Direct Capture and Early Detection of Lyme Disease Spirochete in Skin with a Microneedle Patch
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture of a B. burgdorferi Strain B31
2.2. Design of Microneedles
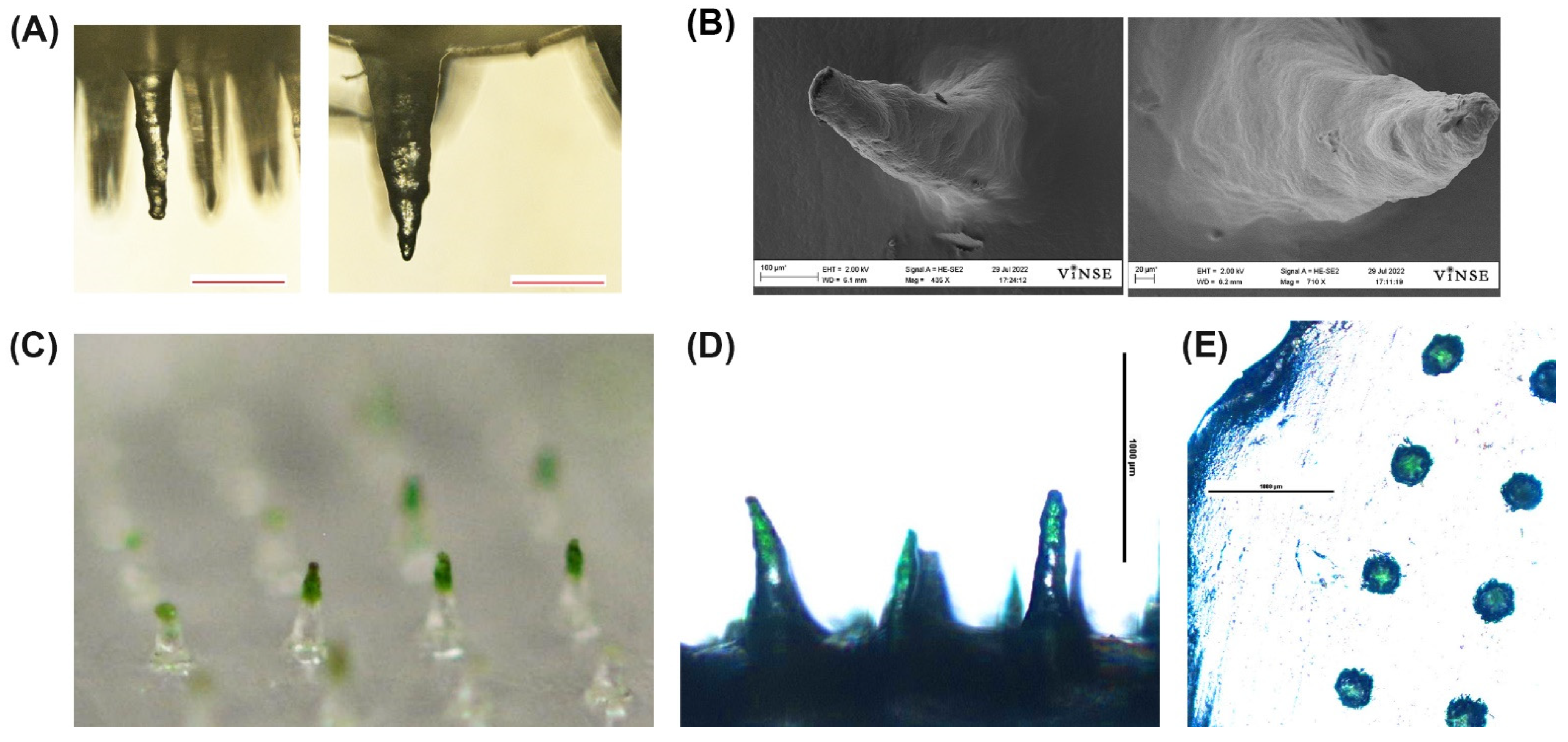
2.3. Fabrication and Imaging of PVA Microneedles
2.4. Penetration Tests
2.5. Mechanical Tests
2.6. Swell and Absorption Test
2.7. Time and Volume to Dissolve MN for PCR Testing
2.8. Preparation of Pig Ear Skin and Penetration of MN in Porcine Ear Skin by OCT
