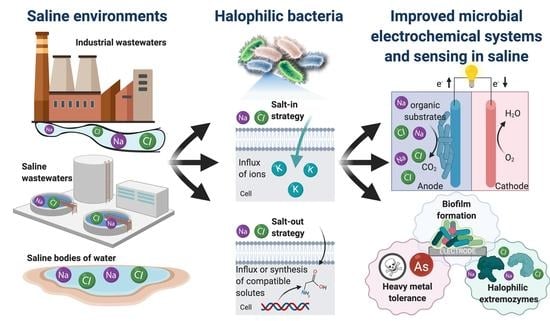
The Use of Electroactive Halophilic Bacteria for Improvements and Advancements in Environmental High Saline Biosensing
Abstract
:1. Introduction
2. Electrifying Halophilic Bacteria
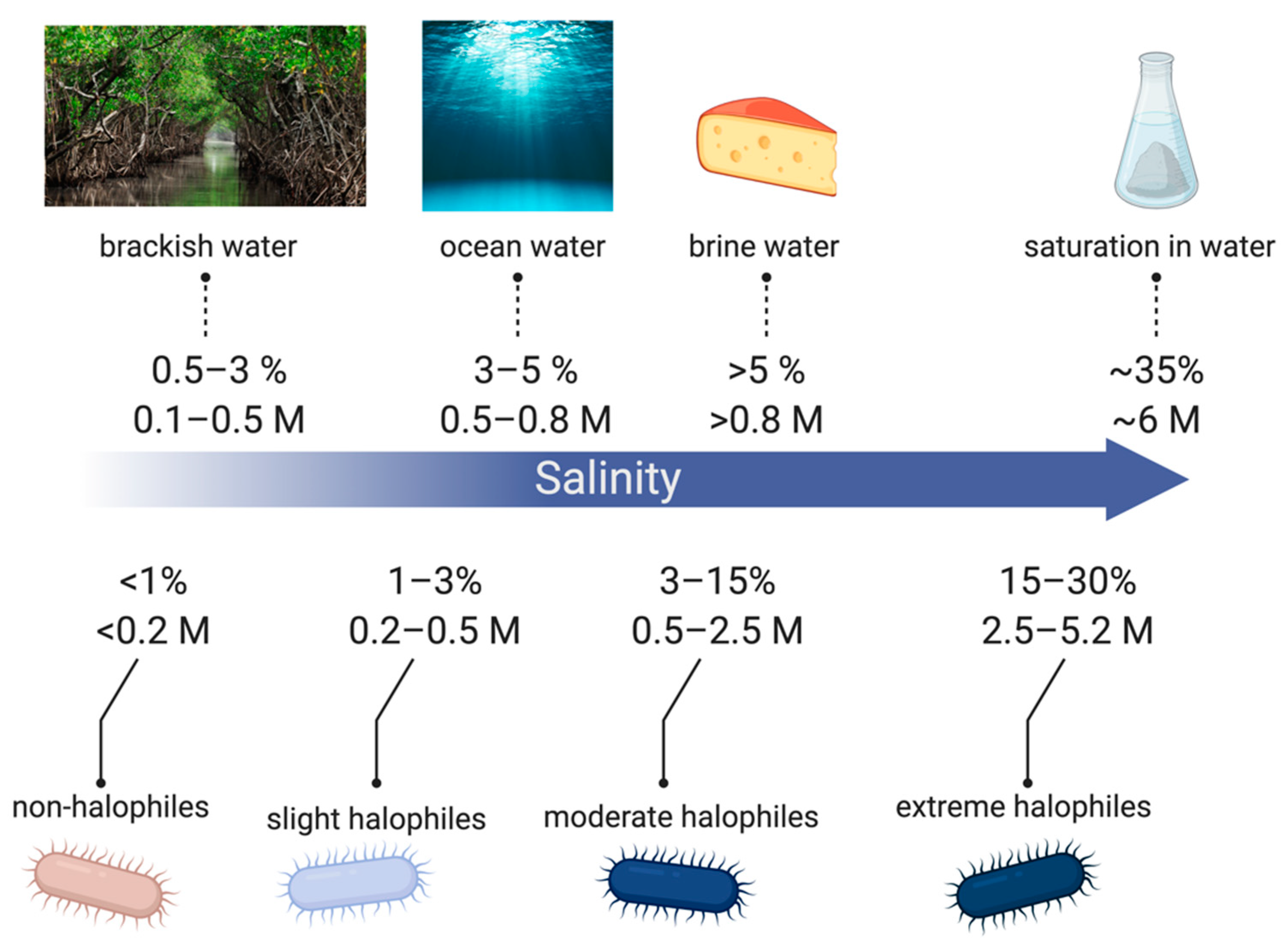
2.1. Mechanisms for Saline Tolerance
2.2. Halophilic Bacteria in Biotechnology
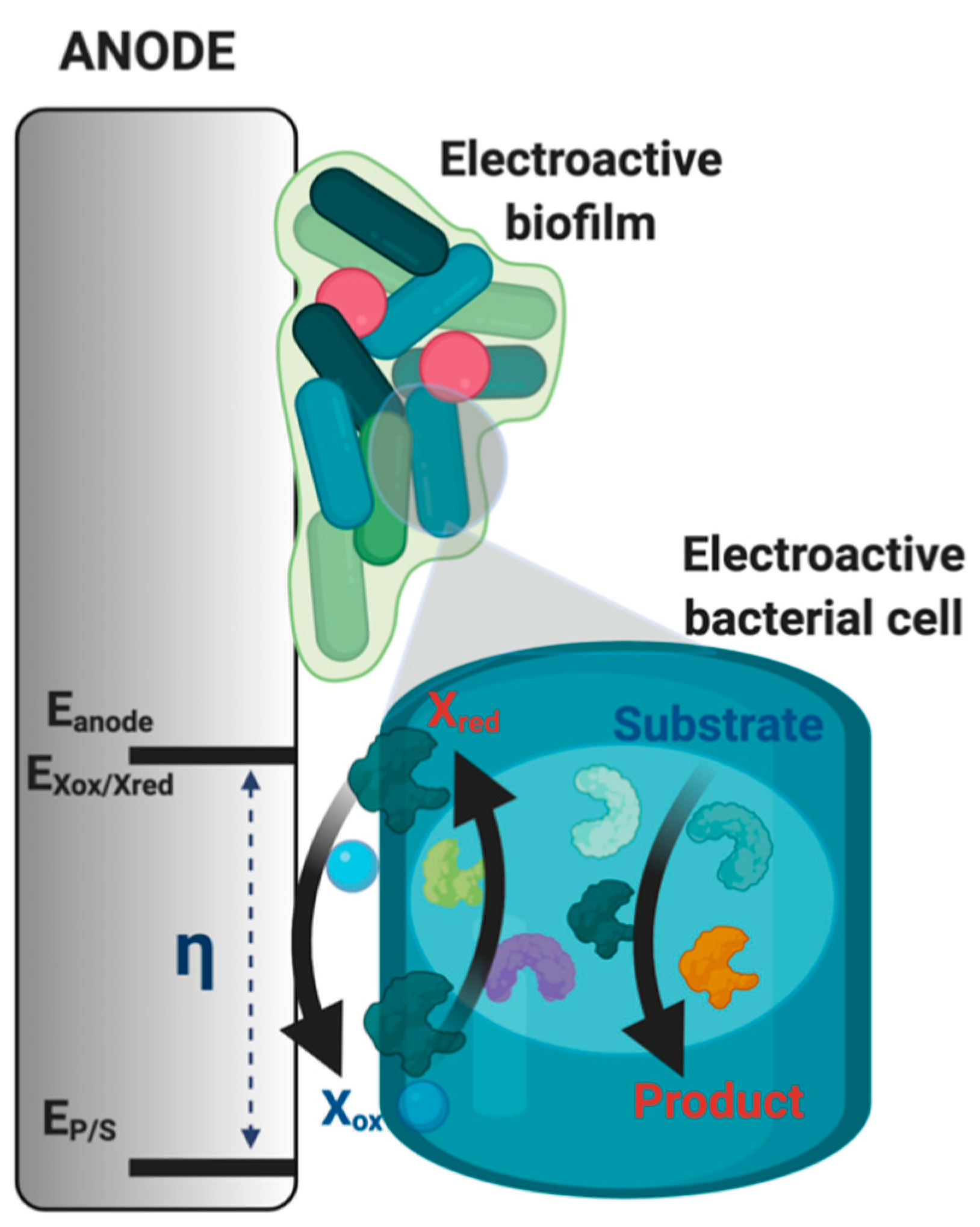
2.3. Electroactive Halophilic Bacteria
3. Microbial Electrochemical Biosensing in High Saline
3.1. Microbial-Based Sensing in High Saline
3.2. Microbial Fuel Cells in High Saline with Potential for Microbial Electrochemical Biosensing
4. Future Directions and Perspectives on Utilizing Halophilic Electroactive Bacteria for Electrochemical Biosensing
4.1. Heavy Metal Sensing with Electroactive Halophilic Bacteria
4.2. Engineering Systems with Electroactive Halophilic Bacteria for Microbial Electrochemistry
4.3. Extremozymes from Halophilic Bacteria for Enzymatic Sensing in High Saline Environments
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, H.; Simoska, O.; Lim, K.; Grattieri, M.; Yuan, M.; Dong, F.; Lee, Y.S.; Beaver, K.; Weliwatte, S.; Gaffney, E.M.; et al. Fundamentals, Applications, and Future Directions of Bioelectrocatalysis. Chem. Rev. 2020, 120, 12903–12993. [Google Scholar] [CrossRef]
- Harnisch, F.; Rabaey, K. Bioelectrochemical Systems. In Materials for Low-Temperature Fuel Cells; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2014; pp. 167–184. [Google Scholar]
- Grattieri, M.; Hasan, K.; Minteer, S.D. Bioelectrochemical Systems as a Multipurpose Biosensing Tool: Present Perspective and Future Outlook. ChemElectroChem 2017, 4, 834–842. [Google Scholar] [CrossRef]
- Zheng, T.; Li, J.; Ji, Y.; Zhang, W.; Fang, Y.; Xin, F.; Dong, W.; Wei, P.; Ma, J.; Jiang, M. Progress and Prospects of Bioelectrochemical Systems: Electron Transfer and Its Applications in the Microbial Metabolism. Front. Bioeng. Biotechnol. 2020, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ren, Z.J. A comprehensive review of microbial electrochemical systems as a platform technology. Biotechnol. Adv. 2013, 31, 1796–1807. [Google Scholar] [CrossRef] [PubMed]
- Schröder, U.; Harnisch, F.; Angenent, L.T. Microbial electrochemistry and technology: Terminology and classification. Energy Environ. Sci. 2015, 8, 513–519. [Google Scholar] [CrossRef]
- Santoro, C.; Arbizzani, C.; Erable, B.; Ieropoulos, I. Microbial fuel cells: From fundamentals to applications. A review. J. Power Sources 2017, 356, 225–244. [Google Scholar] [CrossRef] [PubMed]
