Recent Advances in Electrochemical Biosensors: Applications, Challenges, and Future Scope
Abstract
:1. Introduction
2. Types of Biosensors
2.1. Catalytic Biosensors
2.2. Affinity Biosensors
3. Electrochemical Biosensors
3.1. Amperometric Biosensor
3.2. Voltammetric Methods
3.3. Impedimetric Biosensor
3.4. Potentiometric Biosensors
4. Applications of Electrochemical Biosensors
4.1. Food Industry
4.2. Medical Sciences
4.3. Defence
4.4. Metabolic Engineering and Plant Biology
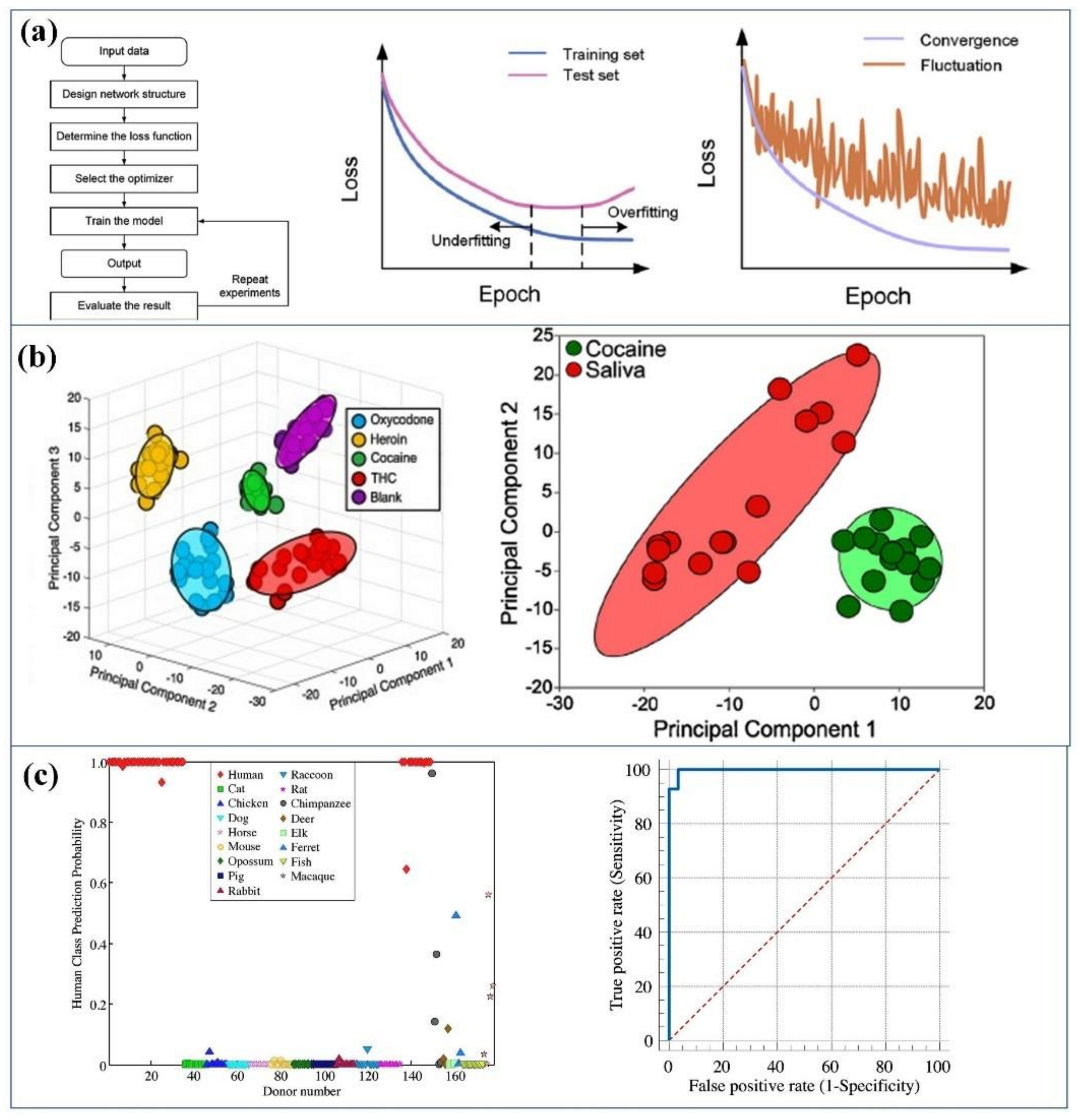
5. Machine Learning for Biosensors
5.1. Improvement in Biosensor by ML
5.2. Various Algorithms in ML
5.3. ML Data Analysis
5.3.1. Support Vector Machine (SVM)
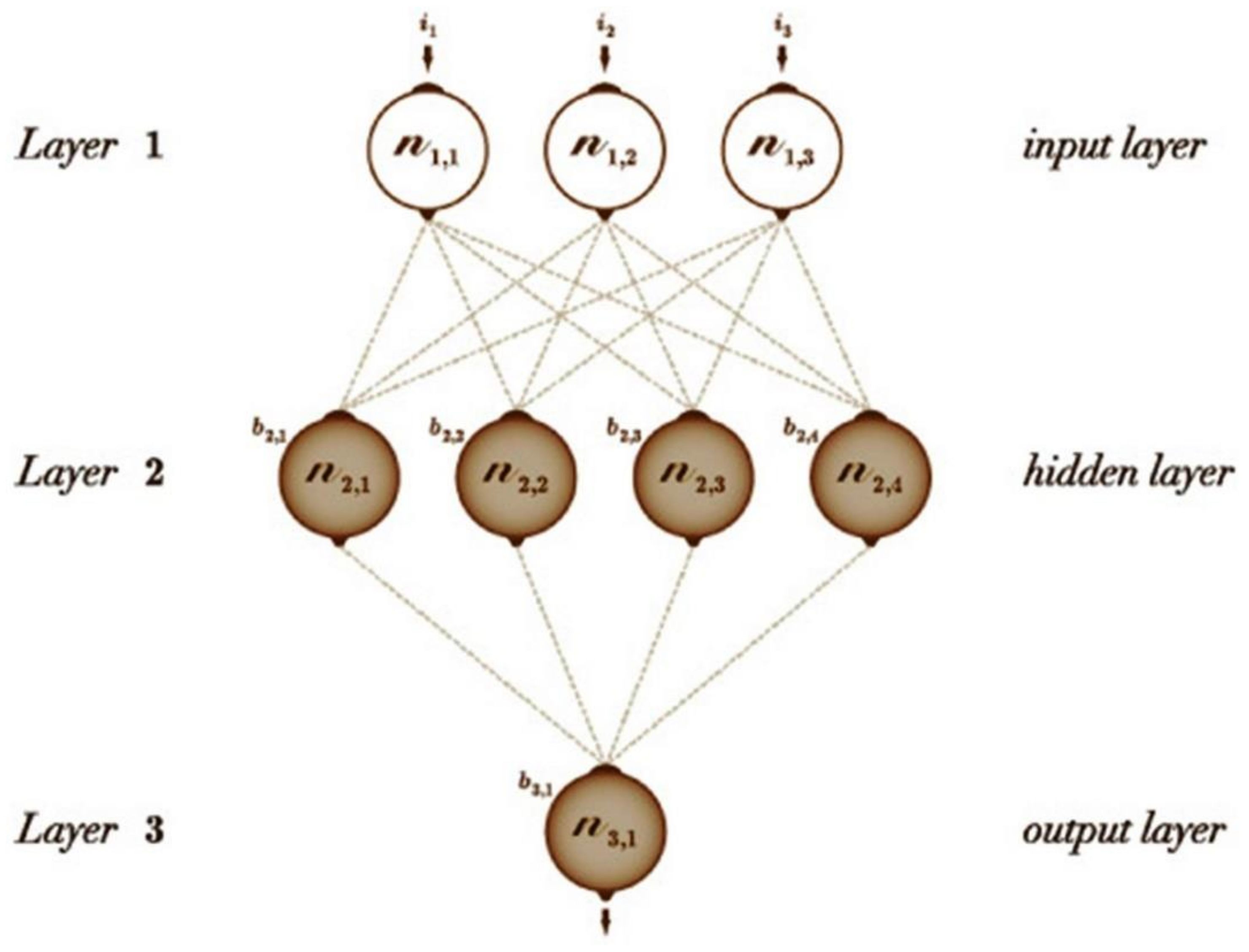
5.3.2. Feedforward Artificial Neural Networks (ANN)
5.3.3. Convolutional Neural Network (CNN)
5.3.4. Recurrent Neural Networks (RNN)
6. Challenges and Solution
6.1. Challenges
- The LOD determines the lowest limit of the analyte that can be detected by a sensor and ideal biosensors must have a very low value of LOD.
- The reproducibility of sensors is very important when it comes to their fabrication and marketing. The results obtained for a particular sensor must be reproducible to all the similar sensors produced, as testing each sensor will not be possible.
- Finally, the most important characteristic of a sensor is its application to real samples. If a sensor is not effective in testing a real sample, it cannot be used in the diagnosis. The real samples that are mostly used for electrochemical biosensors are saliva, blood, urine, sweat, body fluid, tears, etc. The real sample collection is itself a challenge; some factors need to be considered for collecting a real sample for detection.
- The matrix effect in case of electrochemical sensors interferes with the sensor performance. To avoid this matrix effect, the real sample needs to be diluted, but extra dilution may cause deviation from reality. An ideal electrochemical biosensor should sense a real sample without requiring any processing and dilution. Similarly, the samples collected via saliva need dilution before sensing and the pH variation is the problem with the urine samples affecting the peak position. The tear samples due to less complexity have been used for diabetes detection, but the pH variation is again a challenge. Moreover, the concentration of the analytes in the tears produced from irritation and emotion may differ from each other. Moreover, the real samples contain species like protein, fats, etc. that may get adsorbed on the sensor surface and impact the sensitivity and reproducibility of an electrochemical biosensor. The researchers are looking for advanced new materials and techniques (active and passive methods) to address this issue. In the active method, shear forces are produced that prevent the adhesion of the extra species on the sensor surface, whereas in passive methods, polymers are used to make the surface hydrophilic, thus preventing proteins from adsorption. The biosensors developed must be stable under extreme environmental conditions and hence, the stability of the electrochemical biosensors is very important.
6.2. Solutions
- Using nanomaterials might address the stability issue in some cases, but some nanomaterials seem to aggregate and reduce stability.
- The miniaturization of the electrochemical biosensors and using cheap materials in their fabrication is another step that needs to be taken in making them cheap.
- Micro-nano fabrication techniques are effective in reducing the size of the electrochemical biosensor. The smaller biosensors would be easy to use and dispose of, can be transported easily, and their application in extreme conditions would involve fewer efforts.
- The electrochemical biosensors have mostly been confined to the research labs. There needs to be a collaboration between clinics, hospitals, and research labs so that they can be tested in real-life circumstances, which will help in evaluating their performance. Multidisciplinary approach is important for further widespread use and commercialization of biosensors.
