong, X. Detection of sulfadimethoxine using optical images of liquid crystals. Analyst 2019, 144, 1761–1767. [Google Scholar] [CrossRef]
- Lu, C.; Tang, Z.; Liu, C.; Kang, L.; Sun, F. Magnetic-nanobead-based competitive enzyme-linked aptamer assay for the analysis of oxytetracycline in food. Anal. Bioanal. Chem. 2015, 407, 4155–4163. [Google Scholar] [CrossRef]
- Kim, C.H.; Lee, L.P.; Min, J.R.; Lim, M.W.; Jeong, S.H. An indirect competitive assay-based aptasensor for detection of oxytetracycline in milk. Biosens. Bioelectron. 2014, 51, 426–430. [Google Scholar] [CrossRef]
- Wang, S.; Liu, J.; Yong, W.; Chen, Q.; Zhang, L.; Dong, Y.; Su, H.; Tan, T. A direct competitive assay-based aptasensor for sensitive determination of tetracycline residue in Honey. Talanta 2015, 131, 562–569. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Yong, W.; Liu, J.; Zhang, L.; Chen, Q.; Dong, Y. Development of an indirect competitive assay-based aptasensor for highly sensitive detection of tetracycline residue in honey. Biosens. Bioelectron. 2014, 57, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Tian, Y.; Huang, P.; Wu, F.Y. Using target-specific aptamers to enhance the peroxidase-like activity of gold nanoclusters for colorimetric detection of tetracycline antibiotics. Talanta 2020, 208, 120342. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Xu, J.; Li, Y.; Gao, H.; Guo, J.; Shen, F.; Sun, C. A novel colorimetric aptasensor using cysteamine-stabilized gold nanoparticles as probe for rapid and specific detection of tetracycline in raw milk. Food Control 2015, 54, 7–15. [Google Scholar] [CrossRef]
- Ramezani, M.; Danesh, N.M.; Lavaee, P.; Abnous, K.; Taghdisi, S.M. A novel colorimetric triple-helix molecular switch aptasensor for ultrasensitive detection of tetracycline. Biosens. Bioelectron. 2015, 70, 181–187. [Google Scholar] [CrossRef]
- He, L.; Luo, Y.; Zhi, W.; Zhou, P. Colorimetric Sensing of Tetracyclines in Milk Based on the Assembly of Cationic Conjugated Polymer-Aggregated Gold Nanoparticles. Food Anal. Methods 2013, 6, 1704–1711. [Google Scholar] [CrossRef]
- Emrani, A.S.; Danesh, N.M.; Lavaee, P.; Ramezani, M.; Abnous, K.; Taghdisi, S.M. Colorimetric and fluorescence quenching aptasensors for detection of streptomycin in blood serum and milk based on double-stranded DNA and gold nanoparticles. Food Chem. 2016, 190, 115–121. [Google Scholar] [CrossRef]
- Song, K.M.; Jeong, E.; Jeon, W.; Cho, M.; Ban, C. Aptasensor for ampicillin using gold nanoparticle based dual fluorescence–colorimetric methods. Anal. Bioanal. Chem. 2012, 402, 2153–2161. [Google Scholar] [CrossRef]
- Alizadeh, N.; Salimi, A.; Hallaj, R. Hemin/G-Quadruplex Horseradish Peroxidase-Mimicking DNAzyme: Principle and Biosensing Application. Adv. Biochem. Eng. Biotechnol. 2020, 170, 85–106. [Google Scholar]