- Kretzschmar, J.; Böhme, P.; Liebetrau, J.; Mertig, M.; Harnisch, F. Microbial Electrochemical Sensors for Anaerobic Digestion Process Control—Performance of Electroactive Biofilms under Real Conditions. Chem. Eng. Technol. 2018, 41, 687–695. [Google Scholar] [CrossRef]
- Grattieri, M.; Minteer, S.D. Self-Powered Biosensors. ACS Sens. 2018, 3, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Abrevaya, X.C.; Sacco, N.J.; Bonetto, M.C.; Hilding-Ohlsson, A.; Cortón, E. Analytical applications of microbial fuel cells. Part I: Biochemical oxygen demand. Biosens. Bioelectron. 2015, 63, 580–590. [Google Scholar] [CrossRef] [PubMed]
- Lovley, D.R. Bug juice: Harvesting electricity with microorganisms. Nat. Rev. Microbiol. 2006, 4, 497–508. [Google Scholar] [CrossRef] [PubMed]
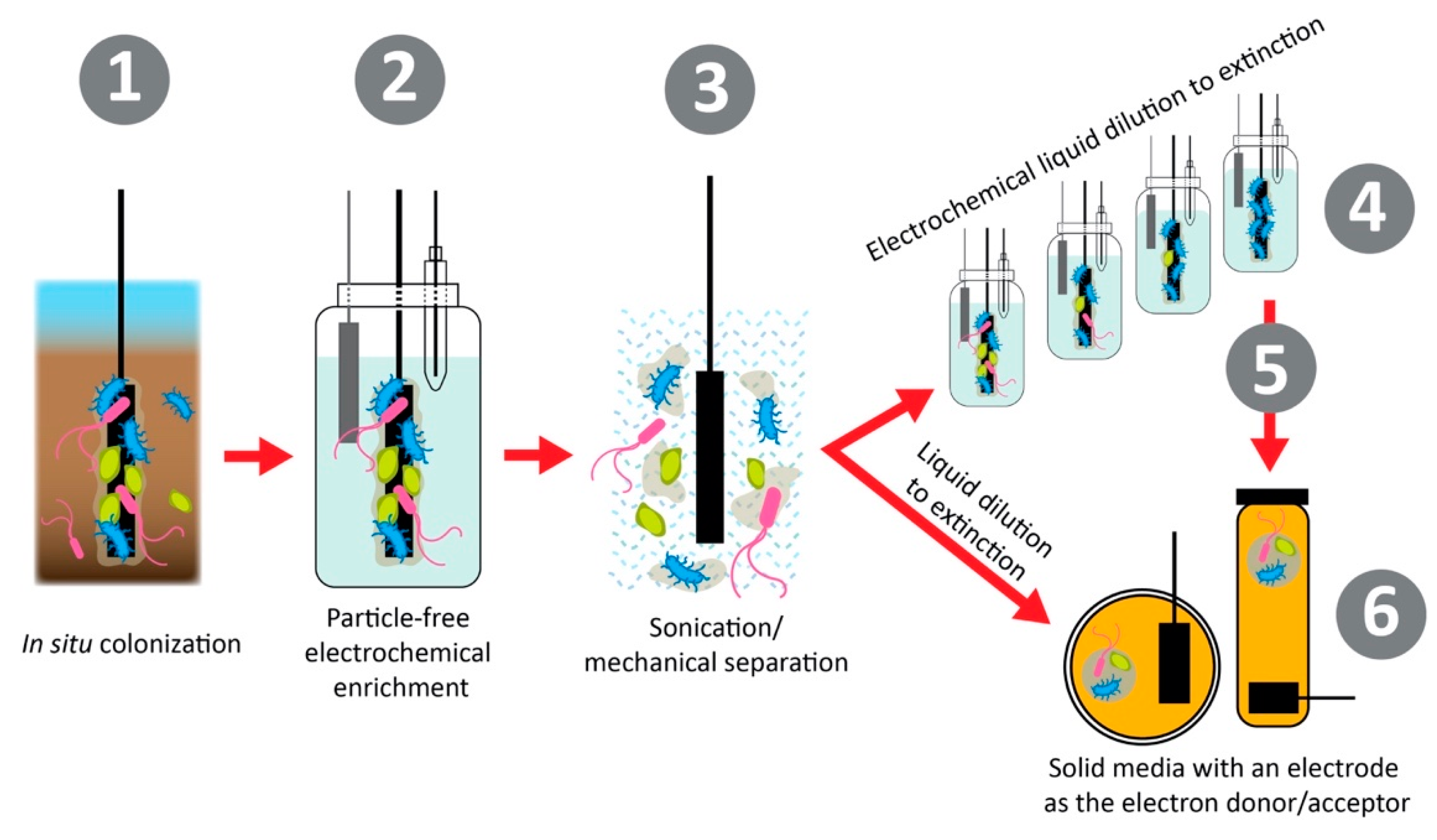
- Koch, C.; Korth, B.; Harnisch, F. Microbial ecology-based engineering of Microbial Electrochemical Technologies. Microb. Biotechnol. 2018, 11, 22–38. [Google Scholar] [CrossRef] [PubMed]
- Yee, M.O.; Deutzmann, J.; Spormann, A.; Rotaru, A.E. Cultivating electroactive microbes-from field to bench. Nanotechnology 2020, 31, 174003. [Google Scholar] [CrossRef]
- Korth, B.; Harnisch, F. Spotlight on the Energy Harvest of Electroactive Microorganisms: The Impact of the Applied Anode Potential. Front. Microbiol. 2019, 10, 1352. [Google Scholar] [CrossRef]
- Logan, B.E.; Rossi, R.; Ragab, A.; Saikaly, P.E. Electroactive microorganisms in bioelectrochemical systems. Nat. Rev. Microbiol. 2019, 17, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Torres, C.I. On the importance of identifying, characterizing, and predicting fundamental phenomena towards microbial electrochemistry applications. Curr. Opin. Biotechnol. 2014, 27, 107–114. [Google Scholar] [CrossRef]
- Gaffney, E.M.; Grattieri, M.; Rhodes, Z.; Minteer, S.D. Editors’ Choice—Review—Exploration of Computational Approaches for Understanding Microbial Electrochemical Systems: Opportunities and Future Directions. J. Electrochem. Soc. 2020, 167, 065502. [Google Scholar] [CrossRef]
- Recio-Garrido, D.; Perrier, M.; Tartakovsky, B. Modeling, optimization and control of bioelectrochemical systems. Chem. Eng. J. 2016, 289, 180–190. [Google Scholar] [CrossRef]
- Bond, D.R.; Lovley, D.R. Electricity production by Geobacter sulfurreducens attached to electrodes. Appl. Environ. Microbiol. 2003, 69, 1548–1555. [Google Scholar] [CrossRef]
- Bouhenni, R.A.; Vora, G.J.; Biffinger, J.C.; Shirodkar, S.; Brockman, K.; Ray, R.; Wu, P.; Johnson, B.J.; Biddle, E.M.; Marshall, M.J.; et al. The Role of Shewanella oneidensis MR-1 Outer Surface Structures in Extracellular Electron Transfer. Electroanalysis 2010, 22, 856–864. [Google Scholar] [CrossRef]
