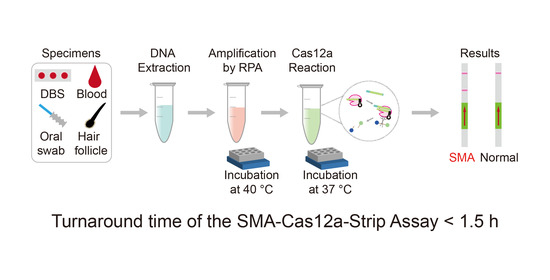
Cas12a and Lateral Flow Strip-Based Test for Rapid and Ultrasensitive Detection of Spinal Muscular Atrophy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Source of Samples
2.2. Primers and crRNAs
2.3. DNA Amplification
2.4. The SMA-Cas12a Assay with a Fluorescence Probe
2.5. Selecting Optimal RPA Primers
2.6. The SMA-Cas12a-Strip Assay with an FITC-Biotin-Labeled Probe
2.7. Evaluation of Analytical Performance of the SMA-Cas12a-Strip Assay
2.8. Exploring Extensible Utility
2.9. Statistical Analysis
3. Results
3.1. Establishment of the SMA-Cas12a Assay with a Fluorescence Probe
3.2. Specific Amplification of SMN1 by RPA
3.3. Establishment of the SMA-Cas12a-Strip Assay
3.4. Analytical Performance of the SMA-Cas12a-Strip Assay
3.5. Extensible Utility of the Cas12a-Strip Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Pearn, J. Classification of spinal muscular atrophies. Lancet 1980, 1, 919–922. [Google Scholar] [CrossRef]
- Verhaart, I.E.C.; Robertson, A.; Wilson, I.J.; Aartsma-Rus, A.; Cameron, S.; Jones, C.C.; Cook, S.F.; Lochmuller, H. Prevalence, incidence and carrier frequency of 5q-linked spinal muscular atrophy—A literature review. Orphanet J. Rare Dis. 2017, 12, 124. [Google Scholar] [CrossRef] [Green Version]
- Lefebvre, S.; Burglen, L.; Reboullet, S.; Clermont, O.; Burlet, P.; Viollet, L.; Benichou, B.; Cruaud, C.; Millasseau, P.; Zeviani, M.; et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 1995, 80, 155–165. [Google Scholar] [CrossRef] [Green Version]
- Wirth, B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA). Hum. Mutat. 2000, 15, 228–237. [Google Scholar] [CrossRef]
- Alias, L.; Bernal, S.; Fuentes-Prior, P.; Barcelo, M.J.; Also, E.; Martinez-Hernandez, R.; Rodriguez-Alvarez, F.J.; Martin, Y.; Aller, E.; Grau, E.; et al. Mutation update of spinal muscular atrophy in Spain: Molecular characterization of 745 unrelated patients and identification of four novel mutations in the SMN1 gene. Hum. Genet. 2009, 125, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Cartegni, L.; Hastings, M.L.; Calarco, J.A.; de Stanchina, E.; Krainer, A.R. Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am. J. Hum. Genet. 2006, 78, 63–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finkel, R.S.; Mercuri, E.; Darras, B.T.; Connolly, A.M.; Kuntz, N.L.; Kirschner, J.; Chiriboga, C.A.; Saito, K.; Servais, L.; Tizzano, E.; et al. Nusinersen versus Sham Control in Infantile-Onset Spinal Muscular Atrophy. N. Engl. J. Med. 2017, 377, 1723–1732. [Google Scholar] [CrossRef] [Green Version]
- Mercuri, E.; Darras, B.T.; Chiriboga, C.A.; Day, J.W.; Campbell, C.; Connolly, A.M.; Iannaccone, S.T.; Kirschner, J.; Kuntz, N.L.; Saito, K.; et al. Nusinersen versus Sham Control in Later-Onset Spinal Muscular Atrophy. N. Engl. J. Med. 2018, 378, 625–635. [Google Scholar] [CrossRef]
- Hoy, S.M. Onasemnogene Abeparvovec: First Global Approval. Drugs 2019, 79, 1255–1262. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, S. Risdiplam: First Approval. Drugs 2020, 80, 1853–1858. [Google Scholar] [CrossRef] [PubMed]
- Chien, Y.H.; Chiang, S.C.; Weng, W.C.; Lee, N.C.; Lin, C.J.; Hsieh, W.S.; Lee, W.T.; Jong, Y.J.; Ko, T.M.; Hwu, W.L. Presymptomatic Diagnosis of Spinal Muscular Atrophy Through Newborn Screening. J. Pediatr. 2017, 190, 124–129.e121. [Google Scholar] [CrossRef]
