Kinetics of Drug Molecule Interactions with a Newly Developed Nano-Gold-Modified Spike Protein Electrochemical Receptor Sensor
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Instruments
2.2. Experimental Method
2.2.1. Pretreatment and Electrochemical Characterization of Glassy Carbon Electrodes
2.2.2. Preparation of the S Protein Receptor Sensor
- Five μL of sulfur Thi-Chit compound solution was added to 2.5 mL of 2% chitosan solution (the chitosan solution used 2 g of chitosan dissolved in 100 mL of 1% volume acetic acid solution). This was stirred until completely dissolved to 0.32 mL of 10% glutaraldehyde solution, mixed well by blowing, then 0.2 mL of 0.01 M thionin solution was added, and finally a 2% volume of acetic acid solution was added to make a total volume of 6 mL. This was mixed well and then the solution was ready to use. The solution was dropcast on the working electrode surface. After drying in the ultra-clean bench, the cyclic voltammetry curve was measured. Then, the electrode was immersed in 0.5 mol/L NaOH solution for 5 min, removed and rinsed three times with ultrapure water, and placed in ultrapure water for 0.5 h after cleaning.
- The above electrode was dried and self-assembled in nano gold–horseradish peroxidase solution (1 mL of gold nanosol [18] was mixed with 1 mL of 2.0 g/L horseradish peroxidase, stirred for 2 h with a magnetic stirrer and allowed to stand for 12 h at 4°C before use) at 4 °C for 24 h; its cyclic voltammetry curves were measured after assembly, and the characterization of gold nanoparticles is provided in the attached document.