2.9. RNAse P Assay for Extraction Control
2.10. Determination of Bacteria Recovery Using qPCR
2.11. MN Application to Pig Ear Skin and Capture Test
3. Results
3.1. Microneedle Designs
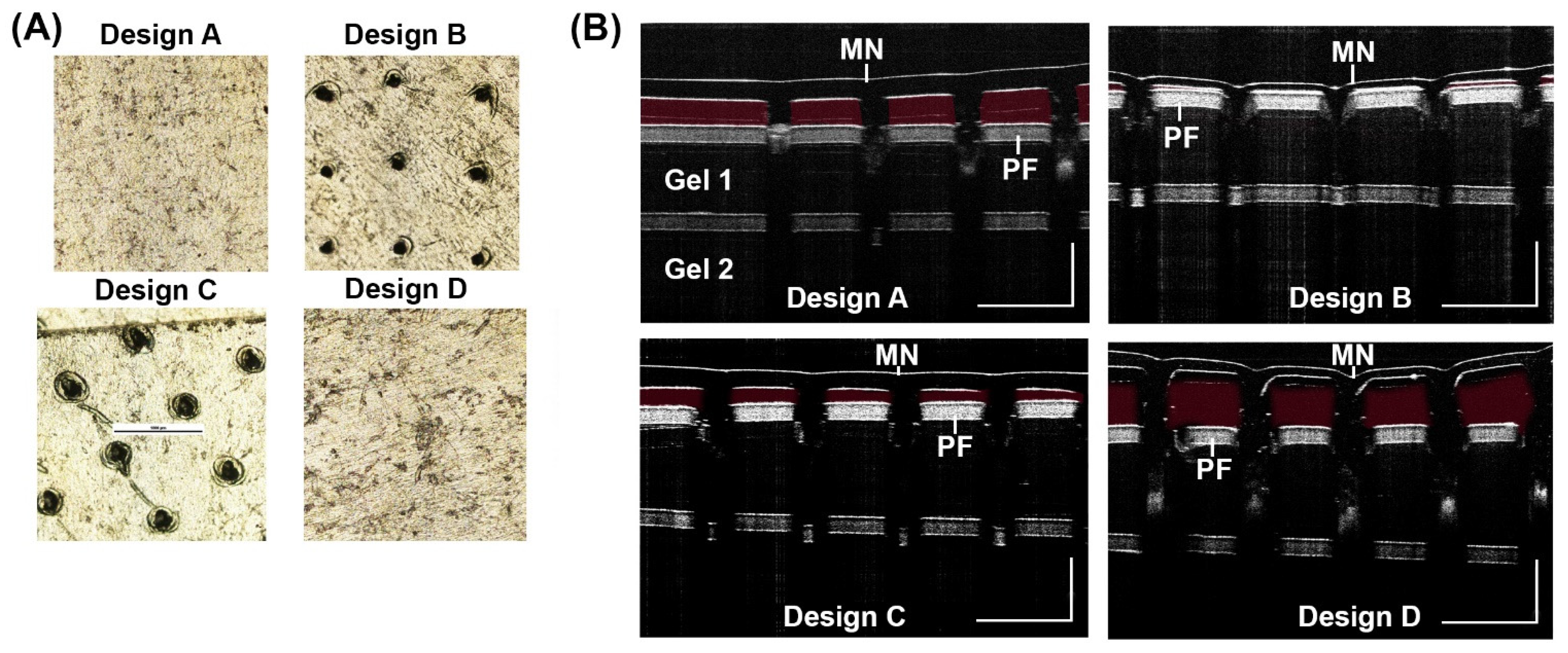
3.2. Penetration Test
3.3. Mechanical Tests
3.4. Swell and Absorption Tests
3.5. Volume Needed to Dissolve PVA MN
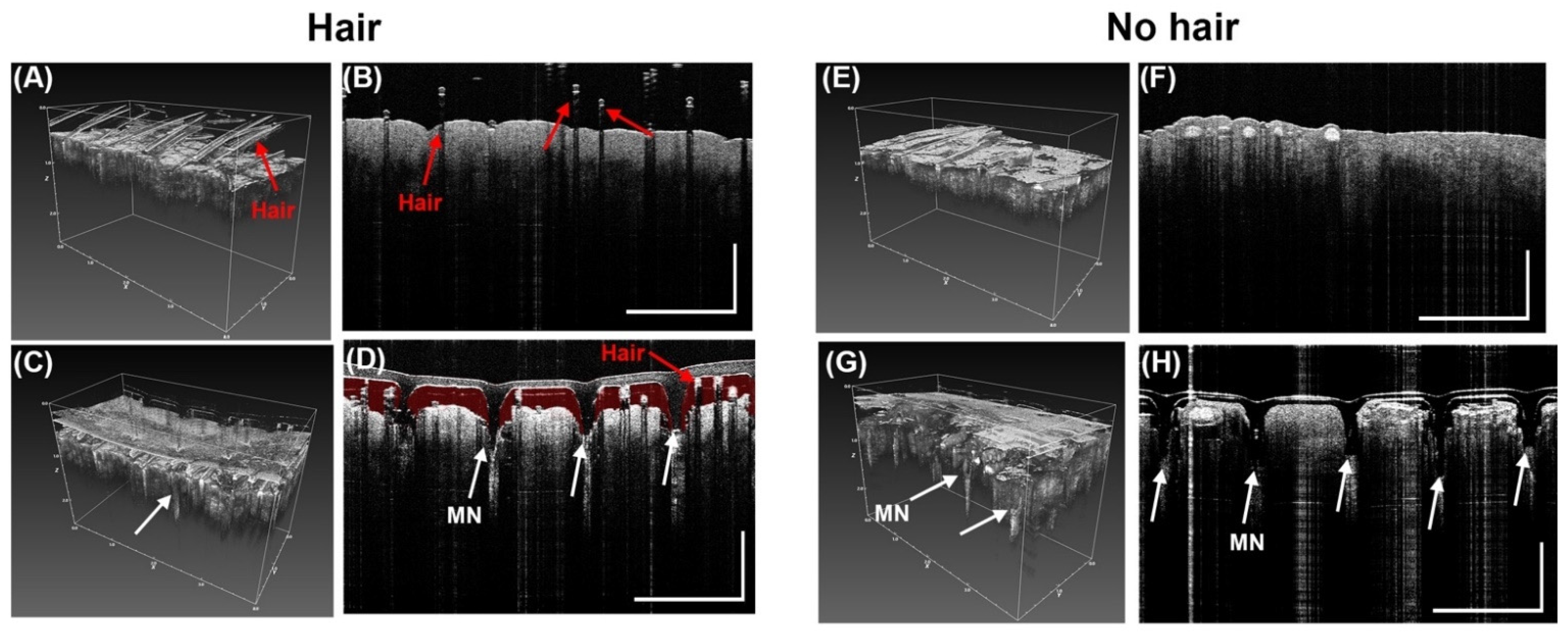
3.6. Penetration of MN in Porcine Ear Skin by OCT
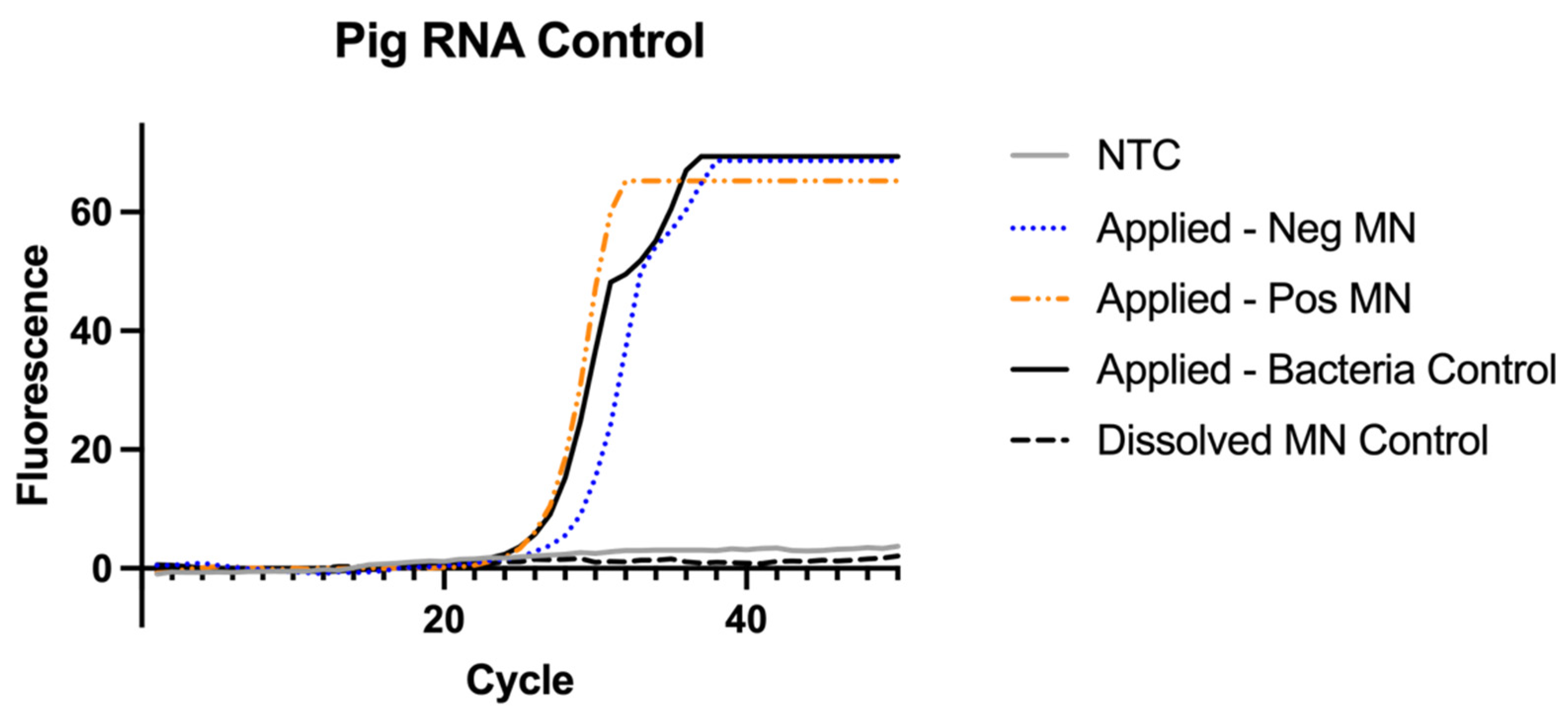
3.7. RNAse P Assay for Sampling Control
