Application of Micropore Device for Accurate, Easy, and Rapid Discrimination of Saccharomyces pastorianus from Dekkera spp.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Preparation
2.2. Cell Size Measurement Using Light Microscope
2.3. Micropore Device and Ionic Current Measurement
2.4. Resistive Pulse Analysis and Cell Discrimination
2.5. Discrimination of Decision Boundary and Discrimination Error
2.6. Zeta Potential Measurement
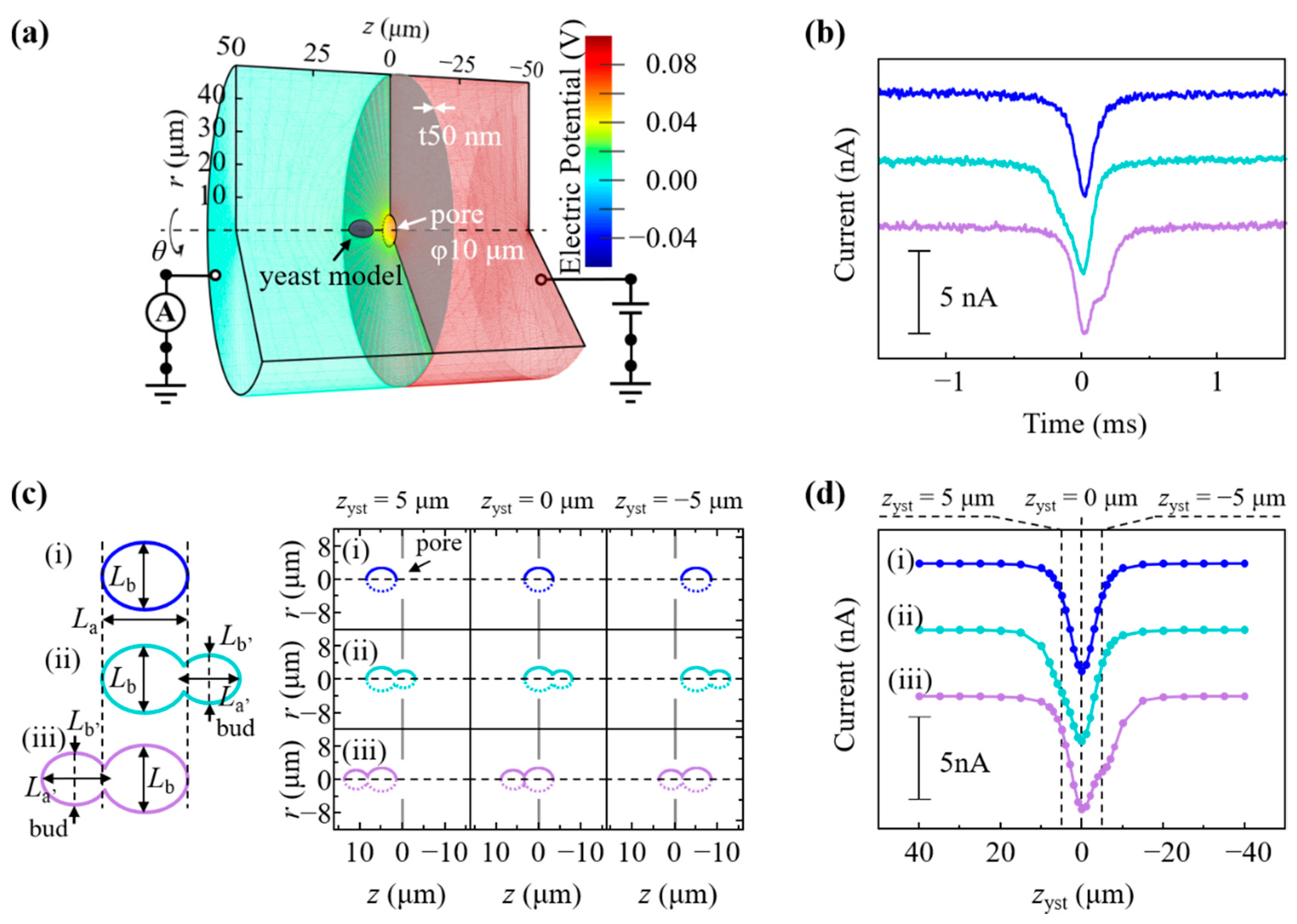
2.7. Multiphysics Simulations of Ionic Current Waveform for a Budding Yeast
3. Results and Discussion
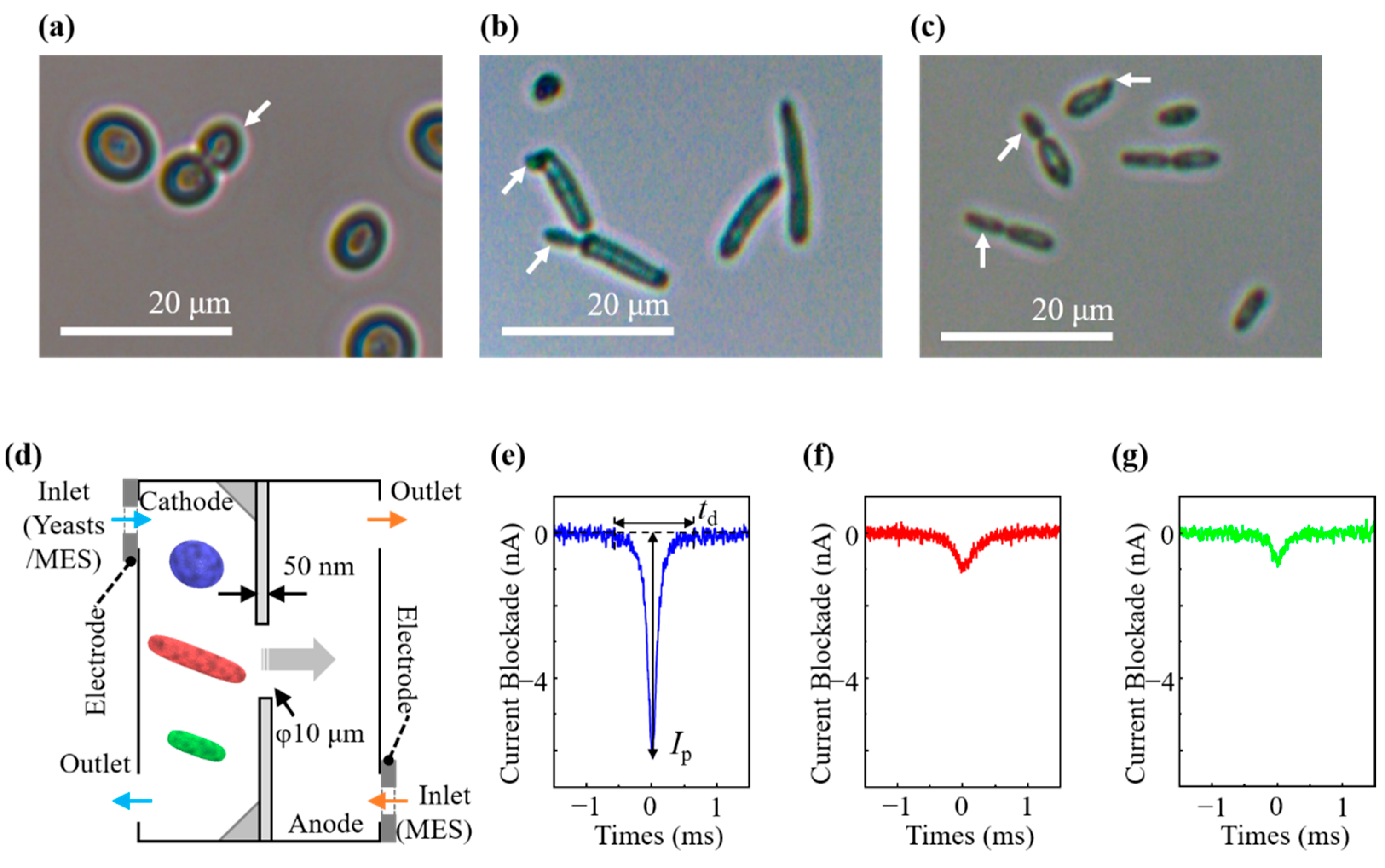
3.1. Morphological Examination by Light Microscopy
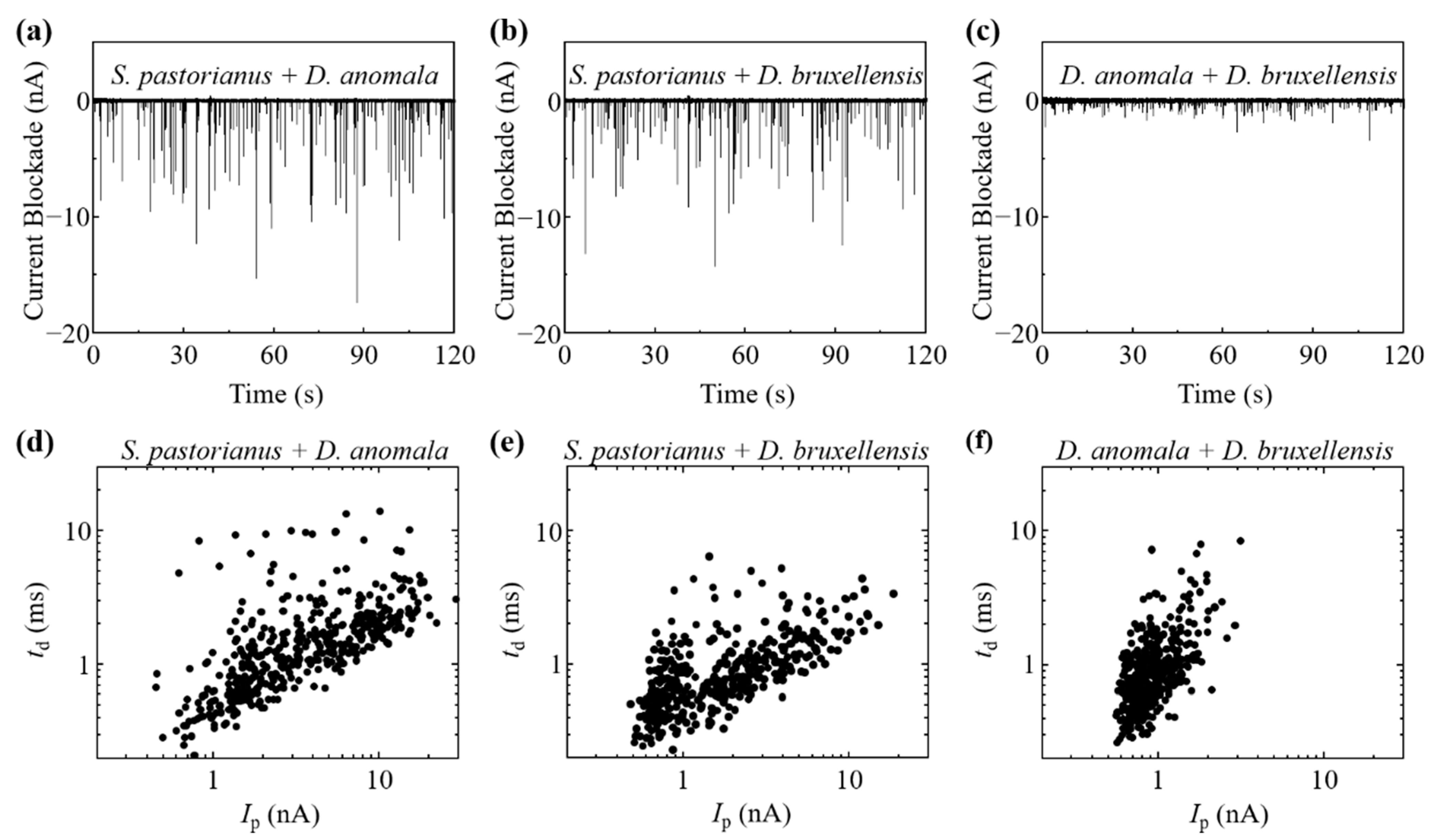
3.2. Ionic Current Measurement of Cells and Cell Discrimination
3.3. Analysis of Budding S. pastorianus
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Song, Y.; Zhang, J.; Li, D. Microfluidic and Nanofluidic Resistive Pulse Sensing: A Review. Micromachines 2017, 8, 204. [Google Scholar] [CrossRef]
- Tsutsui, M.; Yoshida, T.; Yokota, K.; Yasaki, H.; Yasui, T.; Arima, A.; Tonomura, W.; Nagashima, K.; Yanagida, T.; Kaji, N.; et al. Discriminating single-bacterial shape using low-aspect-ratio pores. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Yusko, E.C.; Bruhn, B.R.; Eggenberger, O.; Houghtaling, J.; Rollings, R.C.; Walsh, N.C.; Nandivada, S.; Pindrus, M.; Hall, A.R.; Sept, D.; et al. Real-time shape approximation and fingerprinting of single proteins using a nanopore. Nat. Nanotechnol. 2017, 12, 360–367. [Google Scholar] [CrossRef] [Green Version]
- Ryuzaki, S.; Tsutsui, M.; He, Y.; Yokota, K.; Arima, A.; Morikawa, T.; Taniguchi, M.; Kawai, T. Rapid structural analysis of nanomaterials in aqueous solutions. Nanotechnology 2017, 28, 155501. [Google Scholar] [CrossRef]
