Cholesterol Chip for the Study of Cholesterol–Protein Interactions Using SPR
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of Biotinylated Cholesterol
2.3. Expression and Purification of Hedgehog Protein
2.4. Expression and Purification of PTP1B
2.5. Immobilization of Biotinylated Cholesterol on SA Sensor Chip
2.6. Binding Kinetics and Affinity Measurement of Cholesterol–Protein Interaction
3. Results and Discussion
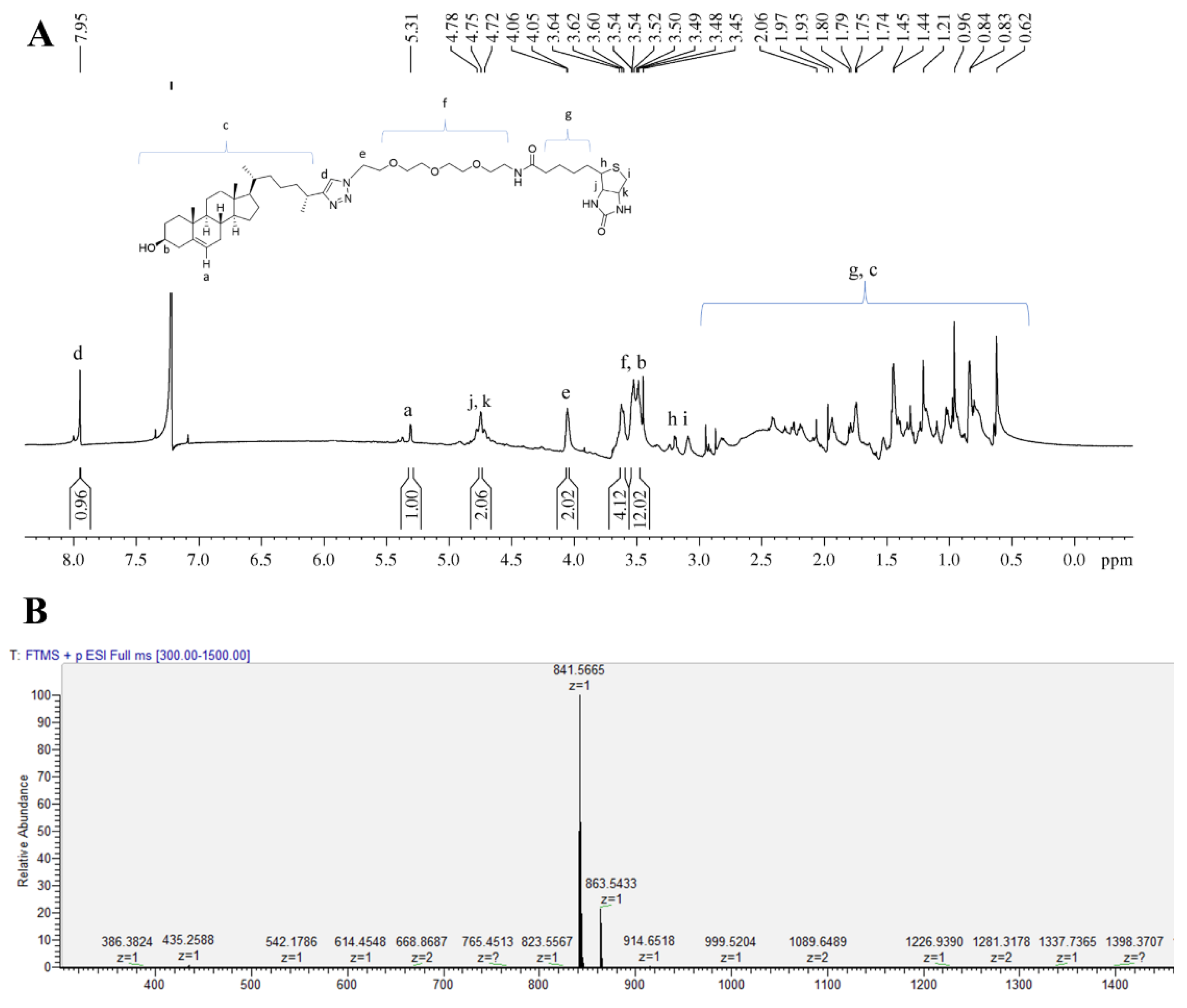
3.1. Synthesis of Biotinylated Cholesterol and Its Characterization
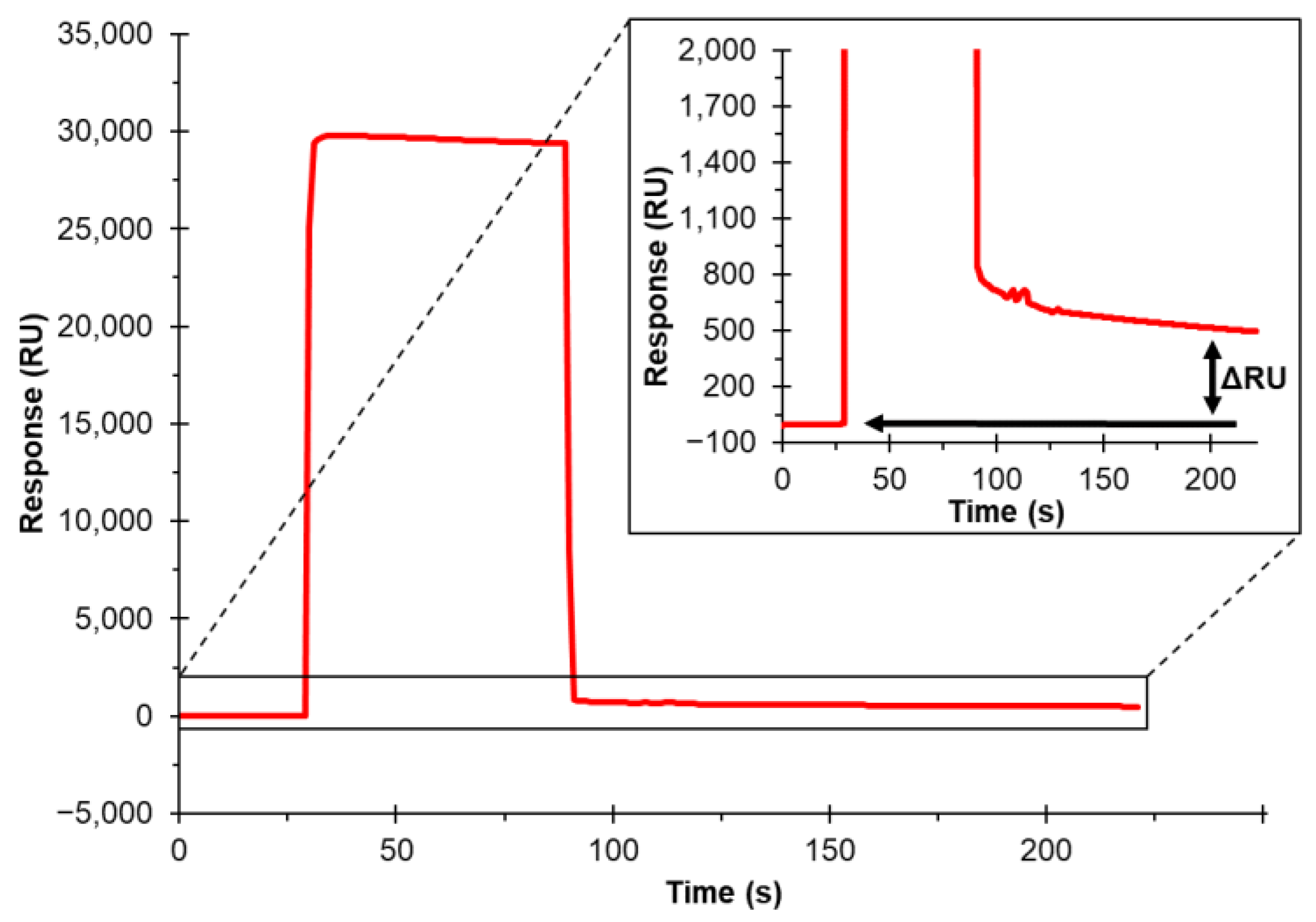
3.2. Immobilization of Biotinylated Cholesterol on SA Sensor Chip
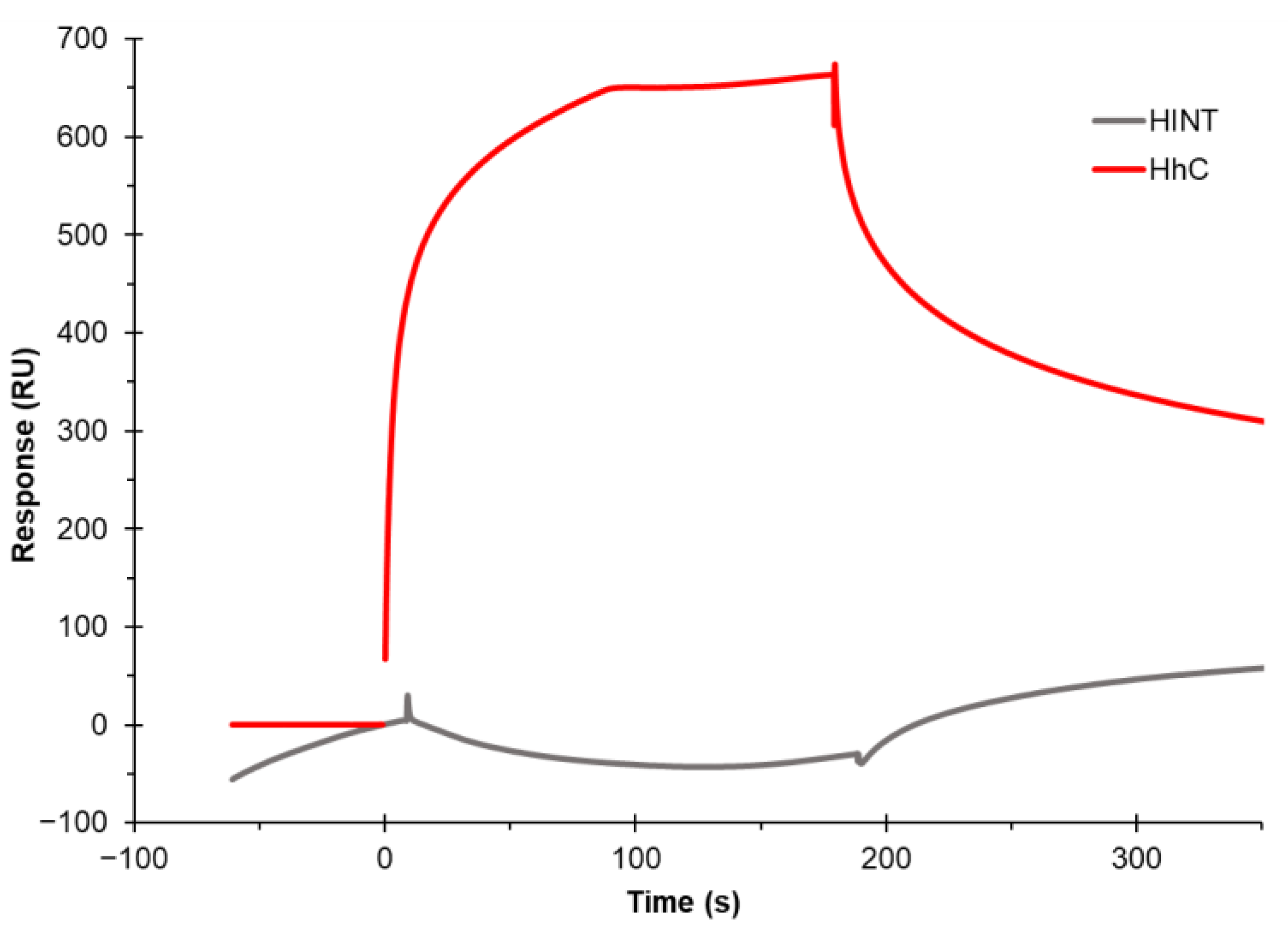
3.3. Establishing Binding to the Cholesterol Chip Is Specific for Cholesterol-Binding Proteins
3.4. Binding Kinetics and Affinity Measurement of Cholesterol–Protein Interaction
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Luo, J.; Yang, H.; Song, B.-L. Mechanisms and regulation of cholesterol homeostasis. Nat. Rev. Mol. Cell Biol. 2020, 21, 225–245. [Google Scholar] [CrossRef] [PubMed]
- Elkins, M.R.; Sergeyev, I.V.; Hong, M. Determining Cholesterol Binding to Membrane Proteins by Cholesterol 13C Labeling in Yeast and Dynamic Nuclear Polarization NMR. J. Am. Chem. Soc. 2018, 140, 15437–15449. [Google Scholar] [CrossRef] [PubMed]
- Bukiya, A.N.; Dopico, A.M. Common structural features of cholesterol binding sites in crystallized soluble proteins. J. Lipid Res. 2017, 58, 1044–1054. [Google Scholar] [CrossRef] [PubMed]