- Heidelberg, J.F.; Paulsen, I.T.; Nelson, K.E.; Gaidos, E.J.; Nelson, W.C.; Read, T.D.; Eisen, J.A.; Seshadri, R.; Ward, N.; Methe, B.; et al. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 2002, 20, 1118–1123. [Google Scholar] [CrossRef]
- Holmes, D.E.; Chaudhuri, S.K.; Nevin, K.P.; Mehta, T.; Methe, B.A.; Liu, A.; Ward, J.E.; Woodard, T.L.; Webster, J.; Lovley, D.R. Microarray and genetic analysis of electron transfer to electrodes in Geobacter sulfurreducens. Environ. Microbiol. 2006, 8, 1805–1815. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, N.; Chilkoor, G.; Vemuri, B.; Rathinam, N.; Sani, R.K.; Gadhamshetty, V. Extremophiles for microbial-electrochemistry applications: A critical review. Bioresour. Technol. 2018, 255, 318–330. [Google Scholar] [CrossRef] [PubMed]
- Dopson, M.; Ni, G.; Sleutels, T.H.J.A. Possibilities for extremophilic microorganisms in microbial electrochemical systems. FEMS Microbiol. Rev. 2016, 40, 164–181. [Google Scholar] [CrossRef]
- Lefebvre, O.; Tan, Z.; Kharkwal, S.; Ng, H.Y. Effect of increasing anodic NaCl concentration on microbial fuel cell performance. Bioresour. Technol. 2012, 112, 336–340. [Google Scholar] [CrossRef]
- Oren, A. Diversity of halophilic microorganisms: Environments, phylogeny, physiology, and applications. J. Ind. Microbiol. Biotechnol. 2002, 28, 56–63. [Google Scholar] [CrossRef]
- Rajendran, N. Environmental Diversity and Biological Survivability of Halophilic Bacteria. In Halophiles: Biodiversity and Sustainable Exploitation; Springer International Publishing: New York, NY, USA, 2015; pp. 173–188. [Google Scholar]
- Hozzein, W.N. Biodiversity of halophilic and halotolerant actinobacteria. In Halophiles: Biodiversity and Sustainable Exploitation; Springer International Publishing: New York, NY, USA, 2015; pp. 1–28. [Google Scholar]
- Saenger, W. Structure and Dynamics of Water Surrounding Biomolecules. Annu. Rev. Biophys. Biophys. Chem. 1987, 16, 93–114. [Google Scholar] [CrossRef]
- Jin, M.; Gai, Y.; Guo, X.; Hou, Y.; Zeng, R. Properties and Applications of Extremozymes from Deep-Sea Extremophilic Microorganisms: A Mini Review. Mar. Drugs 2019, 17, 656. [Google Scholar] [CrossRef] [PubMed]
- Siglioccolo, A.; Paiardini, A.; Piscitelli, M.; Pascarella, S. Structural adaptation of extreme halophilic proteins through decrease of conserved hydrophobic contact surface. BMC Struct. Biol. 2011, 11, 50. [Google Scholar] [CrossRef]