- On a global scale, bacterial diseases are responsible for the greatest number of deaths and illnesses. The electrochemical biosensors can prove effective in sensing these bacterial infections at early stages. These biosensors would also be very useful in detecting new pathogens in the water sources. However, huge efforts on technical and scientific ground will be required to make them more viable. The designing and fabrication process needs to be made more cost-effective. Moreover, the enzymatic electrochemical biosensors are used commonly in the research, but their stability and modification remain a concern. Another challenge is the storage of enzymes.
- The integration of electrochemical biosensors with POC devices would be a great initiative for application in clinics. Such biosensors would not be affected by the interference species and can detect any concentration of the analyte. In addition to this, nano technology will help in improving the LOD and sensitivity of the electrochemical biosensors.
7. Future Outlook
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Thevenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical biosensors: Recommended definitions and classification. Pure Appl. Chem. 1999, 71, 2333–2348. [Google Scholar] [CrossRef] [Green Version]
- Khosla, A. Micropatternable Multifunctional Nanocomposite Polymers for Flexible Soft MEMS Applications. Ph.D. Thesis, Applied Science: School of Engineering Science, Simon Fraser University, Burnaby, BC, Canada, 2011. [Google Scholar]
- Ahmad, R.; Khan, M.; Tripathy, N.; Khan, M.I.R.; Khosla, A. Hydrothermally synthesiz0ed nickel oxide nanosheets for non-enzymatic electrochemical glucose detection. J. Electrochem. Soc. 2020, 167, 107504. [Google Scholar] [CrossRef]
- Sharma, A.; Ahmed, A.; Singh, A.; Oruganti, S.; Khosla, A.; Arya, S. Recent advances in tin oxide nanomaterials as electrochemical/chemiresistive sensors. J. Electrochem. Soc. 2021, 168, 027505. [Google Scholar] [CrossRef]
- Chullasat, K.; Kanatharana, P.; Limbut, W.; Numnuam, A.; Thavarungkul, P. Ultra trace analysis of small molecule by label-free impedi-metric immunosensor using multilayer modified electrode. Biosens. Bioelectron. 2011, 26, 4571–4578. [Google Scholar] [CrossRef]
- Canbaz, M.Ç.; Şimşek, C.S.; Sezgintürk, M.K. Electrochemical biosensor based on self-assembled monolayersmodifi ed with gold nanoparticles for detection of HER-3. Anal Chim. Acta. 2014, 814, 31–38. [Google Scholar] [CrossRef]
- Kim, Y.H.; Park, J.S.; Jung, H.I. An impedimetric biosensor for real-time monitoring of bacterial growth in a microbial fermentor. Sens. Actuators B Chem. 2009, 138, 270–277. [Google Scholar] [CrossRef]
- Ahmad, R.; Mahmoudi, T.; Ahn, M.S.; Hahn, Y.B. Recent advances in nanowires-based field-effect transistors for biological sensor applications. Biosens. Bioelectron. 2018, 100, 312–325. [Google Scholar] [CrossRef]
- Kumar, S.; Pavelyev, V.; Tripathi, N.; Platonov, V.; Sharma, P.; Ahmad, R.; Mishra, P.; Khosla, A. Review—Recent Advances in the Development of Carbon Nanotubes Based Flexible Sensors. J. Electrochem. Soc. 2020, 167, 047506. [Google Scholar] [CrossRef]
- Sheng, Y.; Zhang, T.; Zhang, S.; Johnston, M.; Zheng, X.; Shan, Y.; Liu, T.; Huang, Z.; Qian, F.; Xie, Z. A CRISPR/Cas13a-powered catalytic electrochemical biosensor for successive and highly sensitive RNA diagnostics. Biosens. Bioelectron. 2021, 178, 113027. [Google Scholar] [CrossRef] [PubMed]
- Srisomwat, C.; Teengam, P.; Chuaypen, N.; Tangkijvanich, P.; Vilaivan, T.; Chailapakul, O. Pop-up paper electrochemical device for label-free hepatitis B virus DNA detection. Sens. Actuators B Chem. 2020, 316, 128077. [Google Scholar] [CrossRef]
- Carceller, J.M.; Mifsud, M.; Climent, M.J.; Iborra, S.; Corma, A. Production of chiral alcohols from racemic mixtures by integrated heterogeneous chemoenzymatic catalysis in fixed bed continuous operation. Green Chem. 2020, 22, 2767–2777. [Google Scholar] [CrossRef]
- Asefpour Vakilian, K.; Massah, J. A portable nitrate biosensing device using electrochemistry and spectroscopy. IEEE Sens. J. 2018, 18, 3080–3089. [Google Scholar] [CrossRef]
- Hollon, T.C.; Pandian, B.; Adapa, A.R.; Urias, E.; Save, A.V.; Khalsa, S.S.S.; Eichberg, D.G.; D’Amico, R.S.; Farooq, Z.U.; Lewis, S. Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nat. Med. 2020, 26, 52. [Google Scholar] [CrossRef]
- Moon, G.; Choi, J.-R.; Lee, C.; Oh, Y.; Kim, K.; Kim, D. Machine learning-based design of meta-plasmonic biosensors with negative index metamaterials. Biosens. Bioelectron. 2020, 164, 112335. [Google Scholar] [CrossRef] [PubMed]
- Cinti, S.; Arduini, F.; Moscone, D.; Palleschi, G.; Gonzalez-Macia, L.; Killard, A.J. Cholesterol biosensor based on inkjet-printed Prussian blue nanoparticle-modified screen-printed electrodes. Sens. Actuators B Chem. 2015, 221, 187–190. [Google Scholar] [CrossRef]
- Sannini, A.; Albanese, D.; Malvano, F.; Crescitelli, A.; Di Matteo, M. An amperometric biosensor for the determination of lactic acid during malolactic fermentation. Chem. Eng. Trans. 2015, 44, 283–288. [Google Scholar]
- Boffi, A.; Favero, G.; Federico, R.; Macone, A.; Antiochia, R.; Tortolini, C.B.; Sanzó, G.; Mazzei, F. Amine oxidase-based biosensors for spermine and spermidine determination. Anal. Bioanal. Chem. 2015, 407, 1131–1137. [Google Scholar] [CrossRef]
- Ciriello, R.; Cataldi, T.R.I.; Crispo, F.; Guerrieri, A. Quantification of l-lysine in cheese by a novel amperometric biosensor. Food Chem. 2015, 169, 13–19. [Google Scholar] [CrossRef]
- Strambini, L.M.; Longo, A.; Scarano, S.; Prescimone, T.; Palchetti, I.; Minunni, M.; Giannessi, D.; Barillaro, G. Self-powered microneedle-based biosensors for pain-free high-accuracy measurement of glycaemia in interstitial fluid. Biosens. Bioelectron. 2015, 66, 162–168. [Google Scholar] [CrossRef]
- Habtamu, H.B.; Ugo, P. Miniaturized Enzymatic Biosensor via Biofunctionalization of the Insulator of Nanoelectrode Ensembles. Electroanalysis 2015, 27, 2187–2193. [Google Scholar] [CrossRef]
- Malvano, F.; Albanese, D.; Sannini, A.; Crescitelli, A.; Pilloton, R.; Di Matteo, M. Ethanol content in must grape by alcohol dehydrogenase biosensors based on doped Polyaniline modified screen printed electrodes. Chem. Eng. Trans. 2015, 43, 37–42. [Google Scholar]
- Tomassetti, M.; Serone, M.; Angeloni, R.; Campanella, L.; Mazzone, E. Amperometric enzyme sensor to check the total antioxidant capacity of several mixed berries. Comparison with two other spectrophotometric and fluorimetric methods. Sensors 2015, 15, 3435–3452. [Google Scholar] [CrossRef]
- Barberis, A.; Spissu, Y.; Fadda, A.; Azara, E.; Bazzu, G.; Marceddu, S.; Angioni, A.; Sanna, D.; Schirra, M.; Serra, P.A. Simultaneous amperometric detection of ascorbic acid and antioxidant capacity in orange, blueberry and kiwi juice, by a telemetric system coupled with a fullerene- or nanotubes-modified ascorbate subtractive biosensor. Biosens. Bioelectron. 2015, 67, 214–223. [Google Scholar] [CrossRef]
- Tortolini, C.; Bollella, P.; Antiochia, R.; Favero, G.; Mazzei, F. Inhibition-based biosensor for atrazine detection. Sens. Actuators B Chem. 2016, 224, 552–558. [Google Scholar] [CrossRef]
- Grattieri, M.; Babanova, S.; Santoro, C.; Guerrini, E.; Trasatti, S.P.; Cristiani, P.; Bestetti, M.; Atanassov, P. Enzymatic Oxygen Microsensor Based on Bilirubin Oxidase Applied to Microbial Fuel Cells Analysis. Electroanalysis 2015, 27, 327–335. [Google Scholar] [CrossRef]
- Silletti, S.; Rodio, G.; Pezzotti, G.; Turemis, M.; Dragone, R.; Frazzoli, C.; Giardi, M.T. An optical biosensor based on a multiarray of enzymes for monitoring a large set of chemical classes in milk. Sens. Actuators B Chem. 2015, 215, 607–617. [Google Scholar] [CrossRef]
- Chiavaioli, F.; Biswas, P.; Trono, C.; Jana, S.; Bandyopadhyay, S.; Basumallick, N.; Giannetti, A.; Tombelli, S.; Bera, S.; Mallick, A.; et al. Sol-Gel-Based Titania-Silica Thin Film Overlay for Long Period Fiber Grating-Based Biosensors. Anal. Chem. 2015, 87, 12024–12031. [Google Scholar] [CrossRef] [PubMed]
- Tardivo, M.; Toffoli, V.; Fracasso, G.; Borin, D.; Dal Zilio, S.; Colusso, A.; Carrato, S.; Scoles, G.; Meneghetti, M.; Colombatti, M.; et al. Parallel optical read-out of micromechanical pillars applied to prostate specific membrane antigen detection. Biosens. Bioelectron. 2015, 72, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Casalini, S.; Dumitru, A.C.; Leonardi, F.; Bortolotti, C.A.; Herruzo, E.T.; Campana, A.; De Oliveira, R.F.; Cramer, T.; Garcia, R.; Biscarini, F. Multiscale sensing of antibody-antigen interactions by organic transistors and single-molecule force spectroscopy. ACS Nano 2015, 9, 5051–5062. [Google Scholar] [CrossRef]