- Epand, R.M. Cholesterol and the interaction of proteins with membrane domains. Prog. Lipid Res. 2006, 45, 279–294. [Google Scholar] [CrossRef]
- Peters, C.; Wolf, A.; Wagner, M.; Kuhlmann, J.; Waldmann, H. The cholesterol membrane anchor of the Hedgehog protein confers stable membrane association to lipid-modified proteins. Proc. Natl. Acad. Sci. USA 2004, 101, 8531–8536. [Google Scholar] [CrossRef]
- He, R.-J.; Yu, Z.-H.; Zhang, R.-Y.; Zhang, Z.-Y. Protein tyrosine phosphatases as potential therapeutic targets. Acta Pharmacol. Sin. 2014, 35, 1227–1246. [Google Scholar] [CrossRef]
- Boivin, B.; Sagabala, R.S. Role of Cholesterol and Oxysterols in Regulating PTP1B Activity. Free Radic. Biol. Med. 2017, 112, 146. [Google Scholar] [CrossRef]
- Kumar, A.; Pandey, R.R.; Brantley, B. Tetraethylorthosilicate film modified with protein to fabricate cholesterol biosensor. Talanta 2006, 69, 700–705. [Google Scholar] [CrossRef]
- Fuentes, M.; Santander, N.; Cortés, V. Insulin increases cholesterol uptake, lipid droplet content, and apolipoprotein B secretion in CaCo-2 cells by upregulating SR-BI via a PI3K, AKT, and mTOR-dependent pathway. J. Cell. Biochem. 2019, 120, 1550–1559. [Google Scholar] [CrossRef]
- Heybrock, S.; Kanerva, K.; Meng, Y.; Ing, C.; Liang, A.; Xiong, Z.-J.; Weng, X.; Kim, Y.A.; Collins, R.; Trimble, W.; et al. Lysosomal integral membrane protein-2 (LIMP-2/SCARB2) is involved in lysosomal cholesterol export. Nat. Commun. 2019, 10, 3521. [Google Scholar] [CrossRef] [Green Version]
- Cho, Y.Y.; Kwon, O.-H.; Chung, S. Preferred Endocytosis of Amyloid Precursor Protein from Cholesterol-Enriched Lipid Raft Microdomains. Molecules 2020, 25, 5490. [Google Scholar] [CrossRef]