- After washing the electrode surface with ultrapure water, the S protein solution was added dropwise on the electrode surface and self-assembled at 4 °C for 24 h. The cyclic voltammetry curve was measured after assembling.
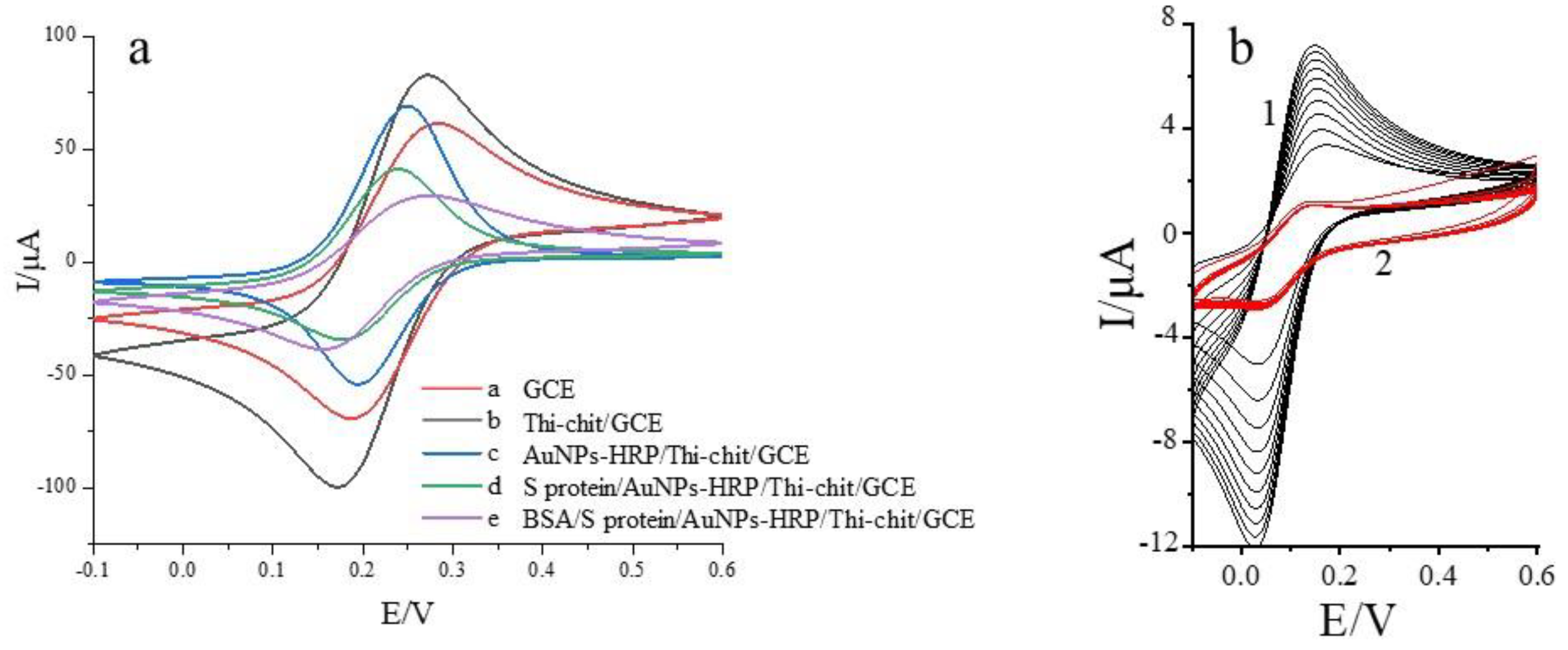
- The electrode was removed and washed with ultrapure water, finally coated with BSA solution (0.5 g/100 mL) and incubated for 1 h at 37 °C to close the non-specific sites, and their cyclic voltammetry curve was measured. The nano-gold-modified receptor spike protein electrochemical biosensor was obtained, and the assembly process is shown in Figure 1: Characterization of electrode surface morphology during electrode assembly using SEM.
2.2.3. Assessment of the Interaction of the S Protein Receptor Sensor with Related Drugs
2.2.4. Simulation of Molecular Docking of the Spike Protein with Six Ligands
3. Results
3.1. Electrochemical Characterization of Spike Protein Receptor Sensors
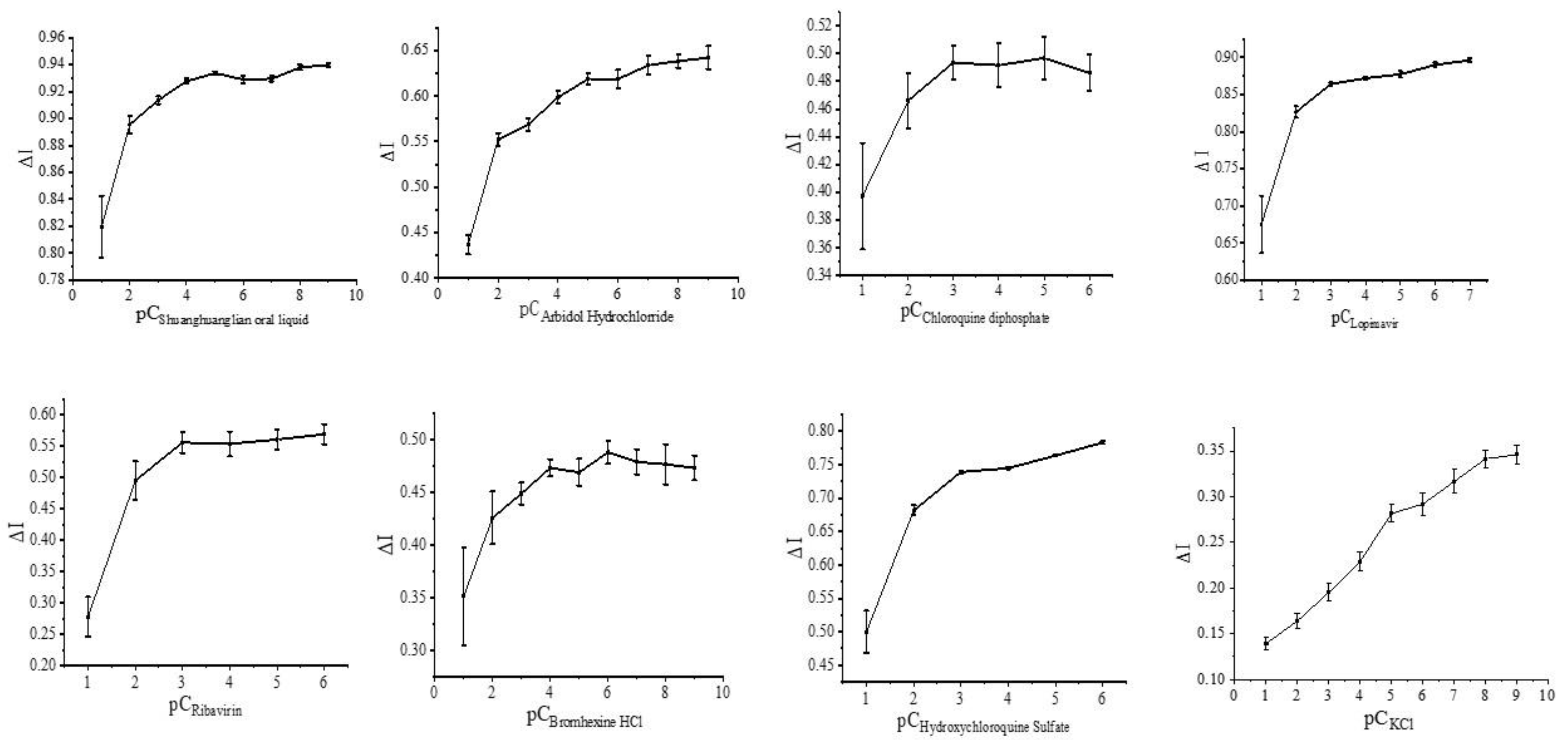
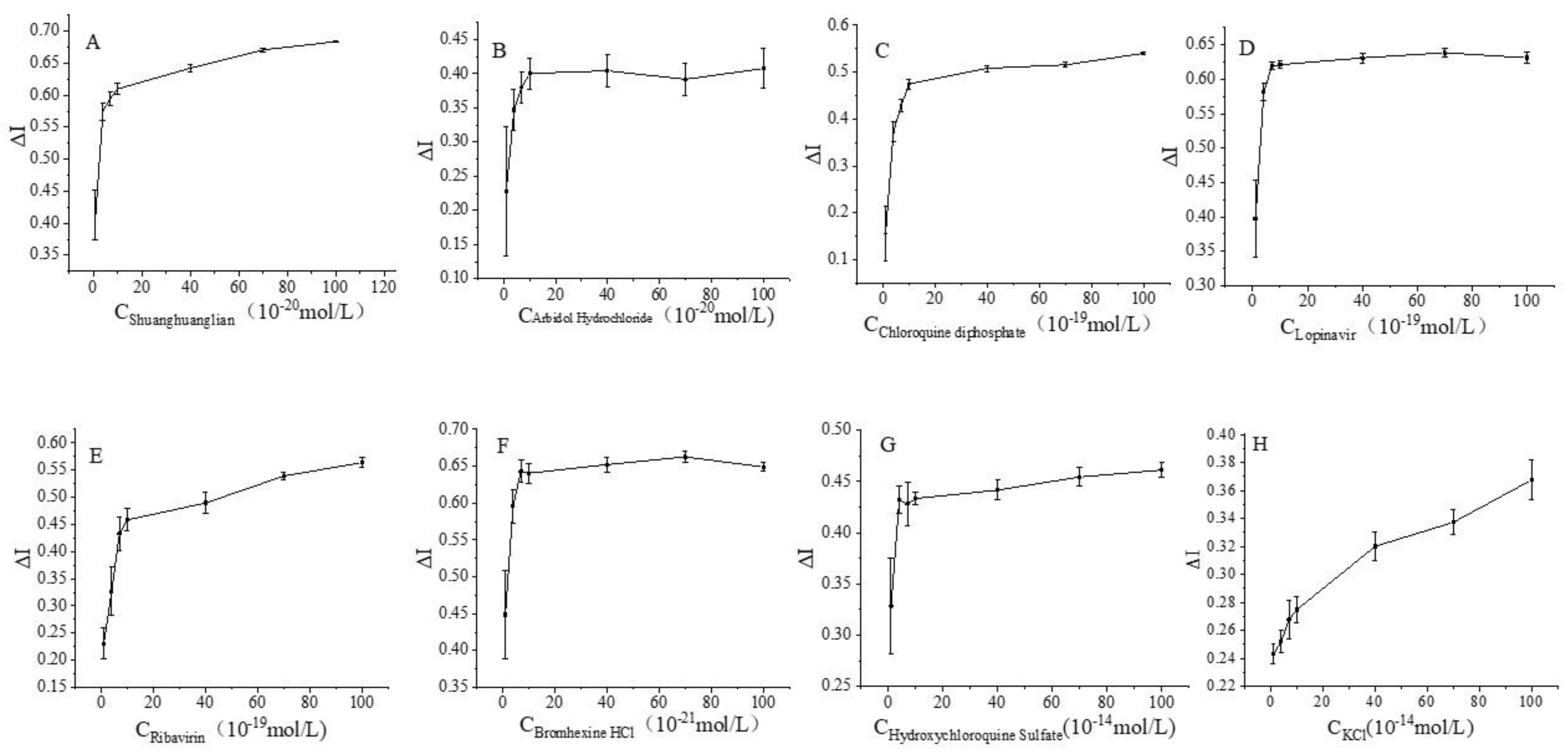
3.2. Regulation of the Spike Protein Receptor Sensor and Related Drugs
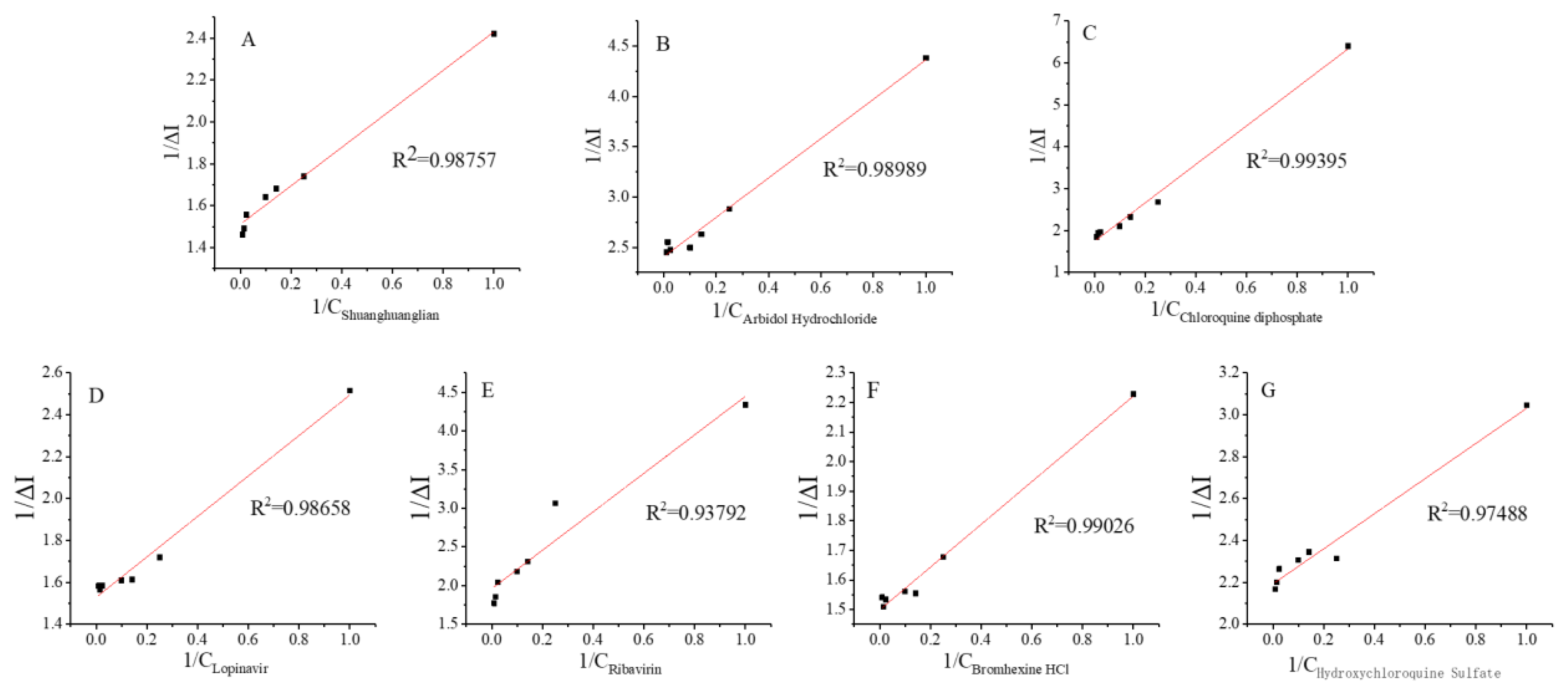
3.3. Kinetics of the Interaction of the Spike Protein Receptor with Different Drug Molecules
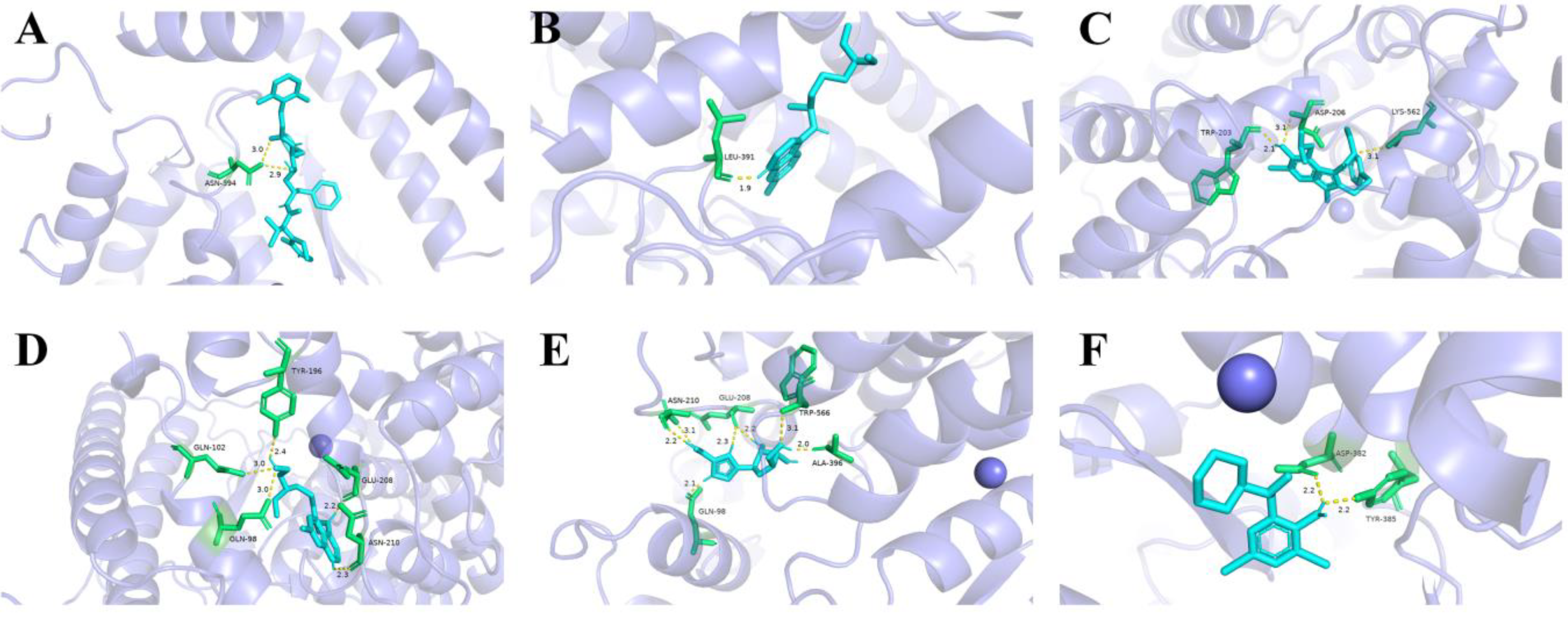
3.4. Molecular Docking Simulation Results