- Zhao, L.; Han, H.; Ma, Z. Improved screen-printed carbon electrode for multiplexed label-free amperometric immuniosensor: Addressing its conductivity and reproducibility challenges. Biosens. Bioelectron. 2018, 101, 304–310. [Google Scholar] [CrossRef]
- Castillo, G.; Spinella, K.; Poturnayová, A.; Šnejdárková, M.; Mosiello, L.; Hianik, T. Detection of aflatoxin B1 by aptamer-based biosensor using PAMAM dendrimers as immobilization platform. Food Control 2015, 52, 9–18. [Google Scholar] [CrossRef]
- Scarano, S.; Dausse, E.; Crispo, F.; Toulmé, J.-J.; Minunni, M. Design of a dual aptamer-based recognition strategy for human matrix metalloproteinase 9 protein by piezoelectric biosensors. Anal. Chim. Acta 2015, 897, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tintoré, M.; Mazzini, S.; Polito, L.; Marelli, M.; Latorre, A.; Somoza, Á.; Aviñó, A.; Fàbrega, C.; Eritja, R. Gold-coated superparamagnetic nanoparticles for single methyl discrimination in DNA aptamers. Int. J. Mol. Sci. 2015, 16, 27625–27639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravalli, A.; da Rocha, C.G.; Yamanaka, H.; Marrazza, G. A label-free electrochemical affisensor for cancer marker detection: The case of HER2. Bioelectrochemistry 2015, 106, 268–275. [Google Scholar] [CrossRef]
- Mulla, Y.; Tuccori, E.; Magliulo, M.; Lattanzi, G.; Palazzo, G.; Persaud, K.; Torsi, L. Capacitance-modulated transistors detects odorant binding protein chiral interactions. Nat Comm. 2015, 6, 6010. [Google Scholar] [CrossRef] [Green Version]
- Di Pietrantonio, F.; Benetti, M.; Cannatà, D.; Verona, E.; Palla-Papavlu, A.; Fernández-Pradas, J.M.; Serra, P.; Staiano, M.; Varriale, A.; D’Auria, S. A surface acoustic wave bio-electronic nose for detection of volatile odorant molecules. Biosens. Bioelectron. 2015, 67, 516–523. [Google Scholar] [CrossRef]
- Cennamo, N.; Giovanni, S.D.; Varriale, A.; Staiano, M.; Di Pietrantonio, F.; Notargiacomo, A.; Zeni, L.; D’Auria, S. Easy to use plastic optical fiber-based biosensor for detection of butanal. PLoS ONE 2015, 10, e0116770. [Google Scholar] [CrossRef] [Green Version]
- Compagnone, D.; Faieta, M.; Pizzoni, D.; di Natale, C.; Paolesse, R.; van Caelenberg, T.; Beheydtd, B.; Pittia, P. Quartz crystal microbalance gas sensor arrays for the quality control of chocolate. Sens. Actuators B Chem. 2015, 207, 1114–1120. [Google Scholar] [CrossRef]
- Zuccaro, L.; Tesauro, C.; Kurkina, T.; Fiorani, P.; Yu, H.K.; Knudsen, B.R.; Kern, K.; Desideri, A.; Balasubramanian, K. Real-Time Label-Free Direct Electronic Monitoring of Topoisomerase Enzyme Binding Kinetics on Graphene. ACS Nano 2015, 9, 11166–11176. [Google Scholar] [CrossRef] [PubMed]
- Mariani, S.; Scarano, S.; Spadavecchia, J.; Minunni, M. A reusable optical biosensor for the ultrasensitive and selective detection of unamplified human genomic DNA with gold nanostars. Biosens. Bioelectron. 2015, 74, 981–988. [Google Scholar] [CrossRef]
- Lacina, K.; Sopoušek, J.; Čunderlová, V.; Hlaváček, A.; Václavek, T.; Lacinová, V. Biosensing based on electrochemical impedance spectroscopy: Influence of the often-ignored molecular charge. Electrochem. Commun. 2018, 93, 183–186. [Google Scholar] [CrossRef]
- Hamidi-Asl, E.; Raoof, J.B.; Hejazi, M.S.; Sharifi, S.; Golabi, S.M.; Palchetti, I.; Mascini, M. Genosensor for Point Mutation Detection of P53 Gene PCR Product Using Magnetic Particles. Electroanalysis 2015, 27, 1378–1386. [Google Scholar] [CrossRef]
- Wu, L.; Lu, X.; Fu, X.; Wu, L.; Liu, H. Gold nanoparticles dotted reduction Graphene oxide Nanocomposite based electrochemical Aptasensor for selective, rapid, sensitive and congener-specific PCB77 detection. Sci. Rep. 2017, 7, 1–7. [Google Scholar] [CrossRef]
- Khosla, A.; Shah, S.; Shiblee, M.N.I.; Mir, S.H.; Nagahara, L.A.; Thundat, T.; Shekar, P.K.; Kawakami, M.; Furukawa, H. Carbon fiber doped thermosetting elastomer for flexible sensors: Physical properties and microfabrication. Sci. Rep. 2018, 8, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, R.; Khan, M.; Mishra, P.; Jahan, N.; Ahsan, M.A.; Ahmad, I.; Khan, M.R.; Watanabe, Y.; Syed, M.A.; Furukawa, H.; et al. Engineered hierarchical CuO nanoleaves based electrochemical nonenzymatic biosensor for glucose detection. J. Electrochem. Soc. 2021, 168, 017501. [Google Scholar] [CrossRef]
- Clark, L.C., Jr.; Lyons, C. Electrode systems for continuous monitoring in cardiovascular surgery. Ann. N. Y. Acad. Sci. 1962, 102, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Gupta, J.; Arya, S.; Verma, S.; Singh, A.; Sharma, A.; Singh, B.; Sharma, R. Performance of template-assisted electrodeposited Copper/Cobalt bilayered nanowires as an efficient glucose and Uric acid senor. Mater. Chem. Phys. 2019, 238, 121969. [Google Scholar] [CrossRef]
- Khan, M.; Nagal, V.; Nakate, U.T.; Khan, M.R.; Khosla, A.; Ahmad, R. Engineered CuO Nanofibers with Boosted Non-Enzymatic Glucose Sensing Performance. J. Electrochem. Soc. 2021, 168, 067507. [Google Scholar] [CrossRef]
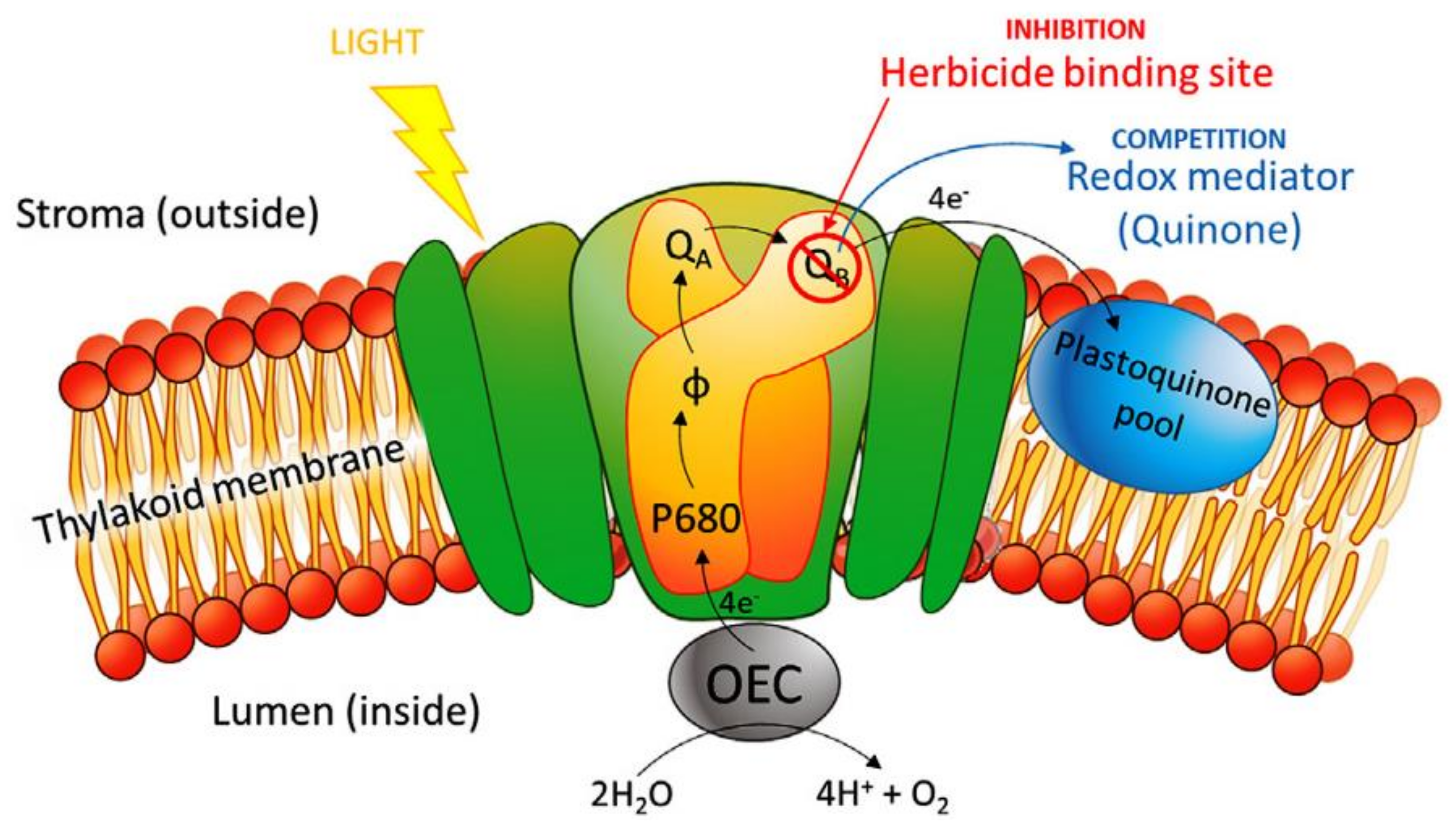
- Tucci, M.; Grattieri, M.; Schievano, A.; Cristiani, P.; Minteer, S.D. Microbial amperometric biosensor for online herbicide detection: Photocurrent inhibition of Anabaena variabilis. Electrochim. Acta 2019, 302, 102–108. [Google Scholar] [CrossRef]
- Chen, X.; Wu, G.; Cai, Z.; Oyama, M.; Chen, X. Advances in enzyme-free electrochemical sensors for hydrogen peroxide, glucose, and uric acid. Microchim. Acta 2014, 181, 689–705. [Google Scholar] [CrossRef]
- Castillo-Ortega, M.M.; Rodriguez, D.E.; Encinas, J.C.; Plascencia, M.; Mendez-Velarde, F.A.; Olayo, R. Conductometric uric acid and urea biosensor prepared from electroconductive polyaniline–poly (n-butyl methacrylate) composites. Sens. Actuators B: Chem. 2002, 85, 19–25. [Google Scholar] [CrossRef]
- Sekhar, P.K.; Moore, Z.; Aravamudhan, S.; Khosla, A. A new low-temperature electrochemical hydrocarbon and NOx sensor. Sensors 2017, 17, 2759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, A.; Khosla, A.; Drewbrook, C.; Gray, B.L. Fabrication and testing of thermally responsive hydrogel-based actuators using polymer heater elements for flexible microvalves. In Microfluidics, BioMEMS, and Medical Microsystems IX; International Society for Optics and Photonics: San Francisco, CA, USA, 2011; Volume 7929, p. 79290G-1. [Google Scholar]
- Ahmad, R.; Tripathy, N.; Kim, S.H.; Umar, A.; Al-Hajry, A.; Hahn, Y.B. High performance cholesterol sensor based on ZnO nanotubes grown on Si/Ag electrodes. Electrochem. Commun. 2014, 38, 4–7. [Google Scholar] [CrossRef]
- Ahmad, R.; Tripathy, N.; Hahn, Y.B. High-performance cholesterol sensor based on the solution-gated field effect transistor fabricated with ZnO nanorods. Biosens. Bioelectron. 2013, 45, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Tan, X.; Chen, S.; Yuan, R.; Hu, F.; Yuan, D.; Xiang, Y. Highly-sensitive cholesterol biosensor based on platinum-gold hybrid functionalized ZnO nanorods. Talanta 2012, 94, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Batra, N.; Tomar, M.; Gupta, V. ZnO-CuO composite matrix based reagentless biosensor for detection of total cholesterol. Biosens. Bioelectron. 2015, 67, 263–271. [Google Scholar] [CrossRef]
- San Fang, C.; Oh, K.H.; Oh, A.; Lee, K.; Park, S.; Kim, S.; Yang, H. An ultrasensitive and incubation-free electrochemicalimmunosensor using a gold-nanocatalyst label mediating outer-spherereaction-philic and inner-sphere-reaction-philic species. Chem. Commun. 2016, 52, 5884–5887. [Google Scholar] [CrossRef]
- Cai, X.; Weng, S.; Guo, R.; Lin, L.; Chen, W.; Zheng, Z.; Lin, X. Ratiometric electrochemical immunoassay based on internalreference value for reproducible and sensitive detection of tumor marker. Biosens. Bioelectron. 2016, 81, 173–180. [Google Scholar] [CrossRef]
- Zhang, H.; Ma, L.; Li, P.; Zheng, J. A novel electrochemical immunosensor based onnonenzymatic Ag@au-Fe3O4 nanoelectrocatalyst for protein biomarkerdetection. Biosens. Bioelectron. 2016, 85, 343–350. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Zhu, Y.; Tang, L.; Yang, X.; Li, C. Biosensor based on self-assembling glucose oxidase and Dendrimer-encapsulated Pt nanoparticles on carbon nanotubes for glucosedetection. Electroanalysis 2007, 19, 717–722. [Google Scholar] [CrossRef]