- Wang, D.; Ge, C.; Zeng, L. Circular strand displacement polymerization reaction: A promising technique? Bioanalysis 2014, 6, 899–901. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Mazumdar, D.; Lu, Y. A simple and sensitive “dipstick” test in serum based on lateral flow separation of aptamer-linked nanostructures. Angew. Chem. Int. Ed. Engl. 2006, 45, 7955–7959. [Google Scholar] [CrossRef] [PubMed]
- Abnous, K.; Danesh, N.M.; Ramezani, M.; Alibolandi, M.; Emrani, A.S.; Lavaee, P.; Taghdisi, S.M. A colorimetric gold nanoparticle aggregation assay for malathion based on target-induced hairpin structure assembly of complementary strands of aptamer. Microchim. Acta 2018, 185, 216. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Ni, H.; Li, C.; Zhang, X.; Wen, K.; Ke, Y.; Yang, H.; Shi, W.; Zhang, S.; Shen, J.; et al. Development of a highly specific chemiluminescence aptasensor for sulfamethazine detection in milk based on in vitro selected aptamers. Sens. Actuators B Chem. 2019, 281, 801–811. [Google Scholar] [CrossRef]
- Yao, Y.; Wang, X.; Duan, W.; Li, F. A label-free, versatile and low-background chemiluminescence aptasensing strategy based on gold nanocluster catalysis combined with the separation of magnetic beads. Analyst 2018, 143, 709–714. [Google Scholar] [CrossRef] [PubMed]
- Leung, K.H.; He, H.Z.; Chan, D.S.; Fu, W.C.; Leung, C.H.; Ma, D.L. An oligonucleotide-based switch-on luminescent probe for the detection of kanamycin in aqueous solution. Sens. Actuators B Chem. 2013, 177, 487–492. [Google Scholar] [CrossRef]
- Cheng, S.; Liu, H.; Zhang, H.; Chu, G.; Guo, Y.; Sun, X. Ultrasensitive electrochemiluminescence aptasensor for kanamycin detection based on silver nanoparticle-catalyzed chemiluminescent reaction between luminol and hydrogen peroxide. Sens. Actuators B Chem. 2020, 304, 127367. [Google Scholar] [CrossRef]
- de-los-Santos-Álvarez, N.; Lobo-Castañón, M.J.; Miranda-Ordieres, A.J.; Tuñón-Blanco, P. SPR sensing of small molecules with modified RNA aptamers: Detection of neomycin B. Biosens. Bioelectron. 2009, 24, 2547–2553. [Google Scholar] [CrossRef]
- Jiang, Y.; Sun, D.W.; Pu, H.; Wei, Q. Ultrasensitive analysis of kanamycin residue in milk by SERS-based aptasensor. Talanta 2019, 197, 151–158. [Google Scholar] [CrossRef]
- Wu, Z. AuNP Tetramer-Based Aptasensor for SERS Sensing of Oxytetracycline. Food Anal. Methods 2019, 12, 1121–1127. [Google Scholar] [CrossRef]
- Meng, F.; Ma, X.; Duan, N.; Wu, S.; Xia, Y.; Wang, Z.; Xu, B. Ultrasensitive SERS aptasensor for the detection of oxytetracycline based on a gold-enhanced nano-assembly. Talanta 2017, 165, 412–418. [Google Scholar] [CrossRef]