3.5. Molecular Docking Simulation and Ka Value
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kashefi-Kheyrabadi, L.; Nguyen, H.V.; Go, A.; Baek, C.; Jang, N.; Lee, J.M.; Nam-Hyuk, C.; Min, J.; Min-Ho, L. Rapid, multiplexed, and nucleic acid amplification-free detection of SARS-CoV-2 RNA using an electrochemical biosensor. Biosens. Bioelectron. 2022, 195, 113649. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, S.; Silakari, O. Scaffold morphing of arbidol (umifenovir) in search of multi-targeting therapy halting the interaction of SARS-CoV-2 with ACE2 and other proteases involved in COVID-19. Virus Res. 2020, 28, 198146. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.; Kim, S.E.; Lee, Y.H.; Hwang, Y.H.; Kim, S.H.; Young, M.; Chung, G.T.; Kim, Y.J.; Lee, J.A. Immunogenicity of candidate SARS-CoV-2 DNA vaccines based on the spike protein. Virology 2022, 573, 118–123. [Google Scholar] [CrossRef] [PubMed]
- WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 22 June 2022).
- Patel, T.K.; Patel, P.B.; Barvaliya, M.; Saurabh, M.K.; Bhalla, H.L.; Khosla, P.P. Efficacy and safety of lopinavir-ritonavir in COVID-19: A systematic review of randomized controlled trials. J. Infect. Public Health 2021, 14, 740–748. [Google Scholar] [CrossRef]