- Cedó, L.; Metso, J.; Santos, D.; García-León, A.; Plana, N.; Sabate-Soler, S.; Rotllan, N.; Rivas-Urbina, A.; Méndez-Lara, K.A.; Tondo, M.; et al. LDL Receptor Regulates the Reverse Transport of Macrophage-Derived Unesterified Cholesterol via Concerted Action of the HDL-LDL Axis. Circ. Res. 2020, 127, 778–792. [Google Scholar] [CrossRef] [PubMed]
- Králová, J.; Jurášek, M.; Mikšátková, L.; Marešová, A.; Fähnrich, J.; Cihlářová, P.; Drašar, P.; Bartůněk, P.; Král, V. Influence of fluorophore and linker length on the localization and trafficking of fluorescent sterol probes. Sci. Rep. 2020, 10, 22053. [Google Scholar] [CrossRef] [PubMed]
- Seigneuret, M.; Favre, E.; Morrot, G.; Devaux, P.F. Strong interactions between a spin-labeled cholesterol analog and erythrocyte proteins in the human erythrocyte membrane. Biochim. Biophys. Acta 1985, 813, 174–182. [Google Scholar] [CrossRef]
- Tukachinsky, H.; Kuzmickas, R.P.; Jao, C.Y.; Liu, J.; Salic, A. Dispatched and Scube Mediate the Efficient Secretion of the Cholesterol-Modified Hedgehog Ligand. Cell Rep. 2012, 2, 308–320. [Google Scholar] [CrossRef]
- Kumari, L.; Shamsher, K.S. Cholesterol oxidase: Role in biotransformation of cholesterol. J. Appl. Biol. Biotechnol. 2015, 3, 053–065. [Google Scholar] [CrossRef]
- Jaipuria, G.; Ukmar-Godec, T.; Zweckstetter, M. Challenges and approaches to understand cholesterol-binding impact on membrane protein function: An NMR view. Cell Mol. Life Sci. 2018, 75, 2137–2151. [Google Scholar] [CrossRef]