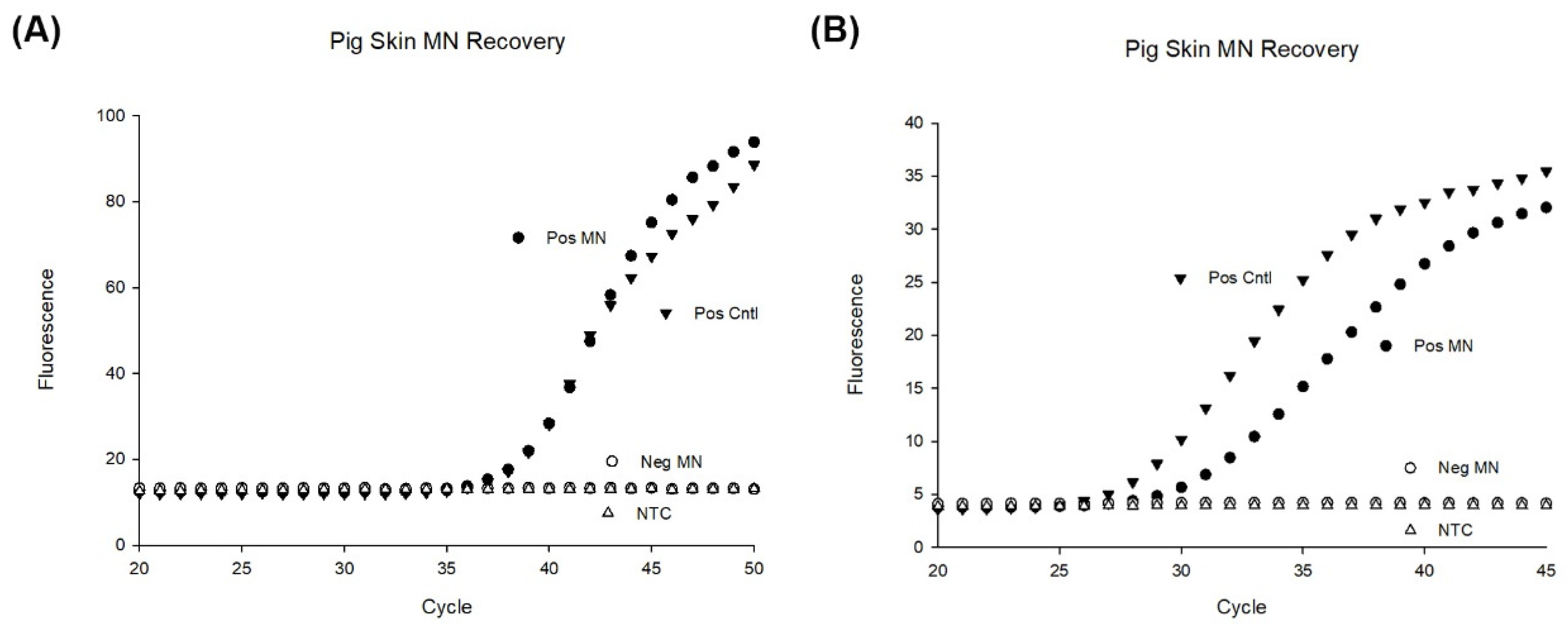
3.8. MN Capture of Inactivated B. burgdorferi
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rochlin, I.; Toledo, A. Emerging tick-borne pathogens of public health importance: A mini-review. J. Med. Microbiol. 2020, 69, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. National Center for Emerging and Zoonotic Infectious Diseases. Available online: http://www.cdc.gov/nczved/divisions/dfbmd/diseases/campylobacter/technical.html (accessed on 16 May 2022).