- Velusamy, P.; Kiruba, K.; Su, C.H.; Arun, V.; Anbu, P.; Gopinath, S.C.B.; Vaseeharan, B. SARS-CoV-2 spike protein: Site-specific breakpoints for the development of COVID-19 vaccines. J. King Saud Univ. Sci. 2021, 33, 101648. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Verma, S.; Patil, V.M.; Gupta, M.K. Mutation informatics: SARS-CoV-2 receptor-binding domain of the spike protein. Drug Discov. Today 2022, 27, 103312. [Google Scholar] [CrossRef]
- Pupovci, H.L. Molecular mechanisms involved in pathogenicity of SARS-CoV-2: Immune evasion and implications for therapeutic strategies. Biomed. Pharmacother. 2022, 153, 113368. [Google Scholar] [CrossRef]
- Chan, W.K.B.; Zhang, Y. Virtual Screening of Human Class-A GPCRs Using Ligand Profiles Built on Multiple Ligand–Receptor Interactions. J. Mol. Biol. 2020, 432, 4872–4890. [Google Scholar] [CrossRef]
- Zhao, Z.; Bourne, P.E. Harnessing systematic protein–ligand interaction fingerprints for drug discovery. Drug Discov. Today 2022, 27, 103319. [Google Scholar] [CrossRef]
- Kuzikov, M.; Woens, J.; Zaliani, A.; Hambach, J.; Eden, T.; Fehse, B.; Ellinger, B.; Riecken, K. High-throughput drug screening allowed identification of entry inhibitors specifically targeting different routes of SARS-CoV-2 Delta and Omicron/BA.1. Biomed. Pharmacother. 2022, 151, 113104. [Google Scholar] [CrossRef]
- Xu, T.; Zheng, W.; Huang, R. High-throughput screening assays for SARS-CoV-2 drug development: Current status and future directions. Drug Discov. Today 2021, 26, 2439–2444. [Google Scholar] [CrossRef]