- Arjmandi, N.; Van Roy, W.; Lagae, L.; Borghs, G. Measuring the Electric Charge and Zeta Potential of Nanometer-Sized Objects Using Pyramidal-Shaped Nanopores. Anal. Chem. 2012, 84, 8490–8496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Y.; Nguyen, J.; Wang, C.; Sun, Y. Electrical measurement of red blood cell deformability on a microfluidic device. Lab. Chip. 2013, 13, 3275–3283. [Google Scholar] [CrossRef] [PubMed]
- Darvish, A.; Goyal, G.; Aneja, R.; Sundaram, R.V.K.; Lee, K.; Ahn, C.W.; Kim, K.-B.; Vlahovska, P.M.; Kim, M.J. Nanoparticle mechanics: Deformation detection via nanopore resistive pulse sensing. Nanoscale 2016, 8, 14420–14431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wanunu, M. Nanopores: A journey towards DNA sequencing. Phys. Life Rev. 2012, 9, 125–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokota, K.; Hashimoto, M.; Kajimoto, K.; Tanaka, M.; Murayama, S.; Tsutsui, M.; Nakajima, Y.; Taniguchi, M.; Kataoka, M. Effect of Electrolyte Concentration on Cell Sensing by Measuring Ionic Current Waveform through Micropores. Biosensors 2021, 11, 78. [Google Scholar] [CrossRef]
- Fusco, V.; Quero, G.M.; Stea, G.; Morea, M.; Visconti, A. Novel PCR-based identification of Weissella confusa using an AFLP-derived marker. Int. J. Food Microbiol. 2011, 145, 437–443. [Google Scholar] [CrossRef]
- Rungrassamee, W.; Tosukhowong, A.; Klanchui, A.; Maibunkaew, S.; Plengvidhya, V.; Karoonuthaisiri, N. Development of bacteria identification array to detect lactobacilli in Thai fermented sausage. J. Microbiol. Methods 2012, 91, 341–353. [Google Scholar] [CrossRef]