- Kugeler, K.J.; Schwartz, A.M.; Delorey, M.J.; Mead, P.S.; Hinckley, A.F. Estimating the frequency of Lyme disease diagnoses, United States, 2010–2018. Emerg. Infect. Dis. 2021, 27, 616. [Google Scholar] [CrossRef] [PubMed]
- Steere, A.C.; Strle, F.; Wormser, G.P.; Hu, L.T.; Branda, J.A.; Hovius, J.W.; Li, X.; Mead, P.S. Lyme borreliosis. Nat. Rev. Dis. Prim. 2016, 2, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, J.A.; Marrotte, R.R.; Desrosiers, N.; Fiset, J.; Gaitan, J.; Gonzalez, A.; Koffi, J.K.; Lapointe, F.J.; Leighton, P.A.; Lindsay, L.R.; et al. Climate change and habitat fragmentation drive the occurrence of Borrelia burgdorferi, the agent of Lyme disease, at the northeastern limit of its distribution. Evol. Appl. 2014, 7, 750–764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zell, R. Global climate change and the emergence/re-emergence of infectious diseases. Int. J. Med. Microbiol. Suppl. 2004, 293, 16–26. [Google Scholar] [CrossRef]
- Dumic, I.; Severnini, E. ‘Ticking bomb’: The impact of climate change on the incidence of Lyme disease. Can. J. Infect. Dis. Med. Microbiol. 2018, 2018, 5719081. [Google Scholar] [CrossRef] [Green Version]
- Wormser, G.P.; Dattwyler, R.; Shapiro, E.D.; Halperin, J.; Steere, A.C.; Klempner, M.S.; Krause, P.J.; Bakken, J.S.; Strle, F.; Stanek, G.; et al. The Clinical Assessment, Treatment, and Prevention of Lyme Disease, Human Granulocytic Anaplasmosis, and Babesiosis: Clinical Practice Guidelines by the Infectious Diseases Society of America. Clin. Infect. Dis. 2006, 43, 1089–1134. [Google Scholar] [CrossRef]
- Marques, A. Chronic Lyme disease: A review. Infect. Dis. Clin. N. Am. 2008, 22, 341–360. [Google Scholar] [CrossRef] [Green Version]
- Centers for Disease Control and Prevention. Signs and Symptoms of Untreated Lyme Disease; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2015.
- Moore, A.; Nelson, C.; Molins, C.; Mead, P.; Schriefer, M. Current guidelines, common clinical pitfalls, and future directions for laboratory diagnosis of Lyme disease, United States. Emerg. Infect. Dis. 2016, 22, 1169. [Google Scholar] [CrossRef] [Green Version]
- US Department of Health & Human Services. Tick-Borne Disease Working Group; US Department of Health & Human Services: Washington, DC, USA, 2018.
- Fix, A.D.; Peña, C.A.; Strickland, G.T. Racial Differences in Reported Lyme Disease Incidence. Am. J. Epidemiol. 2000, 152, 756–759. [Google Scholar] [CrossRef]
- Pearlman, S.I.; Leelawong, M.; Richardson, K.A.; Adams, N.M.; Russ, P.K.; Pask, M.E.; Wolfe, A.E.; Wessely, C.; Haselton, F.R. Low-Resource Nucleic Acid Extraction Method Enabled by High-Gradient Magnetic Separation. ACS Appl. Mater. Interfaces 2020, 12, 12457–12467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowakowski, J.; Schwartz, I.; Liveris, D.; Wang, G.; Aguero-Rosenfeld, M.E.; Girao, G.; McKenna, D.; Nadelman, R.B.; Cavaliere, L.F.; Wormser, G.P.; et al. Laboratory Diagnostic Techniques for Patients with Early Lyme Disease Associated with Erythema Migrans: A Comparison of Different Techniques. Clin. Infect. Dis. 2001, 33, 2023–2027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waddell, L.A.; Greig, J.; Mascarenhas, M.; Harding, S.; Lindsay, R.; Ogden, N. The Accuracy of Diagnostic Tests for Lyme Disease in Humans, A Systematic Review and Meta-Analysis of North American Research. PLoS ONE 2016, 11, e0168613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schutzer, S.E.; Body, B.A.; Boyle, J.; Branson, B.M.; Dattwyler, R.J.; Fikrig, E.; Gerald, N.J.; Gomes-Solecki, M.; Kintrup, M.; Ledizet, M.; et al. Direct Diagnostic Tests for Lyme Disease. Clin. Infect. Dis. 2019, 68, 1052–1057. [Google Scholar] [CrossRef]
- Liveris, D.; Wang, G.; Girao, G.; Byrne, D.W.; Nowakowski, J.; McKenna, D.; Nadelman, R.; Wormser, G.P.; Schwartz, I. Quantitative Detection of Borrelia burgdorferi in 2-Millimeter Skin Samples of Erythema Migrans Lesions: Correlation of Results with Clinical and Laboratory Findings. J. Clin. Microbiol. 2002, 40, 1249–1253. [Google Scholar] [CrossRef] [Green Version]