- Aallaei, M.; Molaakbari, E.; Mostafavi, P.; Salarizaden, N.; Maleksah, R.E.; Afzali, D. Investigation of Cu metal nanoparticles with different morphologies to inhibit SARS-CoV-2 main protease and spike glycoprotein using Molecular Docking and Dynamics Simulation. J. Mol. Struct. 2022, 1253, 132301. [Google Scholar] [CrossRef]
- Kumar, V.; Liu, H.; Wu, C. Drug repurposing against SARS-CoV-2 receptor binding domain using ensemble-based virtual screening and molecular dynamics simulations. Comput. Biol. Med. 2021, 135, 104634. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, C.; Ke, X.; Luo, D.; Liu, Y.; Chen, Q.; Wang, H.; Song, X.; Zheng, Z. Development of a biosensor assessing SARS-CoV-2 main protease proteolytic activity in living cells for antiviral drugs screening. Virol. Sin. 2022, 37, 459–461. [Google Scholar] [CrossRef]
- Dey-Rao, R.; Smith, G.R.; Timilsina, U.; Falls, Z.; Samudrala, R.; Stavrou, S.; Melendy, T. A fluorescence-based, gain-of-signal, live cell system to evaluate SARS-CoV-2 main protease inhibition. Antivir. Res. 2021, 195, 105183. [Google Scholar] [CrossRef]
- Vankadari, N. Arbidol: A potential antiviral drug for the treatment of SARS-CoV-2 by blocking trimerization of the spike glycoprotein. Int. J. Antimicrob. Agents 2020, 56, 105998. [Google Scholar] [CrossRef]
- Kang, X.; Pang, G.; Liang, X.; Wang, M.; Liu, J.; Zhu, W. Study on a hydrogen peroxide biosensor based on horseradish peroxidase/GNPs-thionine/chitosan. Electrochim. Acta 2019, 62, 327–334. [Google Scholar] [CrossRef]