- Shinohara, Y.; Kurniawan, Y.N.; Sakai, H.; Magarifuchi, T.; Suzuki, K. Nanopore based sequencing enables easy and accurate identification of yeasts in breweries. J. Inst. Brew. 2021, 127, 160–166. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Bamforth, C.W. The Microbiology of Malting and Brewing. Microbiol. Mol. Biol. Rev. 2013, 77, 157–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pham, T.; Wimalasena, T.T.; Box, W.G.; Koivuranta, K.; Storgards, E.; Smart, K.A.; Gibson, B.R. Evaluation of ITS PCR and RFLP for Differentiation and Identification of Brewing Yeast and Brewery ‘Wild’ Yeast Contaminants. J. Inst. Brew. 2011, 117, 556–568. [Google Scholar] [CrossRef] [PubMed]
- Kopecká, J.; Němec, M.; Matoulkova, D. Comparison of DNA-based techniques for differentiation of production strains of ale and lager brewing yeast. J. Appl. Microbiol. 2016, 120, 1561–1573. [Google Scholar] [CrossRef]
- Sherman, F. Getting Started with Yeast. Methods Enzymol. 1991, 194, 3–21. [Google Scholar] [PubMed]
- Smeets, R.M.M.; Keyser, U.; Dekker, N.; Dekker, C. Noise in solid-state nanopores. Proc. Natl. Acad. Sci. USA 2008, 105, 417–421. [Google Scholar] [CrossRef] [Green Version]
- Pischel, D.; Buchbinder, J.H.; Sundmacher, K.; Lavrik, I.N.; Flassig, R.J. A guide to automated apoptosis detection: How to make sense of imaging flow cytometry data. PLoS ONE 2018, 13, e0197208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fraley, C.; Raftery, A.E. Model-Based Clustering, Discriminant Analysis, and Density Estimation. J. Am. Stat. Assoc. 2002, 97, 611–631. [Google Scholar] [CrossRef]
- Wernecke, K.-D. Discriminant Analysis. In Wiley Encyclopedia of Clinical Trials; D’Agostino, R.B., Sullivan, L., Massaro, J., Eds.; John Wiley & Sons Inc.: Hoboken, NJ, USA, 2007; pp. 1–19. [Google Scholar]