- Priem, S.; Rittig, M.G.; Kamradt, T.; Burmester, G.R.; Krause, A. An optimized PCR leads to rapid and highly sensitive detection of Borrelia burgdorferi in patients with Lyme borreliosis. J. Clin. Microbiol. 1997, 35, 685–690. [Google Scholar] [CrossRef] [Green Version]
- O’Rourke, M.; Traweger, A.; Lusa, L.; Stupica, D.; Maraspin, V.; Barrett, P.N.; Strle, F.; Livey, I. Quantitative Detection of Borrelia burgdorferi sensu lato in Erythema Migrans Skin Lesions Using Internally Controlled Duplex Real Time PCR. PLoS ONE 2013, 8, e63968. [Google Scholar] [CrossRef]
- Mosel, M.R.; Rebman, A.W.; Carolan, H.E.; Montenegro, T.; Lovari, R.; Schutzer, S.E.; Ecker, D.J.; Yang, T.; Ramadoss, N.S.; Robinson, W.H.; et al. Molecular Microbiological and Immune Characterization of a Cohort of Patients Diagnosed with Early Lyme Disease. J. Clin. Microbiol. 2020, 59, e00615-20. [Google Scholar] [CrossRef]
- Dunaj, J.; Moniuszko, A.; Zajkowska, J.; Pancewicz, S. The role of PCR in diagnostics of Lyme borreliosis. Przeglad Epidemiol. 2013, 67, 35–39. [Google Scholar]
- Bernard, Q.; Grillon, A.; Lenormand, C.; Ehret-Sabatier, L.; Boulanger, N. Skin Interface, a Key Player for Borrelia Multiplication and Persistence in Lyme Borreliosis. Trends Parasitol. 2020, 36, 304–314. [Google Scholar] [CrossRef]
- Shih, C.-M.; Pollack, R.J.; Telford, I.S.R.; Spielman, A. Delayed Dissemination of Lyme Disease Spirochetes from the Site of Deposition in the Skin of Mice. J. Infect. Dis. 1992, 166, 827–831. [Google Scholar] [CrossRef]
- Straubinger, R.K.; Summers, B.A.; Chang, Y.-F.; Appel, M.J. Persistence of Borrelia burgdorferi in experimentally infected dogs after antibiotic treatment. J. Clin. Microbiol. 1997, 35, 111–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samant, P.P.; Niedzwiecki, M.M.; Raviele, N.; Tran, V.; Mena-Lapaix, J.; Walker, D.I.; Felner, E.I.; Jones, D.P.; Miller, G.W.; Prausnitz, M.R. Sampling interstitial fluid from human skin using a microneedle patch. Sci. Transl. Med. 2020, 12, eaaw0285. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, G.; Bian, F.; Cai, L.; Zhao, Y. Encoded Microneedle Arrays for Detection of Skin Interstitial Fluid Biomarkers. Adv. Mater. 2019, 31, e1902825. [Google Scholar] [CrossRef]
- Al Sulaiman, D.; Chang, J.Y.H.; Bennett, N.R.; Topouzi, H.; Higgins, C.A.; Irvine, D.J.; Ladame, S. Hydrogel-Coated Microneedle Arrays for Minimally Invasive Sampling and Sensing of Specific Circulating Nucleic Acids from Skin Interstitial Fluid. ACS Nano 2019, 13, 9620–9628. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.-S.; Kong, Y.; Wang, Y.; Luo, Y.; Fan, X.; Xie, X.; Yang, B.-R.; Wu, M.X. Microneedles for transdermal diagnostics: Recent advances and new horizons. Biomaterials 2019, 232, 119740. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Lillehoj, P.B. Microneedle-based skin patch for blood-free rapid diagnostic testing. Microsyst. Nanoeng. 2020, 6, 96. [Google Scholar] [CrossRef] [PubMed]
- Yi, K.; Wang, Y.; Shi, K.; Chi, J.; Lyu, J.; Zhao, Y. Aptamer-decorated porous microneedles arrays for extraction and detection of skin interstitial fluid biomarkers. Biosens. Bioelectron. 2021, 190, 113404. [Google Scholar] [CrossRef]
- Dixon, R.V.; Skaria, E.; Lau, W.M.; Manning, P.; Birch-Machin, M.A.; Moghimi, S.M.; Ng, K.W. Microneedle-based devices for point-of-care infectious disease diagnostics. Acta Pharm. Sin. B 2021, 11, 2344–2361. [Google Scholar] [CrossRef]
- Park, S.; Kim, Y.J.; Park, S.; Hong, H.; Lee, J.; Kim, S.I.; Lee, K.; Ryu, W. Rapid Extraction and Detection of Biomolecules via a Microneedle Array of Wet-Crosslinked Methacrylated Hyaluronic Acid. Adv. Mater. Technol. 2022, 7, 2100874. [Google Scholar] [CrossRef]
- Samant, P.P.; Prausnitz, M.R. Mechanisms of sampling interstitial fluid from skin using a microneedle patch. Proc. Natl. Acad. Sci. USA 2018, 115, 4583–4588. [Google Scholar] [CrossRef]