- Oren, A. Industrial and environmental applications of halophilic microorganisms. Environ. Technol. 2010, 31, 825–834. [Google Scholar] [CrossRef]
- Margesin, R.; Schinner, F. Potential of halotolerant and halophilic microorganisms for biotechnology. Extremophiles 2001, 5, 73–83. [Google Scholar] [CrossRef]
- Yin, J.; Chen, J.-C.; Wu, Q.; Chen, G.-Q. Halophiles, coming stars for industrial biotechnology. Biotechnol. Adv. 2015, 33, 1433–1442. [Google Scholar] [CrossRef]
- Corral, P.; Amoozegar, M.A.; Ventosa, A. Halophiles and Their Biomolecules: Recent Advances and Future Applications in Biomedicine. Mar. Drugs 2019, 18, 33. [Google Scholar] [CrossRef]
- Amoozegar, M.A.; Safarpour, A.; Noghabi, K.A.; Bakhtiary, T.; Ventosa, A. Halophiles and Their Vast Potential in Biofuel Production. Front. Microbiol. 2019, 10, 1–17. [Google Scholar] [CrossRef]
- Gaffney, E.M.; Grattieri, M.; Beaver, K.; Pham, J.; McCartney, C.; Minteer, S.D. Unveiling salinity effects on photo-bioelectrocatalysis through combination of bioinformatics and electrochemistry. Electrochim. Acta 2020, 337, 135731. [Google Scholar] [CrossRef]
- Grattieri, M.; Beaver, K.; Gaffney, E.M.; Minteer, S.D. Tuning purple bacteria salt-tolerance for photobioelectrochemical systems in saline environments. Faraday Discuss. 2019, 215, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Fukushima, T.; Prior, A.; Baruch, M.; Zajdel, T.J.; Ajo-Franklin, C.M. Modifying cytochrome c maturation can increase the bioelectronic performance of engineered escherichia coli. ACS Synth. Biol. 2020, 9, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Sekar, N.; Jain, R.; Yan, Y.; Ramasamy, R.P. Enhanced photo-bioelectrochemical energy conversion by genetically engineered cyanobacteria. Biotechnol. Bioeng. 2016, 113, 675–679. [Google Scholar] [CrossRef]
- Jensen, H.M.; Albers, A.E.; Malley, K.R.; Londerd, Y.Y.; Cohen, B.E.; Helmsc, B.A.; Weigele, P.; Groves, J.T.; Ajo-Franklin, C.M. Engineering of a synthetic electron conduit in living cells. Proc. Natl. Acad. Sci. USA 2010, 107, 19213–19218. [Google Scholar] [CrossRef] [PubMed]
- Grattieri, M.; Hickey, D.P.; Alkotaini, B.; Robertson, S.J.; Minteer, S.D. Hypersaline microbial self-powered biosensor with increased sensitivity. J. Electrochem. Soc. 2018, 165, H251–H254. [Google Scholar] [CrossRef]
- Grattieri, M.; Minteer, S.D. Microbial fuel cells in saline and hypersaline environments: Advancements, challenges and future perspectives. Bioelectrochemistry 2018, 120, 127–137. [Google Scholar] [CrossRef]