- Romanowski, M.J.; Soccio, R.E.; Breslow, J.L.; Burley, S.K. Crystal structure of the Mus musculus cholesterol-regulated START protein 4 (StarD4) containing a StAR-related lipid transfer domain. Proc. Natl. Acad. Sci. USA 2002, 99, 6949–6954. [Google Scholar] [CrossRef]
- Duo, J.; Bruno, J.; Kozhich, A.; David-Brown, D.; Luo, L.; Kwok, S.; Santockyte, R.; Haulenbeek, J.; Liu, R.; Hamuro, L.; et al. Surface plasmon resonance as a tool for ligand-binding assay reagent characterization in bioanalysis of biotherapeutics. Bioanalysis 2018, 10, 559–576. [Google Scholar] [CrossRef]
- Matharu, Z.; Bee, C.; Schwarz, F.; Chen, H.; Tomlinson, M.; Wu, G.; Rakestraw, G.; Hornsby, M.; Drake, A.; Strop, P.; et al. High-Throughput Surface Plasmon Resonance Biosensors for Identifying Diverse Therapeutic Monoclonal Antibodies. Anal. Chem. 2021, 93, 16474–16480. [Google Scholar] [CrossRef]
- Beseničar, M.; Maček, P.; Lakey, J.H.; Anderluh, G. Surface plasmon resonance in protein–membrane interactions. Chem. Phys. Lipids 2006, 141, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Liu, X.; Malhotra, A.; Li, Q.; Zhang, F.; Linhardt, R.J. Novel method for measurement of heparin anticoagulant activity using SPR. Anal. Biochem. 2017, 526, 39–42. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Jin, W.; Sood, A.; Montgomery, D.W.; Grant, O.C.; Fuster, M.M.; Fu, L.; Dordick, J.S.; Woods, R.J.; Zhang, F.; et al. Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions. Antivir. Res. 2020, 181, 104873. [Google Scholar] [CrossRef]