- Zhang, N.; Zhou, X.; Liu, L.; Zhao, L.; Xie, H.; Yang, Z. Dissolving Polymer Microneedles for Transdermal Delivery of Insulin. Front. Pharmacol. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Paul, R.; Saville, A.C.; Hansel, J.C.; Ye, Y.; Ball, C.; Williams, A.; Chang, X.; Chen, G.; Gu, Z.; Ristaino, J.; et al. Extraction of Plant DNA by Microneedle Patch for Rapid Detection of Plant Diseases. ACS Nano 2019, 13, 6540–6549. [Google Scholar] [CrossRef]
- Li, H.; Feng, J.; Wang, Y.; Liu, G.; Chen, X.; Fu, L. Instant and Multiple DNA Extraction Method by Microneedle Patch for Rapid and on-Site Detection of Food Allergen-Encoding Genes. J. Agric. Food Chem. 2021, 69, 6879–6887. [Google Scholar] [CrossRef] [PubMed]
- Hassan, C.M.; Peppas, N.A. Structure and Morphology of Freeze/Thawed PVA Hydrogels. Macromolecules 2000, 33, 2472–2479. [Google Scholar] [CrossRef]
- Xu, N.; Zhang, M.; Xu, W.; Ling, G.; Yu, J.; Zhang, P. Swellable PVA/PVP hydrogel microneedle patches for the extraction of interstitial skin fluid toward minimally invasive monitoring of blood glucose level. Analyst 2022, 147, 1478–1491. [Google Scholar] [CrossRef]
- Oster, J.; Parker, J.; Brassard, L.À. Polyvinyl-alcohol-based magnetic beads for rapid and efficient separation of specific or unspecific nucleic acid sequences. J. Magn. Magn. Mater. 2001, 225, 145–150. [Google Scholar] [CrossRef]
- Schyma, C.; Huckenbeck, W.; Bonte, W. DNA-PCR analysis of bloodstains sampled by the polyvinyl-alcohol method. J. Forensic Sci. 1999, 44, 95–99. [Google Scholar] [CrossRef]
- Otsuka, E.; Suzuki, A. A simple method to obtain a swollen PVA gel crosslinked by hydrogen bonds. J. Appl. Polym. Sci. 2009, 114, 10–16. [Google Scholar] [CrossRef]
- Patterson, J.W.; Duncan, A.M.; McIntyre, K.C.; Lloyd, V.K. Evidence for genetic hybridization between Ixodes scapularis and Ixodes cookei. Can. J. Zool. 2017, 95, 527–537. [Google Scholar] [CrossRef] [Green Version]
- Bugmyrin, S.V.; Belova, O.A.; Bespyatova, L.A.; Ieshko, E.P.; Karganova, G.G. Morphological features of Ixodes persulcatus and I. ricinus hybrids: Nymphs and adults. Exp. Appl. Acarol. 2016, 69, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Blacklegged Tick Anatomy and Identification. Available online: https://www.tickipedia.org/blacklegged-tick-anatomy-and-identification/ (accessed on 20 May 2022).
- Sharma, A.; Pham, M.N.; Reyes, J.B.; Chana, R.; Yim, W.C.; Heu, C.C.; Kim, D.; Chaverra-Rodriguez, D.; Rasgon, J.L.; Harrell, R.A., II; et al. Cas9-mediated gene editing in the black-legged tick, Ixodes scapularis, by embryo injection and ReMOT Control. iScience 2022, 25, 103781. [Google Scholar] [CrossRef] [PubMed]
- Bockenstedt, L.K.; Gonzalez, D.; Mao, J.; Li, M.; Belperron, A.; Haberman, A. What ticks do under your skin: Two-photon intravital imaging of Ixodes scapularis feeding in the presence of the lyme disease spirochete. Yale J. Biol. Med. 2014, 87, 3. [Google Scholar] [PubMed]
- Kochhar, J.S.; Quek, T.C.; Soon, W.J.; Choi, J.; Zou, S.; Kang, L. Effect of Microneedle Geometry and Supporting Substrate on Microneedle Array Penetration into Skin. J. Pharm. Sci. 2013, 102, 4100–4108. [Google Scholar] [CrossRef] [PubMed]
- Makvandi, P.; Kirkby, M.; Hutton, A.R.; Shabani, M.; Yiu, C.K.; Baghbantaraghdari, Z.; Jamaledin, R.; Carlotti, M.; Mazzolai, B.; Mattoli, V. Engineering microneedle patches for improved penetration: Analysis, skin models and factors affecting needle insertion. Nano-Micro Lett. 2021, 13, 93. [Google Scholar] [CrossRef]
- Larrañeta, E.; Moore, J.; Vicente-Pérez, E.M.; González-Vázquez, P.; Lutton, R.; Woolfson, A.D.; Donnelly, R.F. A proposed model membrane and test method for microneedle insertion studies. Int. J. Pharm. 2014, 472, 65–73. [Google Scholar] [CrossRef] [Green Version]
- Arora, A.; Hakim, I.; Baxter, J.; Rathnasingham, R.; Srinivasan, R.; Fletcher, D.A.; Mitragotri, S. Needle-free delivery of macromolecules across the skin by nanoliter-volume pulsed microjets. Proc. Natl. Acad. Sci. USA 2007, 104, 4255–4260. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Das, D.B.; Rielly, C.D. Microneedle Assisted Micro-Particle Delivery from Gene Guns: Experiments Using Skin-Mimicking Agarose Gel. J. Pharm. Sci. 2014, 103, 613–627. [Google Scholar] [CrossRef] [Green Version]