- Schoch, R.B.; Han, J.; Renaud, P. Transport phenomena in nanofluidics. Rev. Mod. Phys. 2008, 80, 839–883. [Google Scholar] [CrossRef] [Green Version]
- Ng, B.; Barry, P.H. The measurement of ionic conductivities and mobilities of certain less common organic ions needed for junction potential corrections in electrophysiology. J. Neurosci. Methods 1995, 56, 37–41. [Google Scholar] [CrossRef]
- Lide, D.R. CRC Handbook of Chemistry and Physics, 84th ed.; CRC Press Press: Boca Raton, FL, USA, 2003–2004. [Google Scholar]
- Grahame, D.C. The Electrical Double Layer and the Theory of Electrocapillarity. Chem. Rev. 1947, 41, 441–501. [Google Scholar] [CrossRef] [PubMed]
- Hayashida, T.; Yokota, K.; Murayama, S.; Arima, A.; Tsutsui, M.; Taniguchi, M. Tailoring Dielectric Surface Charge via Atomic Layer Thickness. ACS Appl. Mater. Interfaces 2019, 12, 5025–5030. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, H. Approximate analytic expressions for the electrophoretic mobility of spheroidal particles. Electrophoresis 2021, 42, 1003–1009. [Google Scholar] [CrossRef]
- Beckert, S.F.; Domeneghetti, G.; Bond, D. Using historical results obtained in the tensile tests for Type A evaluation of uncertainty. Measurement 2014, 51, 420–428. [Google Scholar] [CrossRef]
- He, Y.; Tsutsui, M.; Fan, C.; Taniguchi, M.; Kawai, T. Controlling DNA Translocation through Gate Modulation of Nanopore Wall Surface Charges. ACS Nano 2011, 5, 5509–5518. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ma, J.; Ni, Z.; Zhang, L.; Hu, G. Effects of access resistance on the resistive-pulse caused by translocating of a nanoparticle through a nanopore. RSC Adv. 2014, 4, 7601–7610. [Google Scholar] [CrossRef] [Green Version]