- Gervais, L.; Gel, M.; Allain, B.; Tolba, M.; Brovko, L.; Zourob, M.; Mandeville, R.; Griffiths, M.; Evoy, S. Immobilization of biotinylated bacteriophages on biosensor surfaces. Sens. Actuators B Chem. 2007, 125, 615–621. [Google Scholar] [CrossRef]
- Chanda, K.; Mm, B. Biosensors for pathogen surveillance. Environ. Chem. Lett. 2018, 16, 1325–1337. [Google Scholar] [CrossRef]
- Erb, E.M.; Chen, X.; Allen, S.; Roberts, C.J.; Tendler, S.J.; Davies, M.C.; Forsén, S. Characterization of the Surfaces Generated by Liposome Binding to the Modified Dextran Matrix of a Surface Plasmon Resonance Sensor Chip. Anal. Biochem. 2000, 280, 29–35. [Google Scholar] [CrossRef]
- Hall, T.M.; Porter, J.A.; Young, K.E.; Koonin, E.V.; Beachy, P.A.; Leahy, D.J. Crystal Structure of a Hedgehog Autoprocessing Domain: Homology between Hedgehog and Self-Splicing Proteins. Cell 1997, 91, 85–97. [Google Scholar] [CrossRef]
- Zhao, J.; Ciulla, D.A.; Xie, J.; Wagner, A.G.; Castillo, D.A.; Zwarycz, A.S.; Lin, Z.; Beadle, S.; Giner, J.-L.; Li, Z.; et al. General Base Swap Preserves Activity and Expands Substrate Tolerance in Hedgehog Autoprocessing. J. Am. Chem. Soc. 2019, 141, 18380–18384. [Google Scholar] [CrossRef] [PubMed]
- Londhe, A.D.; Bergeron, A.; Curley, S.M.; Zhang, F.; Rivera, K.D.; Kannan, A.; Coulis, G.; Rizvi, S.H.M.; Kim, S.J.; Pappin, D.J.; et al. Regulation of PTP1B activation through disruption of redox-complex formation. Nat. Chem. Biol. 2019, 16, 122–125. [Google Scholar] [CrossRef]