- Permana, A.D.; Mir, M.; Utomo, E.; Donnelly, R.F. Withdrawn: Bacterially sensitive nanoparticle-based dissolving microneedles of doxycycline for enhanced treatment of bacterial biofilm skin infection: A proof of concept study. Int. J. Pharm. 2020, 2, 100047. [Google Scholar] [CrossRef]
- Bonfante, G.; Lee, H.; Bao, L.; Park, J.; Takama, N.; Kim, B. Comparison of polymers to enhance mechanical properties of microneedles for bio-medical applications. Micro Nano Syst. Lett. 2020, 8, 13. [Google Scholar] [CrossRef]
- Enfield, J.G.; O’Connell, M.-L.; Lawlor, K.; Jonathan, E.; O’Mahony, C.; Leahy, M.J. In-vivo dynamic characterization of microneedle skin penetration using optical coherence tomography. J. Biomed. Opt. 2010, 15, 46001. [Google Scholar] [CrossRef] [PubMed]
- Shu, B.; Wu, K.-H.; Emery, S.; Villanueva, J.; Johnson, R.; Guthrie, E.; Berman, L.; Warnes, C.; Barnes, N.; Klimov, A.; et al. Design and Performance of the CDC Real-Time Reverse Transcriptase PCR Swine Flu Panel for Detection of 2009 A (H1N1) Pandemic Influenza Virus. J. Clin. Microbiol. 2011, 49, 2614–2619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emery, S.L.; Erdman, D.D.; Bowen, M.D.; Newton, B.R.; Winchell, J.M.; Meyer, R.F.; Tong, S.; Cook, B.T.; Holloway, B.P.; McCaustland, K.A.; et al. Real-Time Reverse Transcription–Polymerase Chain Reaction Assay for SARS-associated Coronavirus. Emerg. Infect. Dis. 2004, 10, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Barbour, A.G.; Hayes, S.F.; Heiland, R.A.; Schrumpf, M.E.; Tessier, S.L. A Borrelia-specific monoclonal antibody binds to a flagellar epitope. Infect. Immun. 1986, 52, 549–554. [Google Scholar] [CrossRef] [Green Version]
- De Leeuw, B.H.; Maraha, B.; Hollemans, L.; Sprong, H.; Brandenburg, A.H.; Westenend, P.J.; Kusters, J.G. Evaluation of Borrelia real time PCR DNA targeting OspA, FlaB and 5S–23S IGS and Borrelia 16S rRNA RT-qPCR. J. Microbiol. Methods 2014, 107, 41–46. [Google Scholar] [CrossRef]
- Wills, M.K.B.; Kirby, A.M.; Lloyd, V.K. Detecting the lyme disease spirochete, Borrelia burgdorferi, in ticks using nested PCR. JoVE J. Vis. Exp. 2018, 132, e56471. [Google Scholar] [CrossRef] [Green Version]
- Johnson, A.R.; Procopio, A.T. Low cost additive manufacturing of microneedle masters. 3D Print. Med. 2019, 5, 2. [Google Scholar] [CrossRef]
- Johnson, A.R.; Caudill, C.L.; Tumbleston, J.R.; Bloomquist, C.; Moga, K.A.; Ermoshkin, A.; Shirvanyants, D.; Mecham, S.J.; Luft, J.C.; DeSimone, J.M. Single-Step Fabrication of Computationally Designed Microneedles by Continuous Liquid Interface Production. PLoS ONE 2016, 11, e0162518. [Google Scholar] [CrossRef] [Green Version]
- Oh, N.G.; Hwang, S.Y.; Na, Y.H. Fabrication of a PVA-Based Hydrogel Microneedle Patch. ACS Omega 2022, 7, 25179–25185. [Google Scholar] [CrossRef]
- Paul, R.; Ostermann, E.; Chen, Y.; Saville, A.C.; Yang, Y.; Gu, Z.; Whitfield, A.E.; Ristaino, J.B.; Wei, Q. Integrated microneedle-smartphone nucleic acid amplification platform for in-field diagnosis of plant diseases. Biosens. Bioelectron. 2021, 187, 113312. [Google Scholar] [CrossRef]
- Flaten, G.E.; Palac, Z.; Engesland, A.; Filipović-Grčić, J.; Vanić, Ž.; Škalko-Basnet, N. In vitro skin models as a tool in optimization of drug formulation. Eur. J. Pharm. Sci. 2015, 75, 10–24. [Google Scholar] [CrossRef]
- Kim, M.; Jung, B.; Park, J.-H. Hydrogel swelling as a trigger to release biodegradable polymer microneedles in skin. Biomaterials 2012, 33, 668–678. [Google Scholar] [CrossRef]
- Dick, I.P.; Scott, R.C. Pig Ear Skin as an In-vitro Model for Human Skin Permeability. J. Pharm. Pharmacol. 1992, 44, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Moniz, T.; Lima, S.A.C.; Reis, S. Human skin models: From healthy to disease-mimetic systems; characteristics and applications. Br. J. Pharmacol. 2020, 177, 4314–4329. [Google Scholar] [CrossRef] [PubMed]