- Xiao, S.; Zhang, Y.; Song, P.; Xie, J.; Pang, J. The investigation of allosteric regulation mechanism of analgesic effect using SD rat taste bud tissue biosensor. Biosens. Bioelectron. 2019, 126, 815–823. [Google Scholar] [CrossRef]
- Wang, X.; Lu, D.; Liu, Y.; Wang, W.; Ren, R.; Li, M.; Pang, G. Electrochemical signal amplification strategies and their use in olfactory and taste evaluation. Biosensors 2022, 12, 566. [Google Scholar] [CrossRef]
- Luo, L.; Qiu, Q.; Huang, F.; Liu, K.; Lan, Y.; Li, X.; Huang, Y.; Cui, L.; Luo, H. Drug repurposing against coronavirus disease 2019 (COVID-19): A review. J. Pharm. Anal. 2021, 11, 683–690. [Google Scholar] [CrossRef] [PubMed]
- Presti, A.L.; Rezza, G.; Stefanelli, P. Selective pressure on SARS-CoV-2 protein coding genes and glycosylation site prediction. Cell 2020, 6, e05001. [Google Scholar] [CrossRef] [PubMed]
- Semper, C.; Watanabe, N.; Savchenko, A. Structural characterization of nonstructural protein 1 from SARS-CoV-2. Cell 2021, 24, 101903. [Google Scholar] [CrossRef] [PubMed]
- Kehinde, I.; Egbejimi, A.; Kaur, M. Inhibitory mechanism of Ambroxol and Bromhexine Hydrochlorides as potent blockers of molecular interaction between SARS-CoV-2 spike protein and human angiotensin-converting Enzyme-2. J. Mol. Graph. Model. 2022, 114, 108201. [Google Scholar] [CrossRef]
- Habtemariam, S.; Nabavi, S.F.; Ghavami, S.; Cismaru, C.A.; Berindan-Neagoe, L.; Nabavi, S.M. Possible use of the mucolytic drug, bromhexine hydrochloride, as a prophylactic agent against SARS-CoV-2 infection based on its action on the Transmembrane Serine Protease 2. Pharmacol. Res. 2020, 157, 104853. [Google Scholar] [CrossRef]