- Dinh, D.; Hui, K.S.; Cho, Y.; Zhou, W.; Hong, X.; Chun, H.-H. Green synthesis of high conductivity silver nanoparticle-reduced graphene oxide composite films. Appl. Surf. Sci. 2014, 298, 62–67. [Google Scholar] [CrossRef]
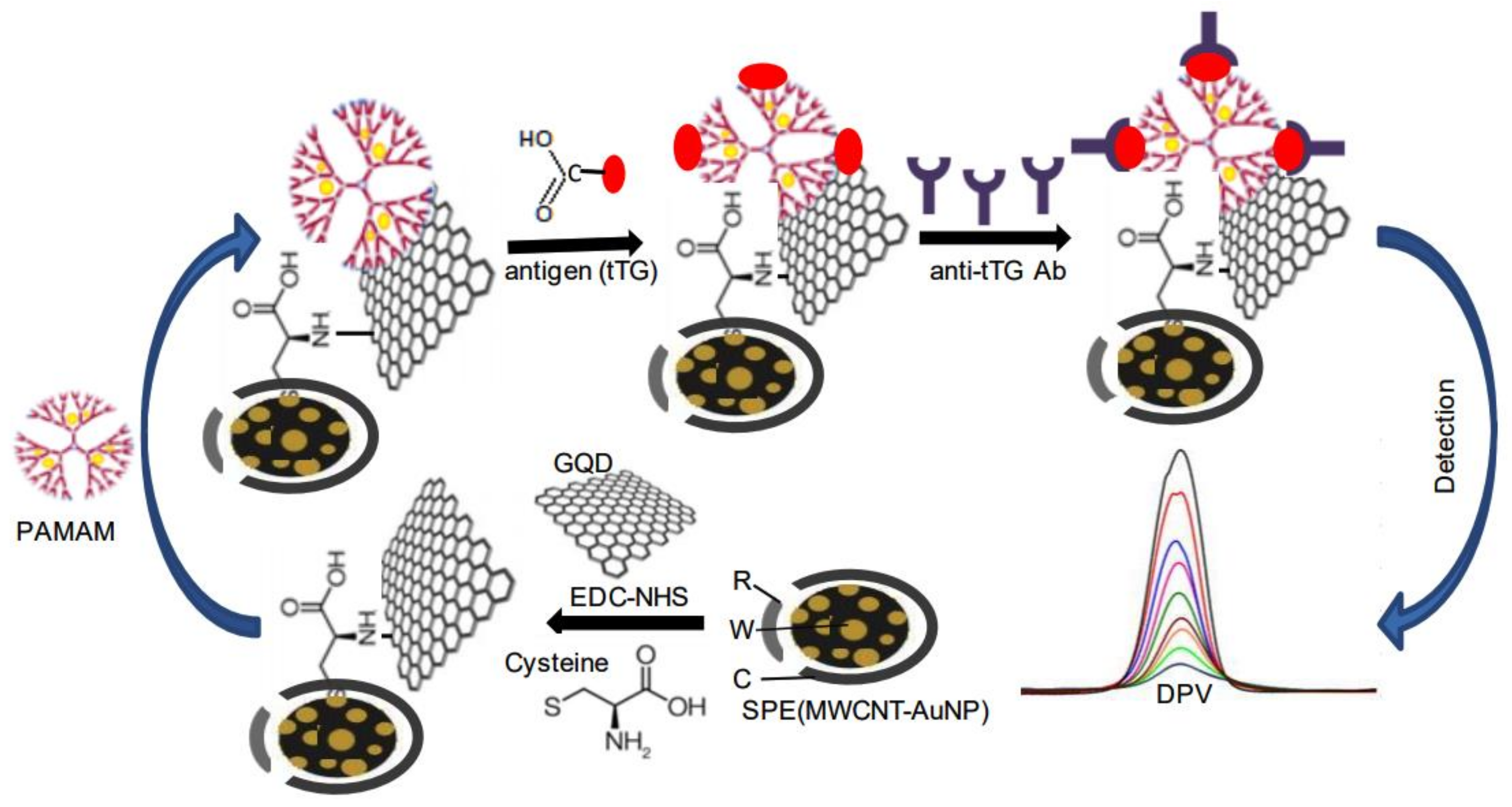
- Gupta, S.; Kaushal, A.; Kumar, A.; Kumar, D. Ultrasensitive transglutaminase based nanosensor for early detection of celiac dis- ease in human. Int. J. Biol. Macromol. 2017, 105, 905–911. [Google Scholar] [CrossRef] [PubMed]
- Neves, M.M.; González-García, M.B.; Nouws, H.P.; Costa-García, A. Celiac disease detection using a transglutaminase electro-chemical immunosensor fabricated on nanohybrid screen-printed carbon electrodes. Biosens. Bioelectron. 2012, 31, 95–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hianik, T.; Ostatná, V.; Zajacová, Z.; Stoikova, E.; Evtugyn, G.; Zajacova, Z.; Ostatna, V. Detection of aptamer—protein interactions using QCM and electrochemical indicator methods. Bioorg. Med. Chem. 2005, 15, 291–295. [Google Scholar] [CrossRef]
- Arya, S.; Singh, A.; Kour, R. Comparative study of CuO, CuO@ Ag and CuO@ Ag: La nanoparticles for their photosensing properties. Mater. Res. Express 2019, 6, 116313. [Google Scholar] [CrossRef]
- Velusamy, V.; Arshak, K.; Korostynska, O.; Oliwa, K.; Adley, C. An overview of foodborne pathogen detection: In the perspective of biosensors. Biotechnol. Adv. 2010, 28, 232–254. [Google Scholar] [CrossRef]
- Hargunani, S.P.; Sonekar, R.P.; Singh, A.; Khosla, A.; Arya, S. Structural and spectral studies of Ce3+ doped Sr3Y (BO3) 3 nano phosphors prepared by combustion synthesis. Mater. Technol. 2020, 1–12. [Google Scholar] [CrossRef]
- Ronkainen, N.J.; Halsall, H.B.; Heineman, W.R. Electrochemical biosensors. Chem. Soc. Rev. 2010, 39, 1747–1763. [Google Scholar] [CrossRef]
- Marks, R.S.; Abdulhalim, I. Nanomaterials for Water Management: Signal Amplification for Biosensing from Nanostructures; CRC Press: Boca Raton, FL, USA, 2015; Volume 4. [Google Scholar]
- Ferreira, A.; Fugivara, C.; Yamanaka, H.; Benedetti, A. Preparation and Characterization of Imunosensors for Disease Diagnosis. Biosens. Health Environ. Biosecurity 2011, 540. [Google Scholar] [CrossRef] [Green Version]
- Ly, S.Y.; Yoo, H.S.; Choa, S.H. Diagnosis of Helicobacter pylori bacterial infections using a voltammetric biosensor. J. Microbiol. Methods 2011, 87, 44–48. [Google Scholar] [CrossRef]
- Maalouf, R.; Chebib, H.; Saïkali, Y.; Vittori, O.; Sigaud, M.; Jaffrezic-Renault, N. Amperometric and impedimetric characterization of a glu-tamate biosensor based on Nafion and a methyl viologen modifi ed glassy carbon electrode. Biosens. Bioelectron. 2007, 22, 2682–2688. [Google Scholar] [CrossRef]
- Wang, J. Analytical Electrochemistry Hoboken; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2006. [Google Scholar]
- Elshafey, R.; Tavares, A.C.; Siaj, M.; Zouro, M. Electrochemical impedance immunosensor based on gold nanoparticles—protein G for the detection of cancer marker epidermal growth factor receptor in human plasma and brain tissue. Biosens. Bioelectron. 2013, 50, 143–149. [Google Scholar] [CrossRef]
- Helali, S.; Martelet, C.; Abdelghani, A.; Maaref, M.A.; Jaffrezic-Renault, N. A disposable immunomagnetic electrochemical sensor based on functionalised magnetic beads on gold surface for the detection of atrazine. Electrochim. Acta 2006, 51, 5182–5186. [Google Scholar] [CrossRef]
- Seven, B.; Bourourou, M.; Elouarzaki, K.; Constant, J.F.; Gondran, C.; Holzinger, M.; Cosnier, S.; Timur, S. Impedimetric biosensor for cancer cell detection. Electrochem. Commun. 2013, 37, 36–39. [Google Scholar] [CrossRef]
- Chowdhury, A.D.; De, A.; Chaudhuri, C.R.; Bandyopadhyay, K.; Sen, P. Label free polyaniline based impedimetric biosensor for detection of E. coli O157: H7 Bacteria. Sens. Actuators B Chem. 2012, 171, 916–923. [Google Scholar] [CrossRef]
- Rushworth, J.V.; Ahmed, A.; Griffiths, H.H.; Pollock, N.M.; Hooper, N.M.; Millner, P.A. A label-free electrical impedimetric biosensor for the specific detection of Alzheimer’s amyloid-beta oligomers. Biosens. Bioelectron. 2014, 56, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Lin, Y.; Yu, P.; Su, L.; Mao, L. Label-free and sequence-specific DNA detection down to apicomolar level with carbon nanotubes as support for probe DNA. Anal. Chim. Acta 2009, 650, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Shirsat, M.; Too, C.O.; Wallace, G. Amperometric glucose biosensor on layer bylayer assembled carbon nanotube and Polypyrrole multilayer film. Electroanalysis 2007, 20, 150–156. [Google Scholar] [CrossRef]
- Koncki, R. Recent developments in potentiometric biosensors for biomedical analysis. Anal. Chim. Acta 2007, 599, 7–15. [Google Scholar] [CrossRef]
- Bandodkar, A.J.; Molinnus, D.; Mirza, O.; Guinovart, T.; Windmiller, J.R.; Valdes-Ramirez, G.; Andrade, F.J.; Schoening, M.J.; Wang, J. Epidermal tattoo potentiometric sodium sensors with wireless signal transduction for continuous non-invasive sweat monitoring. Biosens. Bioelectron. 2014, 54, 603–609. [Google Scholar] [CrossRef]
- Papp, S.; Bojtar, M.; Gyurcsanyi, R.E.; Lindfors, T. Potential reproducibility of potassium-selective electrodes having perfluorinatedalkanoate side chain functionalized poly(3,4-ethylenedioxytiophene) as a hydrophobic solid contact. Anal. Chem. 2019, 91, 9111–9118. [Google Scholar] [CrossRef]
- Parrilla, M.; Cuartero, M.; Sanchez, S.P.; Rajabi, M.; Roxhed, N.; Niklaus, F.; Crespo, G.A. Wearable all-solid-state potentiometric microneedle patch for intradermal potassium detection. Anal. Chem. 2019, 91, 1578–1586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, R.K.; Barfidokht, A.; Karajic, A.; Sempionatto, J.R.; Wang, J.; Wang, J. Wearable potentiometric tattoo biosensor for on-body detection of G-type nerve agents simulants. Sens. Actuators B Chem. 2018, 273, 966–972. [Google Scholar] [CrossRef]
- Cánovas, R.; Blondeau, P.; Andrade, F.J. Modulating the mixed potential for developing biosensors: Direct potentiometric determination of glucose in whole, undiluted blood. Biosens. Bioelectron. 2020, 163, 112302. [Google Scholar] [CrossRef]
- Jakhar, S.; Pundir, C.S. Preparation, characterization and application of urease nanoparticles for construction of an improved potentiometric urea biosensor. Biosens. Bioelectron. 2018, 100, 242–250. [Google Scholar] [CrossRef]
- Scognamiglio, V.; Arduini, F.; Palleschi, G.; Rea, G. Bio sensing technology for sustainable food safety. Trends Anal. Chem. 2014, 62, 1–10. [Google Scholar] [CrossRef]
- Ghasemi-Varnamkhasti, M.; Rodriguez-Mendez, M.L.; Mohtasebi, S.S.; Apetrei, C.; Lozano, J.; Ahmadi, H.; Razavi, S.H.; de Saja, J.A. Monitoring the aging of beers using a bioelectronic tongue. Food Control. 2011, 25, 216–224. [Google Scholar] [CrossRef]
- Arora, P.; Sindhu, A.; Dilbaghi, N.; Chaudhury, A. Biosensors as innovative tools for the detection of food borne pathogens. Biosens. Bioelectron. 2011, 28, 1–12. [Google Scholar] [CrossRef]
- Ercole, C.; Del Gallo, M.; Mosiello, L.; Baccella, S.; Lepidi, A. Escherichia coli detection in vegetable food by a potentiometric biosensor. Sens. Actuators B Chem. 2003, 91, 163–168. [Google Scholar] [CrossRef]
- Torun, O.; Boyaci, I.; Temur, E.; Tamer, U. Comparison of sensing strategies in SPR biosensor for rapid and sensitive enumeration of bacteria. Biosens. Bioelectron. 2012, 37, 53–60. [Google Scholar] [CrossRef]
- Mishra, R.; Dominguez, R.; Bhand, S.; Munoz, R.; Marty, J. A novel Automated flow-based biosensor for the determination of organophosphate pesticides in milk. Biosens. Bioelectron. 2012, 32, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Dong, F.; Chun-yuan, B.; Si-rong, Z.; Jian-guo, S. Recentprogress of commercially available biosensors in china and their applications in fermentation processes. J. Northeast. Agric Univ. 2014, 21, 73–85. [Google Scholar]
- Monosik, R.; Stredansky, M.; Tkac, J.; Sturdik, E. Application of enzyme biosensors in analysis of food and beverages enzyme and microbial technology. Food Anal. Methods 2012, 5, 40–53. [Google Scholar] [CrossRef]
- Arduini, F.; Ricci, F.; Tuta, C.S.; Moscone, D.; Amine, A.; Palleschi, G. Detection of carbamic and organophosphorus pesticides in water samples using cholinesterase biosensor based on Prussian blue modified screen printed electrode. Anal. Chim. Acta 2006, 58, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Ahmed, A.; Sharma, A.; Sharma, C.; Paul, S.; Khosla, A.; Gupta, V.; Arya, S. Promising photocatalytic degradation of methyl orange dye via sol-gel synthesized Ag–CdS@ Pr-TiO2 core/shell nanoparticles. Phys. B Condens. Matter 2021, 616, 413121. [Google Scholar] [CrossRef]