- Jacobi, U.; Kaiser, M.; Toll, R.; Mangelsdorf, S.; Audring, H.; Otberg, N.; Sterry, W.; Lademann, J. Porcine ear skin: An in vitro model for human skin. Ski. Res. Technol. 2007, 13, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Cutler, S.J.; Vayssier-Taussat, M.; Estrada-Peña, A.; Potkonjak, A.; Mihalca, A.D.; Zeller, H. Tick-borne diseases and co-infection: Current considerations. Ticks Tick-Borne Dis. 2021, 12, 101607. [Google Scholar] [CrossRef] [PubMed]
- Nieto, N.C.; Porter, W.T.; Wachara, J.C.; Lowrey, T.J.; Martin, L.; Motyka, P.J.; Salkeld, D.J. Using citizen science to describe the prevalence and distribution of tick bite and exposure to tick-borne diseases in the United States. PLoS ONE 2018, 13, e0199644. [Google Scholar] [CrossRef] [PubMed]
- Marques, A.; Telford, S.R.; Turk, S.-P.; Chung, E.; Williams, C.; Dardick, K.; Krause, P.J.; Brandeburg, C.; Crowder, C.D.; Carolan, H.E.; et al. Xenodiagnosis to Detect Borrelia burgdorferi Infection: A First-in-Human Study. Clin. Infect. Dis. 2014, 58, 937–945. [Google Scholar] [CrossRef]
- Jacobs, M.B.; Grasperge, B.J.; Doyle-Meyers, L.A.; Embers, M.E. Borrelia burgdorferi Migration Assays for Evaluation of Chemoattractants in Tick Saliva. Pathogens 2022, 11, 530. [Google Scholar] [CrossRef]
- Van Gundy, T.J.; Ullmann, A.J.; Brandt, K.S.; Gilmore, R.D. A transwell assay method to evaluate Borrelia burgdorferi sensu stricto migratory chemoattraction toward tick saliva proteins. Ticks Tick-Borne Dis. 2021, 12, 101782. [Google Scholar] [CrossRef]
- Nuttall, P.A. Tick saliva and its role in pathogen transmission. Wien. Klin. Wochenschr. 2019, 1–12. [Google Scholar] [CrossRef]
- Scholl, D.C.; Embers, M.E.; Caskey, J.R.; Kaushal, D.; Mather, T.N.; Buck, W.R.; Morici, L.A.; Philipp, M.T. Immunomodulatory effects of tick saliva on dermal cells exposed to Borrelia burgdorferi, the agent of Lyme disease. Parasites Vectors 2016, 9, 394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Šimo, L.; Kazimirova, M.; Richardson, J.; Bonnet, S.I. The Essential Role of Tick Salivary Glands and Saliva in Tick Feeding and Pathogen Transmission. Front. Cell. Infect. Microbiol. 2017, 7, 281. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Hammond-McKibben, D.; Guralski, D.; Lobo, S.; Fiedler, P.N. Development of a sensitive molecular diagnostic assay for detecting Borrelia burgdorferi DNA from the blood of Lyme disease patients by digital PCR. PLoS ONE 2020, 15, e0235372. [Google Scholar] [CrossRef] [PubMed]
- Liveris, D.; Schwartz, I.; Bittker, S.; Cooper, D.; Iyer, R.; Cox, M.E.; Wormser, G.P. Improving the Yield of Blood Cultures from Patients with Early Lyme Disease. J. Clin. Microbiol. 2011, 49, 2166–2168. [Google Scholar] [CrossRef] [PubMed]


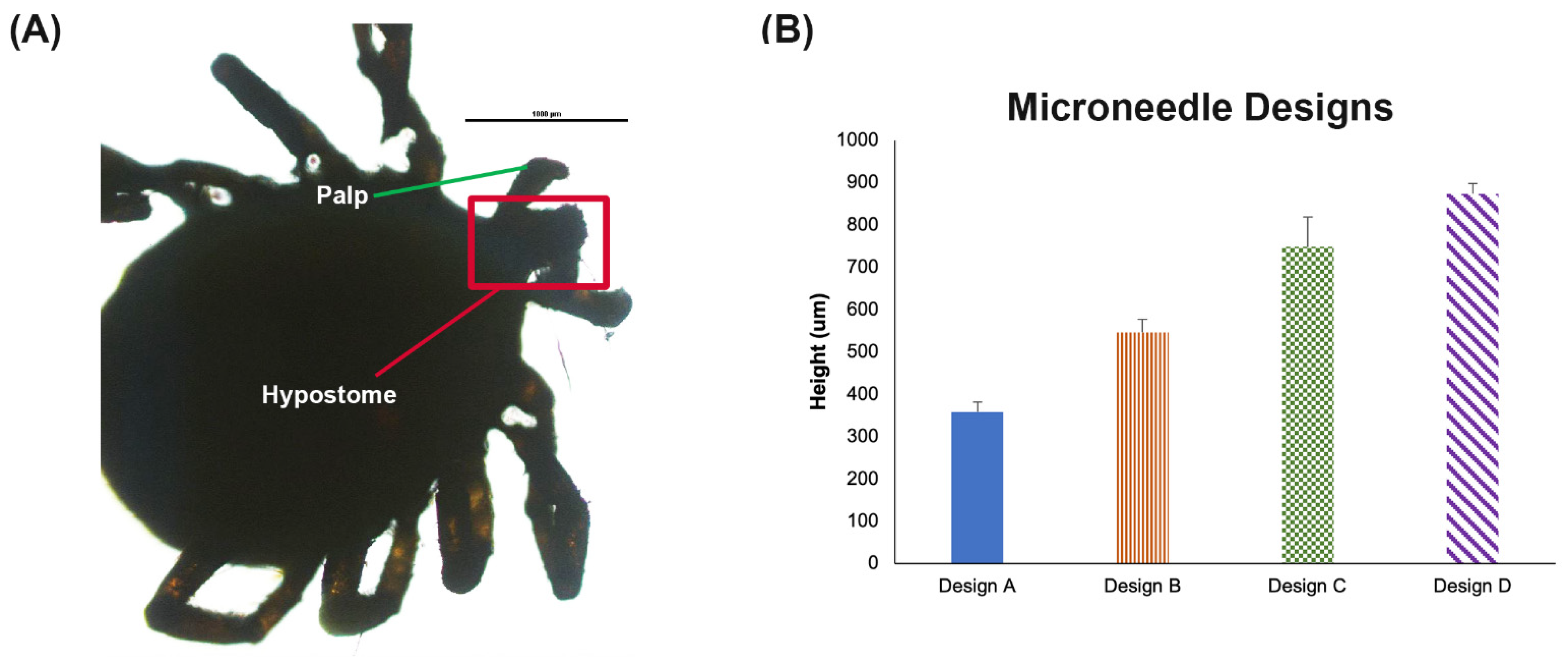

| MN Patch ID | Needle Length (µm) | Base (µm) | Distance between Needles (µm) |
|---|---|---|---|
| Design A | 1000 | 250 | 1000 |
| Design B | 1000 | 350 | 1000 |
| Design C | 1200 | 350 | 1000 |
| Design D | 1600 | 350 | 1000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kight, E.; Alfaro, R.; Gadila, S.K.G.; Chang, S.; Evans, D.; Embers, M.; Haselton, F. Direct Capture and Early Detection of Lyme Disease Spirochete in Skin with a Microneedle Patch. Biosensors 2022, 12, 819. https://doi.org/10.3390/bios12100819
Kight E, Alfaro R, Gadila SKG, Chang S, Evans D, Embers M, Haselton F. Direct Capture and Early Detection of Lyme Disease Spirochete in Skin with a Microneedle Patch. Biosensors. 2022; 12(10):819. https://doi.org/10.3390/bios12100819
Chicago/Turabian StyleKight, Emily, Rosana Alfaro, Shiva Kumar Goud Gadila, Shuang Chang, David Evans, Monica Embers, and Frederick Haselton. 2022. "Direct Capture and Early Detection of Lyme Disease Spirochete in Skin with a Microneedle Patch" Biosensors 12, no. 10: 819. https://doi.org/10.3390/bios12100819
APA StyleKight, E., Alfaro, R., Gadila, S. K. G., Chang, S., Evans, D., Embers, M., & Haselton, F. (2022). Direct Capture and Early Detection of Lyme Disease Spirochete in Skin with a Microneedle Patch. Biosensors, 12(10), 819. https://doi.org/10.3390/bios12100819