- Zhang, K.; Li, T.; Shan, X.; Lu, R.; Zhang, S.; Xu, H. Cholesterol: Bioactivities, Structural Modification, Mechanisms of Action, and Structure-Activity Relationships. Mini-Rev. Med. Chem. 2021, 21, 1830–1848. [Google Scholar] [CrossRef]
- Hofmann, K.; Thiele, C.; Schött, H.-F.; Gaebler, A.; Schoene, M.; Kiver, Y.; Friedrichs, S.; Litjohann, D.; Kuerschner, L. A novel alkyne cholesterol to trace cellular cholesterol metabolism and localization. J. Lipid Res. 2014, 55, 583–591. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, P.; Faris, S.; Sagabala, R.S.; Datta, P.; Xu, Z.; Callahan, B.; Wang, C.; Boivin, B.; Zhang, F.; Linhardt, R.J. Cholesterol Chip for the Study of Cholesterol–Protein Interactions Using SPR. Biosensors 2022, 12, 788. https://doi.org/10.3390/bios12100788
He P, Faris S, Sagabala RS, Datta P, Xu Z, Callahan B, Wang C, Boivin B, Zhang F, Linhardt RJ. Cholesterol Chip for the Study of Cholesterol–Protein Interactions Using SPR. Biosensors. 2022; 12(10):788. https://doi.org/10.3390/bios12100788
Chicago/Turabian StyleHe, Peng, Shannon Faris, Reddy Sudheer Sagabala, Payel Datta, Zihan Xu, Brian Callahan, Chunyu Wang, Benoit Boivin, Fuming Zhang, and Robert J. Linhardt. 2022. "Cholesterol Chip for the Study of Cholesterol–Protein Interactions Using SPR" Biosensors 12, no. 10: 788. https://doi.org/10.3390/bios12100788
APA StyleHe, P., Faris, S., Sagabala, R. S., Datta, P., Xu, Z., Callahan, B., Wang, C., Boivin, B., Zhang, F., & Linhardt, R. J. (2022). Cholesterol Chip for the Study of Cholesterol–Protein Interactions Using SPR. Biosensors, 12(10), 788. https://doi.org/10.3390/bios12100788