- Grattieri, M.; Suvira, M.; Hasan, K.; Minteer, S.D. Halotolerant extremophile bacteria from the Great Salt Lake for recycling pollutants in microbial fuel cells. J. Power Sources 2017, 356, 310–318. [Google Scholar] [CrossRef]
- Ieropoulos, I.A.; Stinchcombe, A.; Gajda, I.; Forbes, S.; Merino-Jimenez, I.; Pasternak, G.; Sanchez-Herranz, D.; Greenman, J. Pee power urinal-microbial fuel cell technology field trials in the context of sanitation. Environ. Sci. Water Res. Technol. 2016, 2, 336–343. [Google Scholar] [CrossRef]
- ElMekawy, A.; Hegab, H.M.; Pant, D.; Saint, C.P. Bio-analytical applications of microbial fuel cell-based biosensors for onsite water quality monitoring. J. Appl. Microbiol. 2018, 124, 302–313. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Jia, W.; Hou, C.; Lei, Y. Microbial biosensors: A review. Biosens. Bioelectron. 2011, 26, 1788–1799. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lu, X.; Chen, J. Development of biosensor technologies for analysis of environmental contaminants. Trends Environ. Anal. Chem. 2014, 2, 25–32. [Google Scholar] [CrossRef]
- Lei, Y.; Chen, W.; Mulchandani, A. Microbial biosensors. Anal. Chim. Acta 2006, 568, 200–210. [Google Scholar] [CrossRef]
- Simoska, O.; Stevenson, K.J. Electrochemical sensors for rapid diagnosis of pathogens in real time. The Analyst 2019, 144, 6461–6478. [Google Scholar] [CrossRef]
- Perumal, V.; Hashim, U. Advances in biosensors: Principle, architecture and applications. J. Appl. Biomed. 2014, 12, 1–15. [Google Scholar] [CrossRef]
- Dong, Y.; Li, G.; Zhou, N.; Wang, R.; Chi, Y.; Chen, G. Graphene Quantum Dot as a Green and Facile Sensor for Free Chlorine in Drinking Water. Anal. Chem. 2012, 84, 8378–8382. [Google Scholar] [CrossRef]
- Woznica, A.; Nowak, A.; Karczewski, J.; Klis, C.; Bernas, T. Automatic biodetector of water toxicity (ABTOW) as a tool for examination of phenol and cyanide contaminated water. Chemosphere 2010, 81, 767–772. [Google Scholar] [CrossRef]
- Hernández-Sánchez, V.; Molina, L.; Ramos, J.L.; Segura, A. New family of biosensors for monitoring BTX in aquatic and edaphic environments. Microb. Biotechnol. 2016, 9, 858–867. [Google Scholar] [CrossRef]
- Chung, T.H.; Meshref, M.N.A.; Dhar, B.R. Microbial electrochemical biosensor for rapid detection of naphthenic acid in aqueous solution. J. Electroanal. Chem. 2020, 873, 114405. [Google Scholar] [CrossRef]
- Jannelli, N.; Anna Nastro, R.; Cigolotti, V.; Minutillo, M.; Falcucci, G. Low pH, high salinity: Too much for microbial fuel cells? Appl. Energy 2017, 192, 543–550. [Google Scholar] [CrossRef]