- Suprun, E.; Evtugyn, G.; Budnikov, H.; Ricci, F.; Moscone, D.; Palleschi, G. Acetylcholinesterase sensor based on screenprinted carbon electrode modified with Prussian blue. Anal. Bioanal. Chem. 2005, 383, 597–604. [Google Scholar] [CrossRef] [Green Version]
- Diesel, E.; Schreiber, M.; van der Meer, J.R. Development of bacteria-based bioassays for arsenic detection in natural waters. Anal. Bioanal. Chem. 2009, 394, 687–693. [Google Scholar] [CrossRef]
- Scognamiglio, V.; Pezzotti, G.; Pezzotti, I.; Cano, J.; Buonasera, K.; Giannini, D.; Giardi, M.T. Biosensors for effective environmental and agrifood protection and commercialization: From research to market. Mikrochim. Acta 2010, 170, 215–225. [Google Scholar] [CrossRef]
- Rea, G.; Polticelli, F.; Antonacci, A.; Scognamiglio, V.; Katiyar, P.; Kulkarni, S.A.; Johanningmeier, U.; Giardi, M.T. Structure-based design of novelChlamydomonas reinhardtiiD1-D2 photosynthetic proteins for herbicide monitoring. Protein Sci. 2009, 18, 2139–2151. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.; Zine, N.; Baraket, A.; Zabala, M.; Campabadal, F.; Caruso, R.; Trivella, M.G.; Jaffrezic-Renault, N.; Errachid, A. A novel biosensor based on hafnium oxide: Application for early stage detection of human interleukin-10. Sens. Actuators B Chem. 2012, 175, 201–207. [Google Scholar] [CrossRef]
- Chen, Y.W.; Liu, M.; Kaneko, T.; McIntyre, P.C. Atomic layer deposited hafnium oxide gate dielectrics for charge-based biosensors. Electrochem. Solid State Lett. 2010, 13, G29–G32. [Google Scholar] [CrossRef]
- Ooi, K.G.J.; Galatowicz, G.; Towler, H.M.A.; Lightman, S.L.; Calder, V.L. Multiplex cytokine detection versus ELISA for aqueous humor: IL-5, IL-10, and IFN profiles in uveitis. Investig. Ophthalmol. Vis. Sci. 2006, 47, 272–277. [Google Scholar] [CrossRef]
- Caruso, R.; Trunfio, S.; Milazzo, F.; Campolo, J.; de Maria, R.; Colombo, T.; Parodi, O. Early expression of proand anti-inflammatory cytokines in left ventricular assist device recipients with multiple organ failure syndrome. Am. Soc. Art. Int. Org. J. 2010, 56, 313–318. [Google Scholar] [CrossRef] [Green Version]
- Caruso, R.; Verde, A.; Cabiati, M.; Milazzo, F.; Boroni, C.; Del Ry, S.; Parodi, O. Association of preoperative interleukin-6 levels with interagency registry for mechanically assisted circulatory support profiles and intensive care unit stay in left ventricular assist device patients. J Heart Lung Transplant. 2012, 31, 625–633. [Google Scholar] [CrossRef] [Green Version]
- Watson, C.J.; Ledwidge, M.T.; Phelan, D.; Collier, P.; Byrne, J.C.; Dunn, M.J.; McDonald, K.M.; Baugh, J.A. Proteomic Analysis of Coronary Sinus Serum Reveals Leucine-Rich α2-Glycoprotein as a Novel Biomarker of Ventricular Dysfunction and Heart Failure. Circ. Hear. Fail. 2011, 4, 188–197. [Google Scholar] [CrossRef] [Green Version]
- Chung, D.; Khosla, A.; Gray, B.L.; Parameswaran, A.M.; Ramaseshan, R.; Kohli, K. Investigations of flexible Ag/AgCl nanocomposite polymer electrodes for suitability in tissue electrical impedance scanning (EIS). J. Electrochem. Soc. 2013, 161, B3071. [Google Scholar] [CrossRef]
- Singh, A.; Arya, S.; Khanuja, M.; Hafiz, A.K.; Datt, R.; Gupta, V.; Khosla, A. Eu doped NaYF 4@ Er: TiO 2 nanoparticles for tunable ultraviolet light based anti-counterfeiting applications. Microsyst. Technol. 2020, 1–10. [Google Scholar] [CrossRef]
- Morris, M.C. Fluorescent biosensors of intracellular targets from genetically encoded reporters to modular polypeptide probes. Cell Biochem. Biophys. 2010, 56, 19–37. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, K.A.; Taylor, D.L. Fluorescent-protein biosensors: New tools for drug discovery. Trends Biotechnol. 1998, 16, 135–140. [Google Scholar] [CrossRef]
- Wolff, M.; Wiedenmann, J.; Nienhaus, G.U.; Valler, M.; Heilker, R. Novel fluorescent proteins for high-content screening. Drug Discov. Today 2006, 11, 1054–1060. [Google Scholar] [CrossRef] [PubMed]
- Gupta, J.; Arya, S.; Singh, A.; Verma, S.; Sharma, A.; Singh, B.; Tomar, A. Template Based Electrochemical Synthesis of Copper (Cu) Nanowires as CH2Cl2 Sensor. Integr. Ferroelectr. 2020, 204, 63–72. [Google Scholar] [CrossRef]
- El-Deiry, W.S.; Sigman, C.C.; Kelloff, G.J. Imaging and oncologic drug development. J Clin. Oncol. 2006, 24, 3261–3273. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Arya, S.; Chauhan, D.; Solanki, P.R.; Khajuria, S.; Khosla, A. Synthesis of Au-SnO2 nanoparticles for electrochemical determination of vitamin B12. J. Mater. Res. Technol. 2020, 9, 14321–14337. [Google Scholar] [CrossRef]
- Morris, M.C. Fluorescent biosensors—probing protein kinase function in cancer and drug discovery. Biochim. Biophys. Acta 2013, 1834, 1387–1395. [Google Scholar] [CrossRef] [PubMed]
- Okumoto, S. Quantitative imaging using genetically encoded sensors for small molecules in plants. Plant J. 2012, 70, 108–117. [Google Scholar] [CrossRef] [Green Version]
- Choi, K.; Seo, W.; Cha, S.; Choi, J. Evaluation of two types of biosensors for immunoassay of botulinum toxin. J. Biochem. Mol. Biol. 1998, 31, 101–105. [Google Scholar]
- Thien, N.T.D.; Wang, H.; Sugiarto, S. Advances in nanomaterials and their applications in point of care (POC) devices for the diagnosis of infectious diseases. Biotechnol. Adv. 2016, 34, 1275–1288. [Google Scholar] [CrossRef]
- Davies, D.; Guo, X.; Musavi, L. Gold nanoparticle-modified carbon electrode biosensor for the detection of listeria monocytogenes. Ind. Biotechnol. 2013, 9, 13–36. [Google Scholar] [CrossRef]
- Yang, G.J.; Huang, J.L.; Menga, W.J. A reusable capacitive immunosensor for detection of Salmonella spp. based on grafted ethylene diamine and self-assembled gold nanoparticle monolayers. Anal. Chim. Acta 2009, 647, 159–166. [Google Scholar] [CrossRef]
- Gaffar, S.; Nurmalasari, R.; Yohan, Y.W. Voltammetric DNA biosensor using gold electrode modified by self assembled monolayer of Thiol for detection of mycobacterium tuberculosis. Procedia Technol. 2017, 27, 74–80. [Google Scholar] [CrossRef]
- Jampasa, S.; Wonsawat, W.; Rodthongkum, N.; Siangproh, W.; Yanatatsaneejit, P.; Vilaivan, T.; Chailapakul, O. Electrochemical detection of human papillomavirus DNA type 16 using a pyrrolidinyl peptide nucleic acid probe immobilized on screen-printed carbon electrodes. Biosens. Bioelectron. 2014, 54, 428–434. [Google Scholar] [CrossRef] [PubMed]
- Topell, S.; Glockshuber, R. Circular permutation of the green fluorescent protein. Methods Mol. Biol. 2002, 183, 31–48. [Google Scholar]
- Tian, L.; Hires, S.A.; Looger, L.L. Imaging neuronal activity with genetically encoded calcium indicators. Cold Spring Harb. Protoc. 2012, 647–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bermejo, C.; Ewald, J.C.; Lanquar, V.; Jones, A.M.; Frommer, W.B. In vivo biochemistry: Quantifying ion and metabolite levels in individual cells or cultures of yeast. Biochem. J. 2011, 438, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bermejo, C.; Haerizadeh, F.; Takanaga, H.; Chermak, D.; Frommer, W.B. Dynamic analysis of cytosolic glucose and ATP levels in yeast using optical sensors. Biochem. J. 2010, 432, 399–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brett, C.L.; Kallay, L.; Hua, Z.; Green, R.; Chyou, A.; Zhang, Y.; Graham, T.R.; Donowitz, M.; Rao, R. Genome-wide analysis reveals the vacuolar pH-state of Saccharomyces cerevisiae. PLoS ONE 2011, 6, e17619. [Google Scholar] [CrossRef] [Green Version]
- Orij, R.; Urbanus, M.L.; Vizeacoumar, F.J.; Giaever, G.; Boone, C.; Nislow, C.; Brul, S.; Smits, G.J. Genome-wide analysis of intracellular pH reveals quantitative control of cell division rate by pHc in Saccharomyces cerevisiae. Genome Biol. 2012, 13, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Liu, P.; Xue, T.; Ge, Y.; Ai, S.; Sheng, Y.; Wu, R.; Xu, L.; Tang, K.; Wen, Y. A novel graphene-like titanium carbide MXene/Au–Ag nanoshuttles bifunctional nanosensor for electrochemical and SERS intelligent analysis of ultra-trace carbendazim coupled with machine learning. Ceram. Int. 2021, 47, 173–184. [Google Scholar] [CrossRef]
- Jamal, M.; Chakrabarty, S.; Shao, H.; McNulty, D.; Yousuf, M.A.; Furukawa, H.; Khosla, A.; Razeeb, K.M. A non enzymatic glutamate sensor based on nickel oxide nanoparticle. Microsyst. Technol. 2018, 24, 4217–4223. [Google Scholar] [CrossRef]