- Millet, J.K.; Whittaker, G.R. Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015, 202, 120–134. [Google Scholar] [CrossRef]
- Ohnishi, T.; Nakamura, T.; Shima, K.; Noguchi, K.; Chiba, N.; Matsuguchi, T. Periodontitis promotes the expression of gingival transmembrane serine protease 2 (TMPRSS2), a priming protease for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). J. Oral Biosci. 2022, 64, 229–236. [Google Scholar] [CrossRef]
- Rahmati, Z.; Roushani, M.; Hosseini, H.; Choobin, H. An electrochemical immunosensor using SARS-CoV-2 spike protein-nickel hydroxide nanoparticles bio-conjugate modified SPCE for ultrasensitive detection of SARS-CoV-2 antibodies. Microchem. J. 2021, 170, 106718. [Google Scholar] [CrossRef]
- Daoudi, K.; Ramachandran, K.; Alawadhi, H.; Boukherroub, R.; Dogheche, E.; El Khakani, M.A.; Gaidi, M. Ultra-sensitive and fast optical detection of the spike protein of the SARS-CoV-2 using AgNPs/SiNWs nanohybrid based sensors. Surf. Interfaces 2021, 27, 101454. [Google Scholar] [CrossRef]
- Abrego-Martinez, J.C.; Jafari, M.; Chergui, S.; Pavel, C.; Che, D.; Siaj, M. Aptamer-based electrochemical biosensor for rapid detection of SARS-CoV-2: Nanoscale electrode-aptamer-SARS-CoV-2 imaging by photo-induced force microscopy. Biosens. Bioelectron. 2022, 195, 113595. [Google Scholar] [CrossRef]