- Modin, O.; Wilén, B.M. A novel bioelectrochemical BOD sensor operating with voltage input. Water Res. 2012, 46, 6113–6120. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Wang, X.; Wang, G.; Ma, L. A BOD biosensor using salt-tolerant Bacillus licheniformis for sea water. In Proceedings of the 3rd International Conference on Bioinformatics and Biomedical Engineering, (iCBBE), Beijing, China, 11–13 June 2009; pp. 12–15. [Google Scholar]
- Alkotaini, B.; Tinucci, S.L.; Robertson, S.J.; Hasan, K.; Minteer, S.D.; Grattieri, M. Alginate-Encapsulated Bacteria for the Treatment of Hypersaline Solutions in Microbial Fuel Cells. ChemBioChem 2018, 19, 1162–1169. [Google Scholar] [CrossRef]
- Grattieri, M.; Shivel, N.D.; Sifat, I.; Bestetti, M.; Minteer, S.D. Sustainable Hypersaline Microbial Fuel Cells: Inexpensive Recyclable Polymer Supports for Carbon Nanotube Conductive Paint Anodes. ChemSusChem 2017, 10, 2053–2058. [Google Scholar] [CrossRef] [PubMed]
- Robertson, S.J.; Grattieri, M.; Behring, J.; Bestetti, M.; Minteer, S.D. Transitioning from batch to flow hypersaline microbial fuel cells. Electrochim. Acta 2019, 317, 494–501. [Google Scholar] [CrossRef]
- Santoro, C.; Walter, X.A.; Soavi, F.; Greenman, J.; Ieropoulos, I. Self-stratified and self-powered micro-supercapacitor integrated into a microbial fuel cell operating in human urine. Electrochim. Acta 2019, 307, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Santoro, C.; Walter, X.A.; Soavi, F.; Greenman, J.; Ieropoulos, I. Air-breathing cathode self-powered supercapacitive microbial fuel cell with human urine as electrolyte. Electrochim. Acta 2020, 353, 136530. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Dou, D.; Zhang, X.; Zhang, L.; Dong, H.; Yu, H. Degradation of Norfloxacin in saline water by synergistic effect of anode and cathode in a novel photo-electrochemical system. J. Cleaner Prod. 2020, 242, 118548. [Google Scholar] [CrossRef]
- Gui, Q.; Lawson, T.; Shan, S.; Yan, L.; Liu, Y. The Application of Whole Cell-Based Biosensors for Use in Environmental Analysis and in Medical Diagnostics. Sensors 2017, 17, 1623. [Google Scholar] [CrossRef] [PubMed]
- Amiri, M.; Bezaatpour, A.; Jafari, H.; Boukherroub, R.; Szunerits, S. Electrochemical Methodologies for the Detection of Pathogens. ACS Sens. 2018, 3, 1069–1086. [Google Scholar] [CrossRef] [PubMed]
- Voica, D.M.; Bartha, L.; Banciu, H.L.; Oren, A. Heavy metal resistance in halophilic Bacteria and Archaea. FEMS Microbiol. Lett. 2016, 363, 146. [Google Scholar] [CrossRef]
- Nakayama, L.; Oshima, T.; Shinmyo, A.; Ogasawara, N. Development of whole-cell biosensor using a moderate halophilic bacterium, Halomonaselongata, for monitoringmet- als in high salinity environments. J. Biotechnol. 2010, 150, 226. [Google Scholar] [CrossRef]