- Lussier, F.; Thibault, V.; Charron, B.; Wallace, G.Q.; Masson, J.-F. Deep learning and artificial intelligence methods for Raman and surface-enhanced Raman scattering. TrAC Trends Anal. Chem. 2020, 124, 115796. [Google Scholar] [CrossRef]
- Dasgupta, A.; Nath, A. Classification of Machine Learning Algorithms. Int. J. Innov. Res. Adv. Eng. 2016, 3, 6–11. [Google Scholar]
- Mahesh, B. Machine Learning Algorithms-A Review. Int. J. Sci. Res. 2020, 9, 381–386. [Google Scholar]
- Yang, J.; Xu, J.; Zhang, X.; Wu, C.; Lin, T.; Ying, Y. Deep learning for vibrational spectral analysis: Recent progress and a practical guide. Anal. Chim. Acta 2019, 1081, 6–17. [Google Scholar] [CrossRef]
- Thrift, W.J.; Ragan, R. Quantification of Analyte Concentration in the Single Molecule Regime Using Convolutional Neural Networks. Anal. Chem. 2019, 91, 13337–13342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dies, H.; Raveendran, J.; Escobedo, C.; Docoslis, A. Rapid identification and quantification of illicit drugs on nanodendritic surface-enhanced Raman scattering substrates. Sens. Actuators B Chem. 2018, 257, 382–388. [Google Scholar] [CrossRef]
- Doty, K.C.; Lednev, I.K. Differentiation of human blood from animal blood using Raman spectroscopy: A survey of forensically relevant species. Forensic Sci. Int. 2018, 282, 204–210. [Google Scholar] [CrossRef]
- Shin, H.; Oh, S.; Hong, S.; Kang, M.; Kang, D.; Ji, Y.-g.; Choi, B.H.; Kang, K.-W.; Jeong, H.; Park, Y. Early-Stage Lung Cancer Diagnosis by Deep Learning-Based Spectroscopic Analysis of Circulating Exosomes. ACS Nano 2020, 14, 5435–5444. [Google Scholar] [CrossRef]
- Xu, Y.; Zomer, S.; Brereton, R.G. Support vector machines: A recent method for classification in chemometrics. Crit. Rev. Anal. Chem. 2006, 36, 177–188. [Google Scholar] [CrossRef]
- Chang, C.-C.; Lin, C.-J. LIBSVM: A library for support vector machines, ACM Trans. Intell. Syst. Technol. 2011, 2, 1–27. [Google Scholar] [CrossRef]
- Vapnik, V. The Nature of Statistical Learning Theory; Springer: Berlin/Heidelberg, Germany, 1995; ISBN 0-387-94559-8. [Google Scholar]
- Burges, C. A tutorial on support vector machines for pattern recognition. In Data Mining and Knowledge Discovery; Kluwer Academic Publishers: Boston, MA, USA, 1998; Volume 2. [Google Scholar]
- Vapnik, V.; Golowich, S.; Smola, A. Support vector method for function approximation, regression estimation, and signal processing. In Advances in Neural Information Processing Systems 9; Mozer, M., Jordan, M., Petsche, T., Eds.; MIT Press: Cambridge, MA, USA, 1997; pp. 281–287. [Google Scholar]
- Wang, C.; Madiyar, F.; Yu, C.; Li, J. Detection of extremely low concentration waterborne pathogen using a multiplexing selfreferencing SERS microfluidic biosensor. J. Biol. Eng. 2017, 11, 9. [Google Scholar] [CrossRef] [Green Version]
- Majumder, S.; Ghosh, N.; Gupta, P. Support vector machine fornoptical diagnosis of cancer. J. Biomed. Opt. 2005, 10, 024034. [Google Scholar] [CrossRef]
- Acharya, U.R.; Oh, S.L.; Hagiwara, Y.; Tan, J.H.; Adeli, H. Deep convolutional neural network for the automated detection and diagnosis of seizure using EEG signals. Comput. Biol. Med. 2018, 100, 270–278. [Google Scholar] [CrossRef]
- Olano, X.; Sallaberry, F.; García De Jalón, A.; Gastón, M. The influence of sky conditions on the standardized calibration of pyranometers and on the measurement of global solar irradiation. Sol. Energy 2015, 121, 116–122. [Google Scholar] [CrossRef]
- Liu, J.; Fang, W.; Zhang, X.; Yang, C. An improved photovoltaic power forecasting model with the assistance of aerosol index data. IEEE Trans. Sustain. Energy 2015, 6, 434–442. [Google Scholar] [CrossRef]
- Asefpour Vakilian, K.; Massah, J. An artificial neural network approach to identify fungal diseases of cucumber (Cucumis sativus L.) plants using digital image processing. Arch. Phytopathol. Plant Protect. 2013, 46, 1580–1588. [Google Scholar] [CrossRef]
- Kallenberg, M.; Petersen, K.; Nielsen, M.; Ng, A.Y.; Diao, P.; Igel, C.; Vachon, C.M.; Holland, K.; Winkel, R.R.; Karssemeijer, N. Unsupervised deep learning applied to breast density segmentation and mammographic risk scoring. IEEE Trans. Med. Imaging 2016, 35, 1322–1331. [Google Scholar] [CrossRef] [PubMed]
- Pereira, S.; Pinto, A.; Alves, V.; Silva, C.A. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans. Med. Imaging 2016, 35, 1240–1251. [Google Scholar] [CrossRef] [PubMed]
- Setio, A.A.A.; Ciompi, F.; Litjens, G.; Gerke, P.; Jacobs, C.; Van Riel, S.J.; Wille, M.M.W.; Naqibullah, M.; Sánchez, C.I.; van Ginneken, B. Pulmonary nodule detection in CT images: False positive reduction using multi-view convolutional networks. IEEE Trans. Med. Imaging 2016, 35, 1160–1169. [Google Scholar] [CrossRef]
- Tahir, A.M.; Chowdhury, M.E.H.; Khandakar, A.; Al-Hamouz, S.; Abdalla, M.; Awadallah, S.; Reaz, M.B.I.; Al-Emadi, N. A Systematic Approach to the Design and Characterization of a Smart Insole for Detecting Vertical Ground Reaction Force (vGRF) in Gait Analysis. Sensors 2020, 20, 957. [Google Scholar] [CrossRef] [Green Version]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Pdf ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Ho, T.K.K.; Gwak, J. Multiple Feature Integration for Classification of Thoracic Disease in Chest Radiography. Appl. Sci. 2019, 9, 4130. [Google Scholar] [CrossRef] [Green Version]
- Liang, G.; Zheng, L. A transfer learning method with deep residual network for pediatric pneumonia diagnosis. Comput. Methods Programs Biomed. 2020, 187, 104964. [Google Scholar] [CrossRef]
- Virkki, R.; Juvén, T.; Rikalainen, H.; Svedstrom, E.; Mertsola, J.; Ruuskanen, O. Differentiation of bacterial and viral pneumonia in children. Thorax 2002, 57, 438–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goyal, M.; Goyal, R.; Lall, B. Learning Activation Functions: A new paradigm of understanding Neural Networks. arXiv 2019, arXiv:1906.09529. [Google Scholar]
- Bailer, C.; Habtegebrial, T.; Varanasi, K.; Stricker, D. Fast Feature Extraction with CNNs with Pooling Layers. arXiv 2018, arXiv:1805.03096. [Google Scholar]
- Lipton, Z.C.; Berkowitz, J.; Elkan, C. A critical review of recurrent neural networks for sequence learning. arXiv 2015, arXiv:1506.00019. [Google Scholar]
- Ordóñez, F.J.; Roggen, D. Deep convolutional and lstm recurrent neural networks for multimodal wearable activity recognition. Sensors 2016, 16, 115. [Google Scholar] [CrossRef] [Green Version]
- Kiddon, C.; Zettlemoyer, L.; Choi, Y. Globally coherent text generation with neural checklist models. In Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing, Austin, TX, USA, 1–5 November 2016; pp. 329–339. [Google Scholar]
- Takeuchi, D.; Yatabe, K.; Koizumi, Y.; Oikawa, Y.; Harada, N. Real-time speech enhancement using equilibriated RNN. In Proceedings of the ICASSP 2020−2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4−8 May 2020. [Google Scholar]
- Doetsch, P.; Kozielski, M.; Ney, H. Fast and robust training of recurrent NEURAL networks for offline handwriting recognition. In Proceedings of the 2014 14th International Conference on Frontiers in Handwriting Recognition, Hersonissos, Greece, 1−4 September 2014; pp. 279–284. [Google Scholar]
- Williams, J.D.; Zweig, G. End-to-end lstm-based dialog control optimized with supervised and reinforcement learning. arXiv 2016, arXiv:1606.01269. [Google Scholar]
- Corbett, P.; Boyle, J. Improving the learning of chemicalprotein interactions from literature using transfer learning and specialized word embeddings. Database 2018, 2018, 1. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wu, Y.; Song, G.; Li, Z.; Fan, Y.; Zhang, Y. Brain Tumor Segmentation Using a Fully Convolutional Neural Network with Conditional Random Fields, International Workshop on Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Springer: Berlin/Heidelberg, Germany, 2016; pp. 75–87. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Fang, L.; Yu, G.; Wang, D.; Xiao, C.-L.; Wang, K. Detection of DNA base modifications by deep recurrent neural network on Oxford Nanopore sequencing data. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rang, F.J.; Kloosterman, W.P.; de Ridder, J. From squiggle to basepair: Computational approaches for improving nanopore sequencing read accuracy. Genome Biol. 2018, 19, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Available online: https://www.dimensions.ai/ (accessed on 12 August 2021).