- Kraszewski, J.N.; Kay, D.M.; Stevens, C.F.; Koval, C.; Haser, B.; Ortiz, V.; Albertorio, A.; Cohen, L.L.; Jain, R.; Andrew, S.P.; et al. Pilot study of population-based newborn screening for spinal muscular atrophy in New York state. Genet. Med. 2018, 20, 608–613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.H.; Chang, Y.Y.; Chen, C.H.; Kuo, Y.S.; Hwu, W.L.; Gerdes, T.; Ko, T.M. Copy number analysis of survival motor neuron genes by multiplex ligation-dependent probe amplification. Genet. Med. 2007, 9, 241–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, J.L.; Lee, F.K.; Yazdanpanah, G.K.; Staropoli, J.F.; Liu, M.; Carulli, J.P.; Sun, C.; Dobrowolski, S.F.; Hannon, W.H.; Vogt, R.F. Newborn blood spot screening test using multiplexed real-time PCR to simultaneously screen for spinal muscular atrophy and severe combined immunodeficiency. Clin. Chem. 2015, 61, 412–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobrowolski, S.F.; Pham, H.T.; Downes, F.P.; Prior, T.W.; Naylor, E.W.; Swoboda, K.J. Newborn screening for spinal muscular atrophy by calibrated short-amplicon melt profiling. Clin. Chem. 2012, 58, 1033–1039. [Google Scholar] [CrossRef] [Green Version]
- Sutomo, R.; Akutsu, T.; Takeshima, Y.; Nishio, H.; Sadewa, A.H.; Harada, Y.; Matsuo, M. Rapid SMN1 deletion test using DHPLC to screen patients with spinal muscular atrophy. Am. J. Med. Genet. 2002, 113, 225–226. [Google Scholar] [CrossRef]
- Vidal-Folch, N.; Gavrilov, D.; Raymond, K.; Rinaldo, P.; Tortorelli, S.; Matern, D.; Oglesbee, D. Multiplex Droplet Digital PCR Method Applicable to Newborn Screening, Carrier Status, and Assessment of Spinal Muscular Atrophy. Clin. Chem. 2018, 64, 1753–1761. [Google Scholar] [CrossRef] [Green Version]
- Park, S.; Lee, H.; Shin, S.; Lee, S.T.; Lee, K.A.; Choi, J.R. Analytical validation of the droplet digital PCR assay for diagnosis of spinal muscular atrophy. Clin. Chim. Acta 2020, 510, 787–789. [Google Scholar] [CrossRef]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef] [Green Version]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef] [Green Version]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.Y.; Cheng, Q.X.; Wang, J.M.; Li, X.Y.; Zhang, Z.L.; Gao, S.; Cao, R.B.; Zhao, G.P.; Wang, J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018, 4, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukama, O.; Wu, J.; Li, Z.; Liang, Q.; Yi, Z.; Lu, X.; Liu, Y.; Liu, Y.; Hussain, M.; Makafe, G.G.; et al. An ultrasensitive and specific point-of-care CRISPR/Cas12 based lateral flow biosensor for the rapid detection of nucleic acids. Biosens. Bioelectron. 2020, 159, 112143. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Tsai, S.Q.; Prew, M.S.; Nguyen, N.T.; Welch, M.M.; Lopez, J.M.; McCaw, Z.R.; Aryee, M.J.; Joung, J.K. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 2016, 34, 869–874. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Kim, J.; Hur, J.K.; Been, K.W.; Yoon, S.H.; Kim, J.S. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 2016, 34, 863–868. [Google Scholar] [CrossRef] [PubMed]
- Labun, K.; Montague, T.G.; Gagnon, J.A.; Thyme, S.B.; Valen, E. CHOPCHOP v2: A web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016, 44, W272–W276. [Google Scholar] [CrossRef] [PubMed]