- Cui, Z.; Luan, X.; Jiang, H.; Li, Q.; Xu, G.; Sun, C.; Zheng, L.; Song, Y.; Davison, P.A.; Huang, W.E. Application of a bacterial whole cell biosensor for the rapid detection of cytotoxicity in heavy metal contaminated seawater. Chemosphere 2018, 200, 322–329. [Google Scholar] [CrossRef]
- Bereza-Malcolm, L.T.; Mann, G.; Franks, A.E. Environmental Sensing of Heavy Metals Through Whole Cell Microbial Biosensors: A Synthetic Biology Approach. ACS Synth. Biol. 2015, 4, 535–546. [Google Scholar] [CrossRef]
- Lindquist, H.D.A. Microbial biosensors for recreational and source waters. J. Microbiol. Methods 2020, 177, 106059. [Google Scholar] [CrossRef]
- Koch, C.; Harnisch, F. Is there a Specific Ecological Niche for Electroactive Microorganisms? ChemElectroChem 2016, 3, 1282–1295. [Google Scholar] [CrossRef]
- Gaffney, E.M.; Grattieri, M.; Minteer, S.D. Draft Genome Sequence of Salinivibrio sp. Strain EAGSL, a Biotechnologically Relevant Halophilic Microorganism. Microbiol. Resour. Announce. 2020, 9, 1–2. [Google Scholar] [CrossRef]
- Chen, Y.H.; Lu, C.W.; Shyu, Y.T.; Lin, S.S. Revealing the Saline Adaptation Strategies of the Halophilic Bacterium Halomonas beimenensis through High-throughput Omics and Transposon Mutagenesis Approaches. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Jensen, H.M.; TerAvest, M.A.; Kokish, M.G.; Ajo-Franklin, C.M. CymA and Exogenous Flavins Improve Extracellular Electron Transfer and Couple It to Cell Growth in Mtr-Expressing Escherichia coli. ACS Synth. Biol. 2016, 5, 679–688. [Google Scholar] [CrossRef] [PubMed]
- Schuergers, N.; Werlang, C.; Ajo-Franklin, C.M.; Boghossian, A.A. A synthetic biology approach to engineering living photovoltaics. Energy Environ. Sci. 2017, 10, 1102–1115. [Google Scholar] [CrossRef]
- Feng, J.; Qian, Y.; Wang, Z.; Wang, X.; Xu, S.; Chen, K.; Ouyang, P. Enhancing the performance of Escherichia coli-inoculated microbial fuel cells by introduction of the phenazine-1-carboxylic acid pathway. J. Biotechnol. 2018, 275, 1–6. [Google Scholar] [CrossRef]
- Raval, V.H.; Bhatt, H.B.; Singh, S.P. Adaptation Strategies in Halophilic Bacteria. Extremophiles 2018, 137–164. [Google Scholar]
- Angelaalincy, M.J.; Navanietha Krishnaraj, R.; Shakambari, G.; Ashokkumar, B.; Kathiresan, S.; Varalakshmi, P. Biofilm Engineering Approaches for Improving the Performance of Microbial Fuel Cells and Bioelectrochemical Systems. Front. Energy Res. 2018, 6, 63. [Google Scholar] [CrossRef]