- Brauer, M.J.; Huttenhower, C.; Airoldi, E.M.; Rosenstein, R.; Matese, J.C.; Gresham, D.; Boer, V.M.; Troyanskaya, O.G.; Botstein, D. Coordination of Growth Rate, Cell Cycle, Stress Response, and Metabolic Activity in Yeast. Mol. Biol. Cell 2008, 19, 352–367. [Google Scholar] [CrossRef] [Green Version]
- Porro, D.; Vai, M.; Vanoni, M.; Alberghina, L.; Hatzis, C. Analysis and modeling of growing budding yeast populations at the single cell level. Cytom. Part A 2009, 75, 114–120. [Google Scholar] [CrossRef]
- Ohnuki, S.; Enomoto, K.; Yoshimoto, H.; Ohya, Y. Dynamic changes in brewing yeast cells in culture revealed by statistical analyses of yeast morphological data. J. Biosci. Bioeng. 2014, 117, 278–284. [Google Scholar] [CrossRef]

| Predicted Classification | |||||
|---|---|---|---|---|---|
| S. pastorianus | D. anomala | D. bruxellensis | Total | ||
| Actual classification | S. pastorianus | 376 372 361 | 22 28 39 | 2 0 0 | 400 400 400 |
| Recall | 92.4 ± 1.9% | ||||
| D. anomala | 19 25 23 | 242 209 203 | 139 166 174 | 400 400 400 | |
| Recall | 54.5 ± 5.3% | ||||
| D. bruxellensis | 10 10 8 | 138 128 155 | 252 262 237 | 400 400 400 | |
| Recall | 62.6 ± 3.1% | ||||
| Discrimination of investigated yeasts based on log10td acquired by RPM. | |||||
| Predicted Classification | |||||
| S. pastorianus | D. anomala | D. bruxellensis | Total | ||
| Actual classification | S. pastorianus | 204 268 209 | 88 15 32 | 108 117 159 | 400 400 400 |
| Recall | 56.8 ± 8.9% | ||||
| D. anomala | 100 178 113 | 106 17 111 | 194 205 176 | 400 400 400 | |
| Recall | 19.5 ± 13.2% | ||||
| D. bruxellensis | 73 97 101 | 80 13 34 | 247 290 265 | 400 400 400 | |
| Recall | 66.8 ± 5.4% | ||||
| Discrimination of investigated yeasts based on log10Ip–log10td acquired by RPM. | |||||
| Predicted Classification | |||||
| S. pastorianus | D. anomala | D. bruxellensis | Total | ||