- Viswanathan, S.; Rani, C.; Anand, A.V.; Ho, J.A.A. Disposable electrochemical immunosensor forcarcinoembryonic antigen using ferrocene liposomes and MWCNT screenprinted electrode. Biosens. Bioelectron. 2009, 24, 1984–1989. [Google Scholar] [CrossRef] [PubMed]

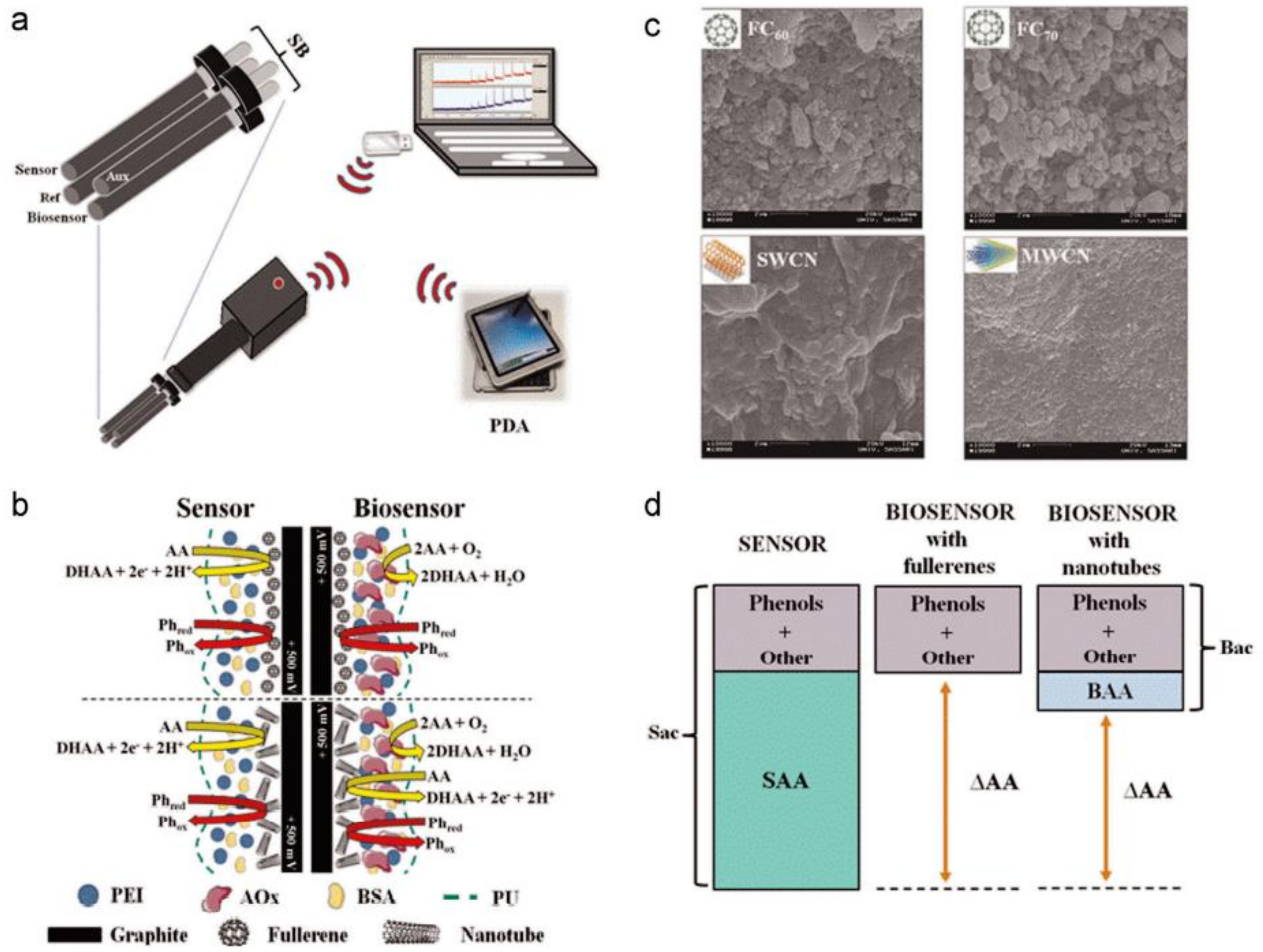
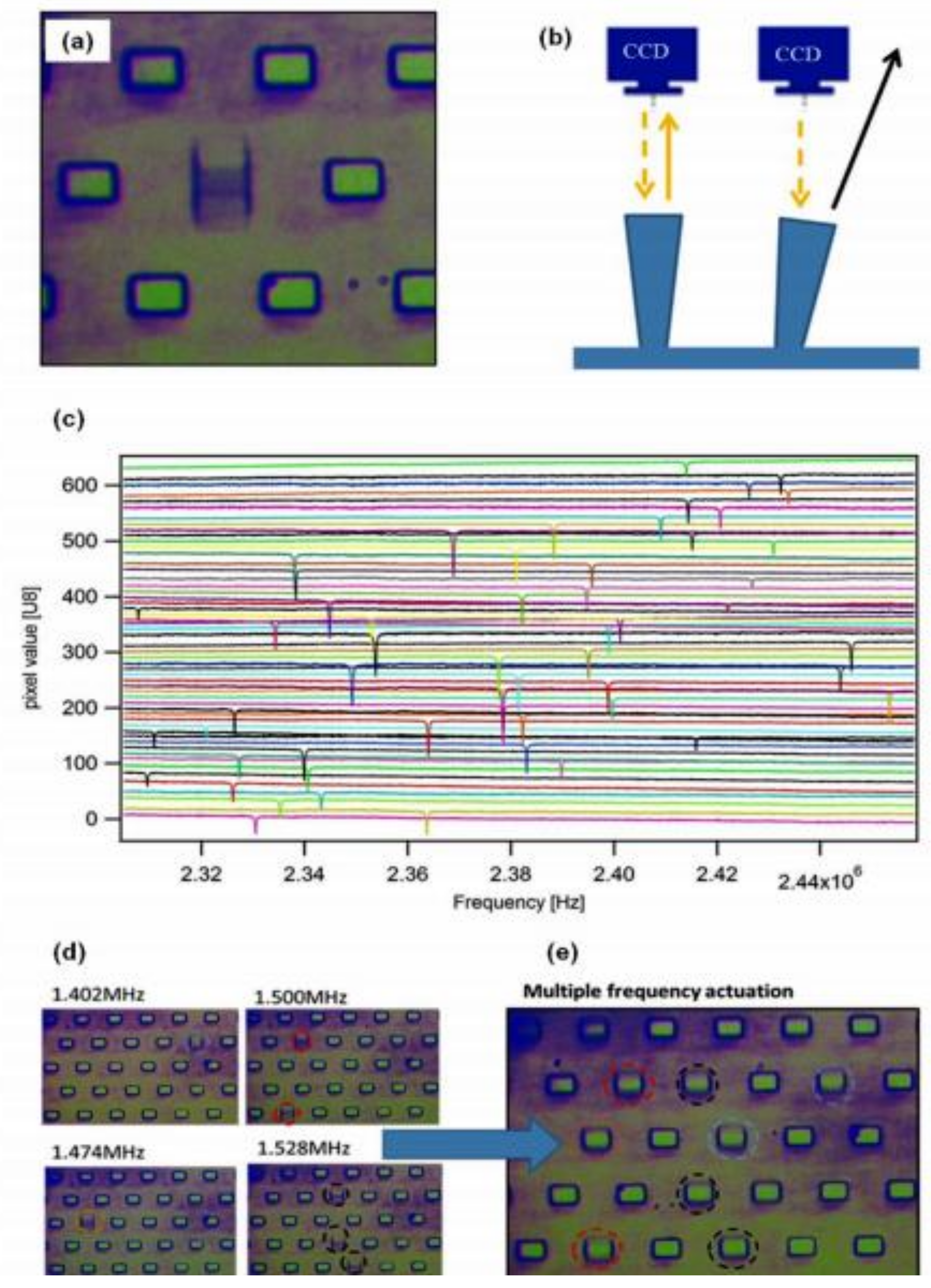
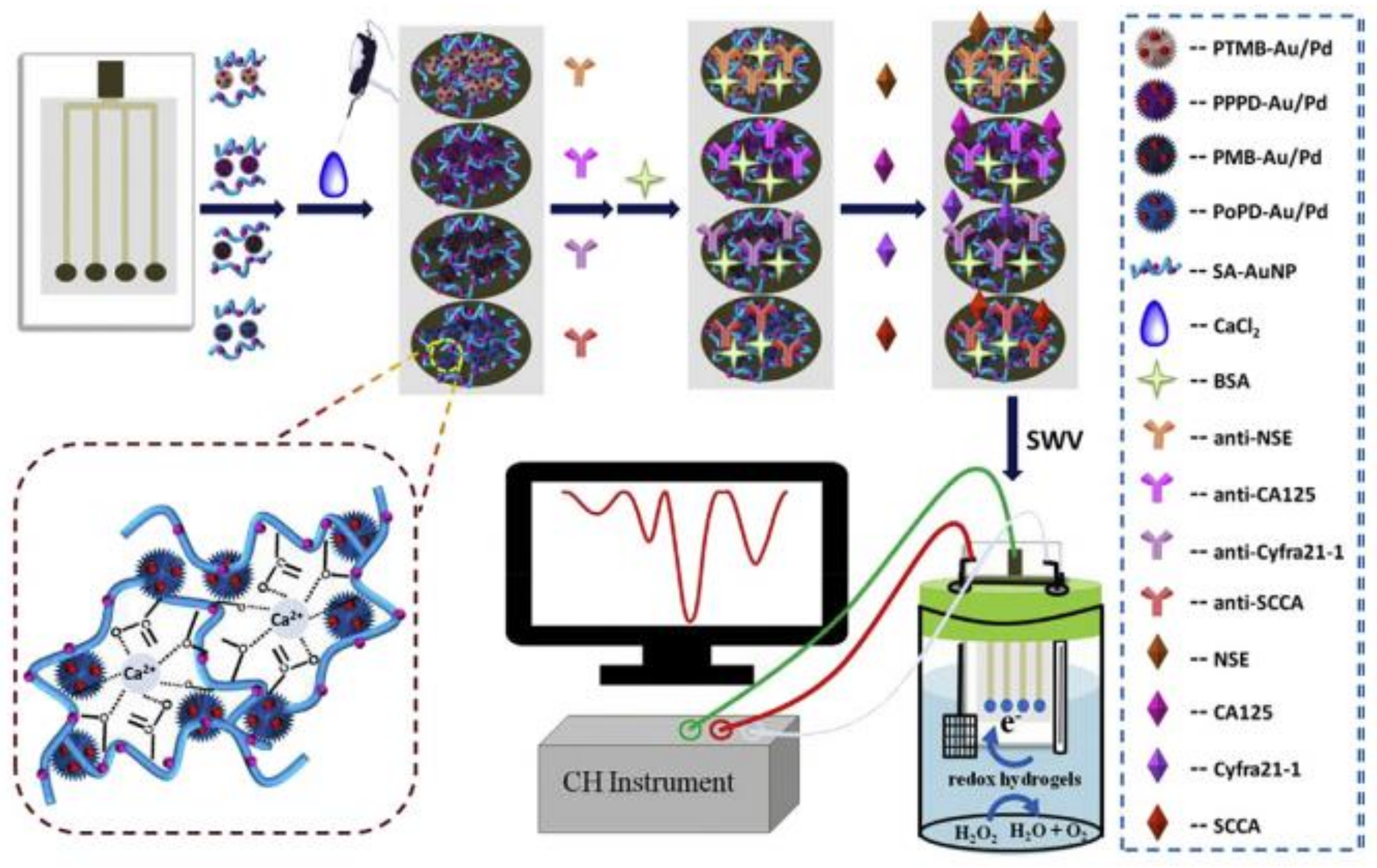
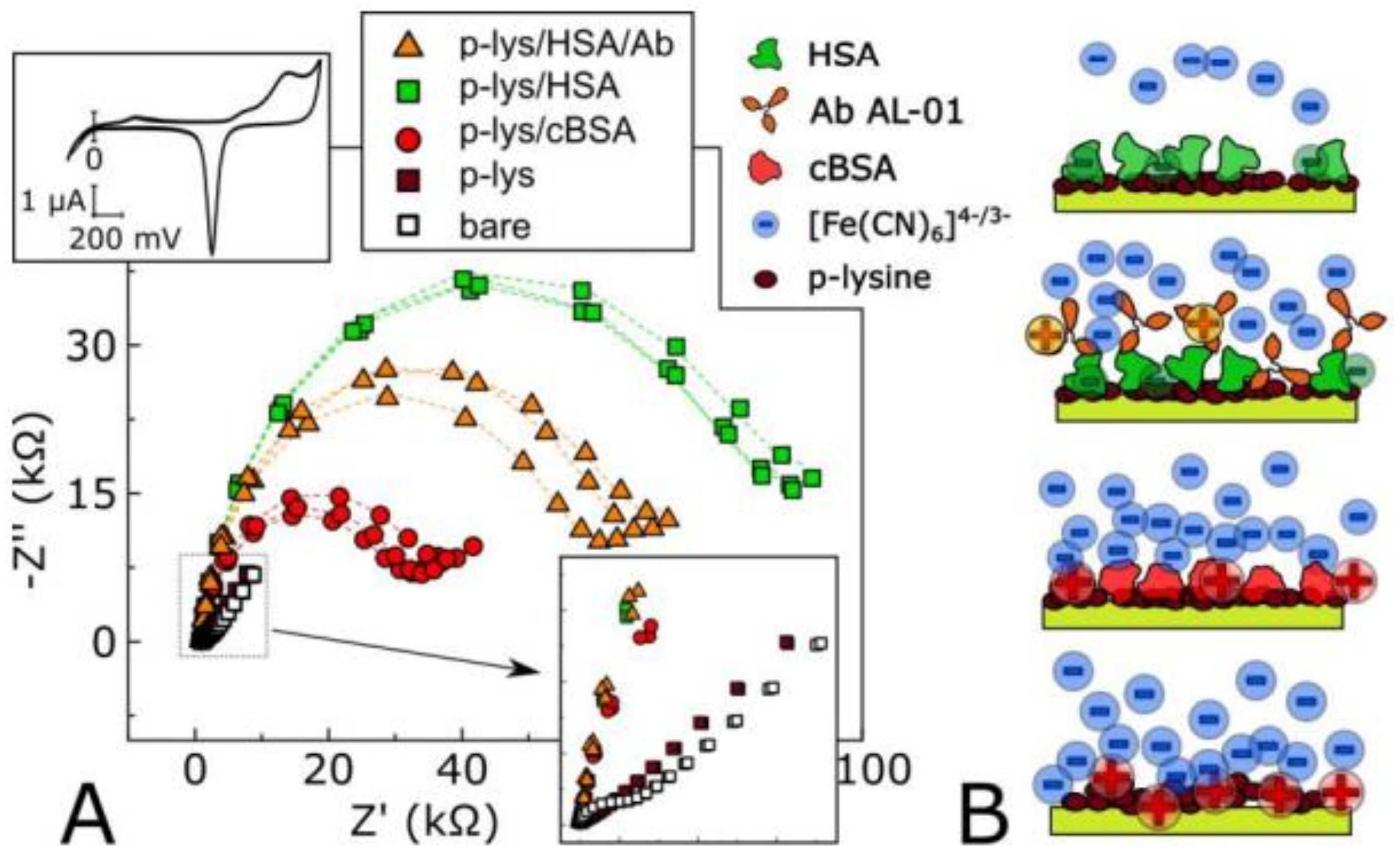




| Method | Target | Biological Element | Target Matrix | Transducer Element | Ref. |
|---|---|---|---|---|---|
| Amperometric | Cholesterol | Cholesterol oxidase | Human serum | Prussian Blue modified SPE | [16] |
| Amperometric | Lactate | Lactate oxidase | Wine | Prussian Blue modified SPE | [17] |
| Amperometric | Polyamines | Polyamine oxidase, spermine oxidase | Food | Prussian Blue modified SPE | [18] |
| Amperometric | Lysine | Lysine oxidase | Cheese | Pt electrode | [19] |
| Amperometric | Glucose | Glucose oxidase | Transdermal fluid | Transdermal microneedles | [20] |
| Amperometric | Glucose | Glucose oxidase | - | Gold nanoelectrode | [21] |
| Amperometric | Ethanol | Alcohol dehydrogenase | wine | Polyaniline doped modified SPE | [22] |
| Amperometric | Antioxidant capacity | Superoxide disumlase | Fruit juice and berries | Pt electrode | [23] |
| Amperometric differential | Antioxidant capacity+ ascorbate | Ascorbate oxidase | Fruit juice | Fullerene modified graphite | [24] |
| Amperometric inhibition | Atrazine | Tyrosinase | Drinking water | Carbon modified SPE | [25] |
| Amperometric | Oxygen profile | Biliribine oxidase | Microbial fuel cell | Pt electrode | [26] |
| Label-free evanescent wave | IgG | Antibody | Human serum | Titania–silica-coated long period gratings optical fibers | [28] |