- Li, H.; Chen, Q.; Hassan, M.M.; Chen, X.; Ouyang, Q.; Guo, Z.; Zhao, J. A magnetite/PMAA nanospheres-targeting SERS aptasensor for tetracycline sensing using mercapto molecules embedded core/shell nanoparticles for signal amplification. Biosens. Bioelectron. 2017, 92, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Yang, L.; Zhuang, H.; Wu, H.; Zhang, J. Engineered “hot” core–shell nanostructures for patterned detection of chloramphenicol. Biosens. Bioelectron. 2016, 78, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Lai, X.; Wang, Y.; Ye, N.; Xiang, Y. Label free aptasensor for ultrasensitive detection of tobramycin residue in pasteurized cow’s milk based on resonance scattering spectra and nanogold catalytic amplification. Food Chem. 2019, 295, 36–41. [Google Scholar] [CrossRef]
- Wang, D.M.; Lin, K.L.; Huang, C.Z. Carbon dots-involved chemiluminescence: Recent advances and developments. Luminescence 2019, 34, 4–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Lee, J.H.; Lu, Y. Quantum Dot Encoding of Aptamer-Linked Nanostructures for One-Pot Simultaneous Detection of Multiple Analytes. Anal. Chem. 2007, 79, 4120–4125. [Google Scholar] [CrossRef]
- Li, S.; Liu, J.; Chen, Z.; Lu, Y.; Low, S.S.; Zhu, L.; Cheng, C.; He, Y.; Chen, Q.; Su, B.; et al. Electrogenerated chemiluminescence on smartphone with graphene quantum dots nanocomposites for Escherichia Coli detection. Sens. Actuators B Chem. 2019, 297, 126811. [Google Scholar] [CrossRef]
- Li, P.; Yu, J.; Zhao, K.; Deng, A.; Li, J. Efficient enhancement of electrochemiluminescence from tin disulfide quantum dots by hollow titanium dioxide spherical shell for highly sensitive detection of chloramphenicol. Biosens. Bioelectron. 2020, 147, 111790. [Google Scholar] [CrossRef]
- Tang, H.; Zhu, C.; Meng, G.; Wu, N. Review—Surface-Enhanced Raman Scattering Sensors for Food Safety and Environmental Monitoring. J. Electrochem. Soc. 2018, 165, B3098–B3118. [Google Scholar] [CrossRef]
- Andrews, D.L. Rayleigh Scattering and Raman Effect, Theory. In Encyclopedia of Spectroscopy and Spectrometry, 3rd ed.; Lindon, J.C., Tranter, G.E., Koppenaal, D.W., Eds.; Academic Press: Oxford, UK, 2017; pp. 924–930. [Google Scholar]
- Stanton, S.G.; Pecora, R.; Hudson, B.S. Resonance enhanced dynamic Rayleigh scattering. J. Chem. Phys. 1981, 75, 5615–5626. [Google Scholar] [CrossRef]
- Ouyang, H.; Liang, A.; Jiang, Z. A simple and selective resonance Rayleigh scattering-energy transfer spectral method for determination of trace neomycin sulfate using Cu2O particle as probe. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 190, 268–273. [Google Scholar] [CrossRef]
- Zhu, J.; Liu, S.; Liu, Z.; Li, Y.; Tian, J.; Hu, X. A highly sensitive and selective assay of doxycycline by dualwavelength overlapping resonance Rayleigh scattering. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 124, 237–242. [Google Scholar] [CrossRef] [PubMed]



















| Detection Technique | Antimicrobial Family | Analyte | Food Product | Fluorophore/Quencher or Intercalating Dye * or Signal Labels | Limit of detection (LOD) (nM or ng/mL) | References |