- Hajian, R.; Balderston, S.; Tran, T.; deBoer, T.; Etienne, J.; Sandhu, M.; Wauford, N.A.; Chung, J.Y.; Nokes, J.; Athaiya, M.; et al. Detection of unamplified target genes via CRISPR-Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 2019, 3, 427–437. [Google Scholar] [CrossRef]
- D’Agata, R.; Breveglieri, G.; Zanoli, L.M.; Borgatti, M.; Spoto, G.; Gambari, R. Direct detection of point mutations in nonamplified human genomic DNA. Anal. Chem. 2011, 83, 8711–8717. [Google Scholar] [CrossRef]
- Kariyawasam, D.S.T.; Russell, J.S.; Wiley, V.; Alexander, I.E.; Farrar, M.A. The implementation of newborn screening for spinal muscular atrophy: The Australian experience. Genet. Med. 2019. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020. [Google Scholar] [CrossRef] [Green Version]
- Tsou, J.H.; Leng, Q.; Jiang, F. A CRISPR Test for Detection of Circulating Nuclei Acids. Transl. Oncol. 2019, 12, 1566–1573. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhang, W.; Qiu, X.; Mei, Q.; Luo, Y.; Fu, W. Rapid and sensitive exosome detection with CRISPR/Cas12a. Anal. Bioanal. Chem. 2020. [Google Scholar] [CrossRef] [PubMed]
- Shao, N.; Han, X.; Song, Y.; Zhang, P.; Qin, L. CRISPR-Cas12a Coupled with Platinum Nanoreporter for Visual Quantification of SNVs on a Volumetric Bar-Chart Chip. Anal. Chem. 2019, 91, 12384–12391. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Mukama, O.; Wu, W.; Li, Z.; Habimana, J.D.; Zhang, Y.; Zeng, R.; Nie, C.; Zeng, L. A CRISPR/Cas12a Based Universal Lateral Flow Biosensor for the Sensitive and Specific Detection of African Swine-Fever Viruses in Whole Blood. Biosensors 2020, 10, 203. [Google Scholar] [CrossRef]
- Curti, L.A.; Pereyra-Bonnet, F.; Repizo, G.D.; Fay, J.V.; Salvatierra, K.; Blariza, M.J.; Ibanez-Alegre, D.; Rinflerch, A.R.; Miretti, M.; Gimenez, C.A. CRISPR-based platform for carbapenemases and emerging viruses detection using Cas12a (Cpf1) effector nuclease. Emerg. Microbes Infect. 2020, 9, 1140–1148. [Google Scholar] [CrossRef]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA detection using recombination proteins. PLoS Biol. 2006, 4, e204. [Google Scholar] [CrossRef]




| Confirmed by MLPA 1/qPCR 2 | |||
|---|---|---|---|
| SMA Patients | Non-SMA Individuals | ||
| Tested by the SMA-Cas12a-Strip Assay | Positive | 90 (TP) 3 | 0 (FP) 4 |
| Negative | 0 (FN) 5 | 78 (TN) 6 | |
| Sensitivity 7: 100% | Specificity 8: 100% | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Li, Z.; Chen, M.; Hu, Z.; Wu, L.; Zhou, M.; Liang, D. Cas12a and Lateral Flow Strip-Based Test for Rapid and Ultrasensitive Detection of Spinal Muscular Atrophy. Biosensors 2021, 11, 154. https://doi.org/10.3390/bios11050154
Zhang C, Li Z, Chen M, Hu Z, Wu L, Zhou M, Liang D. Cas12a and Lateral Flow Strip-Based Test for Rapid and Ultrasensitive Detection of Spinal Muscular Atrophy. Biosensors. 2021; 11(5):154. https://doi.org/10.3390/bios11050154
Chicago/Turabian StyleZhang, Chunhua, Zhuo Li, Miaomiao Chen, Zhiqing Hu, Lingqian Wu, Miaojin Zhou, and Desheng Liang. 2021. "Cas12a and Lateral Flow Strip-Based Test for Rapid and Ultrasensitive Detection of Spinal Muscular Atrophy" Biosensors 11, no. 5: 154. https://doi.org/10.3390/bios11050154
APA StyleZhang, C., Li, Z., Chen, M., Hu, Z., Wu, L., Zhou, M., & Liang, D. (2021). Cas12a and Lateral Flow Strip-Based Test for Rapid and Ultrasensitive Detection of Spinal Muscular Atrophy. Biosensors, 11(5), 154. https://doi.org/10.3390/bios11050154