| Label-free CCD + software for imaging | Prostate specific antigen | Antibody | Human serum | Dense arrays of micropillars | [29] |
| Label-free field effect transistor | Interleukin 4 | Antibody | Human serum | Organic transistor | [30] |
| Voltametric/ impedimetric | Aflatoxin B1 | Aptamer | Peanuts and peanuts corn snacks | Dendrimer- modified gold electrode | [32] |
| Label-free, piezoelectric using 2 different aptamers | Metalloproteinase 9 | Aptamers | Human serum | Quartz crystal microbalance | [33] |
| Colorimetric, aggregation using 2 aptamers | DNA methylation | Aptamers for α-thrombin | DNA | Au coated magnetic nanoparticles | [34] |
| Impedimetric | Human epidermal growth factor receptor 2 | Antibody | Human serum | Au–nano-particles on SPE | [35] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, A.; Sharma, A.; Ahmed, A.; Sundramoorthy, A.K.; Furukawa, H.; Arya, S.; Khosla, A. Recent Advances in Electrochemical Biosensors: Applications, Challenges, and Future Scope. Biosensors 2021, 11, 336. https://doi.org/10.3390/bios11090336
Singh A, Sharma A, Ahmed A, Sundramoorthy AK, Furukawa H, Arya S, Khosla A. Recent Advances in Electrochemical Biosensors: Applications, Challenges, and Future Scope. Biosensors. 2021; 11(9):336. https://doi.org/10.3390/bios11090336
Chicago/Turabian StyleSingh, Anoop, Asha Sharma, Aamir Ahmed, Ashok K. Sundramoorthy, Hidemitsu Furukawa, Sandeep Arya, and Ajit Khosla. 2021. "Recent Advances in Electrochemical Biosensors: Applications, Challenges, and Future Scope" Biosensors 11, no. 9: 336. https://doi.org/10.3390/bios11090336
APA StyleSingh, A., Sharma, A., Ahmed, A., Sundramoorthy, A. K., Furukawa, H., Arya, S., & Khosla, A. (2021). Recent Advances in Electrochemical Biosensors: Applications, Challenges, and Future Scope. Biosensors, 11(9), 336. https://doi.org/10.3390/bios11090336