|---|---|---|---|---|---|---|
| FRET | AMGL | Kanamycin | Milk | CDs/AuNPs | 18 nM (10.5 ng/mL) | [72] |
| FRET | Kanamycin | Milk | CDs/l-MoS2 NSs | 1.1 μM (640.9 ng/mL) | [73] | |
| FRET | Kanamycin | Milk | FAM/rGO | 1 pM (0.583 pg/mL) | [42] | |
| FRET | Kanamycin | Milk | FAM/AuNPs (+ exonuclease III) | 321 pM (0.19 ng/mL) | [74] | |
| FRET | Kanamycin | Milk | BHQ1/GO QDs | 6 pg/mL | [75] | |
| FRET | Kanamycin | Milk | FAM/AuNPs | 0.3 nM (0.175 ng/mL) | [76] | |
| FRET | Neomycin B | Milk | FAM/AuNPs | 0.01 μM (6.15 ng/mL) | [77] | |
| FRET | BL | Benzylpenicillin | Milk | FAM/rGO | 9.2 nM (3.1 ng/mL) | [78] |
| FRET | PHENI | CAP | Honey | FAM/BHQ1 | 1.2 nM (0.39 ng/mL) | [79] |
| FRET | CAP | Milk | FAM/BHQ1 | 0.70 ng/mL | [80] | |
| FRET | CAP | Honey | FAM/BHQ1 | 0.285 pg/mL | [81] | |
| FRET | CAP | Milk | DSAC2N/GO | 1.26 pg/mL | [82] | |
| FRET | CAP | Milk | CdTe QDs/GO | 0.2 ng/mL | [83] | |
| FRET | CAP | Milk, shrimp | FAM/ZrP MOF (PCN-222) | 0.08 pg/mL | [84] | |
| FRET | CAP | Milk, fish | SYBR Green I/Cu-TCPP MOFs NSs | 0.3 pg/mL | [85] | |
| FRET | CAP | Milk | QDs/AuNPs | 3 pg/mL | [86] | |
| FRET | CAP | Milk | QDs/AuNPs | 0.3 pM (0.097 pg/mL) | [87] | |
| FRET | Florfenicol | Milk | ATTO 647N/GO | 5.75 nM (2.06 ng/mL) | [58] | |
| FRET | QUINO | Ofloxacin | Milk | RB/AuNPs | 4.61 nM (1.66 ng/mL) | [88] |
| FRET | Enrofloxacin | Milk | Enrofloxacin/GO | 3.7 nM (1.33 ng/mL) | [89] | |
| FRET | TETRA | OTC | Milk | FAM/BHQ1 | 6.44 nM (2.96 ng/mL) | [90] |
| FRET | SULFA | SDMX | Milk | FAM/CPNBs | 10 ng/mL | [91] |
| FRET | MULTI | SDMX, kanamycin, ampicillin | Milk | Apt1-Cy3, Apt2-FAM, Apt3-Cy5/GO (+ DNase I) | 1.997, 2.664, 2.337 ng/mL | [47] |
| Fluorescence | AMGL | Kanamycin | Milk | TO * | 59 nM (34.4 ng/mL) | [92] |
| Fluorescence | Kanamycin | Milk | Thioflavin T + Apt + cDNA1 + cDNA2 | 0.33 nM (0.19 ng/mL) | [93] | |
| Fluorescence | Streptomycin | Milk | SYBR Gold dye * (+ exonuclease III) | 54.5 nM (31.7 ng/mL) | [94] | |
| Fluorescence | Streptomycin | Milk | QDs (self-quenching) | 0.03 ng/mL | [95] | |
| Fluorescence | PHENI | CAP | Milk, fish | SYBR green * | 0.033 pg/mL | [96] |
| Fluorescence | CAP | Fish | SSB/DIL-Lip vesicle | 1 pM (0.32 pg/mL) | [97] | |
| Fluorescence | CAP | Milk | BSA-AuNCs/CAP | 33 nM (10.7 ng/mL) | [98] | |
| Fluorescence | CAP | Milk | Apt- Fe3O4 MNPs + cDNA-UCNPs | 0.01 ng/mL | [99] | |
| Fluorescence | TETRA | OTC | Milk | Apt- Fe3O4 MNPs + cDNA-UCNPs | 0.036 ng/mL | [100] |
| Fluorescence | Tetracycline | Milk | TO * | 29 ng/mL | [101] | |
| Fluorescence | Tetracycline | Milk, pork meat | UCNPs (+ Fe3O4 MNPs) | 0.0062 ng/mL | [102] | |
| Fluorescence | TTC, CTC, OTC, doxycycline | Milk | NMM + Apt + cDNA1 + cDNA2 | 4.6 ng/mL | [103] | |
| Fluorescence | SULFA | Sulfadimethoxine | Fish | MNPs-Apt + NaYF4: Yb, Tm UCNPs-cDNA | 0.11 ng/mL | [104] |