- Montgomery, K.; Charlesworth, J.; LeBard, R.; Visscher, P.; Burns, B. Quorum Sensing in Extreme Environments. Life 2013, 3, 131–148. [Google Scholar] [CrossRef]
- Chen, S.; Jing, X.; Tang, J.; Fang, Y.; Zhou, S. Quorum sensing signals enhance the electrochemical activity and energy recovery of mixed-culture electroactive biofilms. Biosens. Bioelectron. 2017, 97, 369–376. [Google Scholar] [CrossRef]
- Llamas, I.; Quesada, E.; Martínez-Cánovas, M.J.; Gronquist, M.; Eberhard, A.; González, J.E. Quorum sensing in halophilic bacteria: Detection of N-acyl-homoserine lactones in the exopolysaccharide-producing species of Halomonas. Extremophiles 2005, 9, 333–341. [Google Scholar] [CrossRef]
- Monzon, O.; Yang, Y.; Li, Q.; Alvarez, P.J.J. Quorum sensing autoinducers enhance biofilm formation and power production in a hypersaline microbial fuel cell. Biochem. Eng. J. 2016, 109, 222–227. [Google Scholar] [CrossRef]
- Van den Burg, B. Extremophiles as a source for novel enzymes. Curr. Opin. Microbiol. 2003, 6, 213–218. [Google Scholar] [CrossRef]
- Li, P.-Y.; Zhang, Y.; Xie, B.-B.; Zhang, Y.-Q.; Hao, J.; Wang, Y.; Wang, P.; Li, C.-Y.; Qin, Q.-L.; Zhang, X.-Y.; et al. Structural and Mechanistic Insights into the Improvement of the Halotolerance of a Marine Microbial Esterase by Increasing Intra- and Interdomain Hydrophobic Interactions. Appl. Environ. Microbiol. 2017, 83, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, N.; Jayshree, A. Extremozymes and Extremoproteins in Biosensor Applications. In Encyclopedia of Marine Biotechnology; Wiley: Hoboken, NJ, USA, 2020; pp. 1711–1736. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaffney, E.M.; Simoska, O.; Minteer, S.D. The Use of Electroactive Halophilic Bacteria for Improvements and Advancements in Environmental High Saline Biosensing. Biosensors 2021, 11, 48. https://doi.org/10.3390/bios11020048
Gaffney EM, Simoska O, Minteer SD. The Use of Electroactive Halophilic Bacteria for Improvements and Advancements in Environmental High Saline Biosensing. Biosensors. 2021; 11(2):48. https://doi.org/10.3390/bios11020048
Chicago/Turabian StyleGaffney, Erin M., Olja Simoska, and Shelley D. Minteer. 2021. "The Use of Electroactive Halophilic Bacteria for Improvements and Advancements in Environmental High Saline Biosensing" Biosensors 11, no. 2: 48. https://doi.org/10.3390/bios11020048
APA StyleGaffney, E. M., Simoska, O., & Minteer, S. D. (2021). The Use of Electroactive Halophilic Bacteria for Improvements and Advancements in Environmental High Saline Biosensing. Biosensors, 11(2), 48. https://doi.org/10.3390/bios11020048