| Actual classification | S. pastorianus | 383 389 384 | 15 11 14 | 2 0 2 | 400 400 400 |
| Recall | 96.3 ± 0.8% | ||||
| D. anomala | 11 12 5 | 252 247 243 | 137 141 152 | 400 400 400 | |
| Recall | 61.8 ± 1.1% | ||||
| D. bruxellensis | 3 4 1 | 139 139 62 | 258 257 337 | 400 400 400 | |
| Recall | 71.0 ± 11.5% | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yokota, K.; Takeo, A.; Abe, H.; Kurokawa, Y.; Hashimoto, M.; Kajimoto, K.; Tanaka, M.; Murayama, S.; Nakajima, Y.; Taniguchi, M.; et al. Application of Micropore Device for Accurate, Easy, and Rapid Discrimination of Saccharomyces pastorianus from Dekkera spp. Biosensors 2021, 11, 272. https://doi.org/10.3390/bios11080272
Yokota K, Takeo A, Abe H, Kurokawa Y, Hashimoto M, Kajimoto K, Tanaka M, Murayama S, Nakajima Y, Taniguchi M, et al. Application of Micropore Device for Accurate, Easy, and Rapid Discrimination of Saccharomyces pastorianus from Dekkera spp. Biosensors. 2021; 11(8):272. https://doi.org/10.3390/bios11080272
Chicago/Turabian StyleYokota, Kazumichi, Asae Takeo, Hiroko Abe, Yuji Kurokawa, Muneaki Hashimoto, Kazuaki Kajimoto, Masato Tanaka, Sanae Murayama, Yoshihiro Nakajima, Masateru Taniguchi, and et al. 2021. "Application of Micropore Device for Accurate, Easy, and Rapid Discrimination of Saccharomyces pastorianus from Dekkera spp." Biosensors 11, no. 8: 272. https://doi.org/10.3390/bios11080272
APA StyleYokota, K., Takeo, A., Abe, H., Kurokawa, Y., Hashimoto, M., Kajimoto, K., Tanaka, M., Murayama, S., Nakajima, Y., Taniguchi, M., & Kataoka, M. (2021). Application of Micropore Device for Accurate, Easy, and Rapid Discrimination of Saccharomyces pastorianus from Dekkera spp. Biosensors, 11(8), 272. https://doi.org/10.3390/bios11080272