| Fluorescence | MULTI | CAP, kanamycin | Milk, fish | SYBR gold * | 0.52 pg/mL, 0.41 pg/mL | [46] |
| Fluorescence | OTC, kanamycin | Pig muscle, milk, honey | Apt-MNPs + cDNA1-FAM + cDNA2-ROX | 0.85 ng/mL, 0.92 ng/mL | [105] | |
| FPIA | PHENI | CAP | Honey | FAM/GO/Streptavidin | 0.5 pM (0.162 pg/mL) | [106] |
| FALIA | AMGL | Kanamycin | Milk | CNPs-Apt | 5.10−8 ng/mL | [107] |
| SPEET + fluorescence | AMGL | Kanamycin | Milk | AuNPs-Apt + AgNCs | 1 nM (0.58 ng/mL) | [108] |
| Labeled or Label Free Detection | Antimicrobial Family | Analyte | Food Product | Detection Principle | Limit of detection (LOD) (ng/mL or nM) | References |
|---|---|---|---|---|---|---|
| Labeled | AMGL | Kanamycin | Milk | Apt-MBs + NMOF-Pt-sDNA | 0.2 pg/mL | [122] |
| Label-free | Kanamycin | Milk | Hemin/G-quadruplex DNAzyme | 14.7 pM (8.6 pg/mL) | [123] | |
| Label-free | Kanamycin | Milk, meat | Intrinsic peroxidase-like activity of AuNPs | 0.1 nM (58.2 pg/mL) | [124] | |
| Labeled | Streptomycin | Milk | Apt-Au-PV | 1 pg/mL | [121] | |
| Label-free | Streptomycin | Milk | Intrinsic peroxidase-like activity of AuNPs | 86 nM (50 ng/mL) | [125] | |
| Labeled | Streptomycin | Milk, honey | AuNPs aggregation + Apt | 100, 125 nM (58.2, 72.7 ng/mL) | [126] | |
| Label-free | Tobramycin | Milk and chicken eggs | AuNPs aggregation + Apt + NaCl | 23.3 nM (10.9 ng/mL) | [127] | |
| Label-free | DYES | Malachite green | Fish | AuNPs aggregation + Apt + NaCl | 15.95 nM (5.82 ng/mL) | [128] |
| Labeled | MULTI | OTC, kanamycin | Milk | HRP-AuNPs | 1 ag/mL | [119] |
| Label-free | CAP, tetracycline | Chicken meat, milk | AuNPs aggregation + Apt | 32.9 nM, 7.0 nM (10.6 ng/mL, 3.11 ng/mL) | [50] | |
| Labeled | PHENI | CAP | Honey, fish | Aptamer-HRP | 0.0031ng/mL | [44] |
| Labeled | CAP | Milk | Au MNPs-SSB + Apt-SiO2@Au-HRP | 0.02 ng/mL | [129] | |
| Labeled | CAP | Fish | Au MNPs-Apt-SSB + ds-DNA Ab/EV-AuNPs-HRP | 0.015 ng/mL | [120] | |
| Labeled | CAP | Milk | 3 HRP-mimicking DNAzymes: Fe-MIL-88 (MOFs)-Pt NPs-sDNA | 0.03 pM (0.0097 pg/mL) | [130] | |
| Labeled | CAP | Milk powder | Hemin/G-quadruplex DNAzyme-Au NPs | 0.13 pg/mL | [131] | |
| Label-free | CAP | Milk | AuNPs aggregation + Apt | 0.03 nM (0.0097 ng/mL) | [132] | |
| Label-free | SULFA | SDMX | Raw milk, honey, egg | LCs | 10 ng/mL | [133] |
| Labeled (dc-ELAA) | TETRA | OTC | Chicken muscle, milk, honey | Aptamer + OTC-HRP | 0.88 ng/mL | [134] |
| Labeled (ic-ELAA) | OTC | Milk | Aptamer-HRP | 12.3 ng/mL | [135] | |
| Labeled (dc-ELAA) | Tetracycline | Honey | Aptamer + TTC-HRP | 0.0978 ng/mL | [136] | |
| Labeled (ic-ELAA) | Tetracycline | Honey | Aptamer-HRP | 9.6 × 10−3 ng/mL | [137] | |
| Label-free | Tetracycline | Milk | Intrinsic peroxidase-like activity of AuNCs | 46 nM (20.4 ng/mL) | [138] | |
| Label-free | Tetracycline | Milk | CS-AuNPs aggregation+ Apt | 39 ng/mL | [139] | |
| Label-free | Tetracycline | Milk | AuNPs aggregation + Apt | 266 pM (0.12 ng/mL) | [140] | |
| Label-free | Tetracycline | Milk | AuNPs aggregation + Apt + PDADMAC | 1 μM (444.4 ng/mL) (naked eyes), 45.8 nM (20.4 ng/mL) (detector) | [141] | |
| Dual colorimetric (Label free) and fluorescence (Labeled) | AMGL | Streptomycin | Milk | AuNPs aggregation + Apt | 73.1 nM (42.5 ng/mL) (colorimetric) and 47.6 nM (27.7 ng/mL) (fluorescence) | [142] |
| Dual colorimetric and fluorescenceLabel-free | BL | Ampicillin | Milk | AuNPs aggregation + FAM-Apt | 10 ng/mL (colorimetric) and 2 ng/mL (fluorescence) | [143] |
| Detection Technique | Analyte | Food Product | Detection Principle | Limit of detection (LOD) (ng/mL or nM) | References |
|---|---|---|---|---|---|
| Chemiluminescence | Sulfamethazine | Milk | Luminol-H2O2 + SMZ-HRP + Apt | 0.92 ng/mL | [148] |
| Kanamycin | Milk | Luminol-H2O2 + cDNA-AuNCs + Apt-MBs | 0.035 nM (0.02 ng/mL) | [149] | |
| OTC, TTC, Kanamycin | Milk | ABEI-H2O2-PIP + cDNA-AuNFs + Apt-MNPs | 0.02, 0.02 and 0.002 ng/mL | [49] | |
| CAP | Milk | ABEI-H2O2-PIP + cDNA-AuNFs + Apt-MNPs | 1 ng/mL | [40] | |
| Luminescence | Kanamycin | Fish | SPLPt(II) + Apt | 143 nM (83.3 ng/mL) | [150] |
| ECL | Kanamycin | Milk | Luminol-H2O2 + AgNPs-Apt (catalyser) | 0.06 ng/mL | [151] |
| FQ-EWA | Kanamycin | Milk | Cy3-Apt + GO + AAP | 26 nM (15.1 ng/mL) | [43] |
| SPR | Neomycin B | Buffer | Apt | 5 nM (3.1 ng/mL) | [152] |
| SERS | Kanamycin | Milk | Au@AgNPs-cDNA + Cy3-Apt | 0.90 pg/mL | [153] |
| OTC | Milk | AuNPs-Apt + AuNPs-cDNA1 + AuNPs-cDNA2 + AuNPs-cDNA3 | 4.35 × 10−3 fg/mL | [154] | |
| OTC | Fishmeal | AuNPs (13 nm)-cDNA-AuNPs (80 nm) + Apt + 4-MBA | 4.35 × 10−3 fg/mL | [155] | |
| Tetracycline | Milk | MCNCs-PMAA-MNs-Apt + Au/PATP/SiO2-cDNA | 0.001 ng/mL | [156] | |
| CAP | Milk | Au@Ag NSs-cDNA-Cy5-Apt | 0.19 pg/mL | [157] | |
| RRSS | Tobramycin | Milk | AuNPs-Apt + CuSO4 | 0.19 nM (0.09 ng/mL) | [158] |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaudin, V. The Growing Interest in Development of Innovative Optical Aptasensors for the Detection of Antimicrobial Residues in Food Products. Biosensors 2020, 10, 21. https://doi.org/10.3390/bios10030021
Gaudin V. The Growing Interest in Development of Innovative Optical Aptasensors for the Detection of Antimicrobial Residues in Food Products. Biosensors. 2020; 10(3):21. https://doi.org/10.3390/bios10030021
Chicago/Turabian StyleGaudin, Valérie. 2020. "The Growing Interest in Development of Innovative Optical Aptasensors for the Detection of Antimicrobial Residues in Food Products" Biosensors 10, no. 3: 21. https://doi.org/10.3390/bios10030021
APA StyleGaudin, V. (2020). The Growing Interest in Development of Innovative Optical Aptasensors for the Detection of Antimicrobial Residues in Food Products. Biosensors, 10(3), 21. https://doi.org/10.3390/bios10030021