- Liv, L.; Çoban, G.; Nakiboğlu, N. A rapid, ultrasensitive voltammetric biosensor for determining SARS-CoV-2 spike protein in real samples. Biosens. Bioelectron. 2021, 192, 113497. [Google Scholar] [CrossRef]
- Layqah, L.; Eissa, S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchim Acta 2019, 186, 224–234. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Li, X.; Pan, J.; Zhang, Y.; Zhang, L.; Wang, C.; Yan, X.; Liu, X.; Lu, G. Ultrasensitive detection of SARS-CoV-2 spike protein in untreated saliva using SERS-based biosensor. Biosens. Bioelectron. 2021, 190, 113421. [Google Scholar] [CrossRef]
- Kim, H.E.; Schuck, A.; Lee, S.H.; Lee, Y.; Kang, M.; Kim, Y.-S. Sensitive electrochemical biosensor combined with isothermal amplification for point-of-care COVID-19 tests. Biosens. Bioelectron. 2021, 182, 113168. [Google Scholar] [CrossRef]
- Raziq, A.; Kidakova, A.; Boroznjak, R.; Reut, J.; Öpik, A.; Syritski, V. Development of a portable MIP-based electrochemical sensor for detection of SARS-CoV-2 antigen. Biosens. Bioelectron. 2021, 178, 113029. [Google Scholar] [CrossRef]



| Mode | Affinity (kcal/mol) | Dist From rmsd l.b. | Best Mode rmsd u.b. |
|---|---|---|---|
| 1 | −8.9 | 0.000 | 0.000 |
| 2 | −8.7 | 3.837 | 7.715 |
| 3 | −8.5 | 3.917 | 7.819 |
| 4 | −8.2 | 3.689 | 8.673 |
| 5 | −8.1 | 3.782 | 7.807 |
| 6 | −8.1 | 21.586 | 29.164 |
| 7 | −8.0 | 14.650 | 19.782 |
| 8 | −7.8 | 3.704 | 9.997 |
| Drug Molecule | Chemical Structure | Ka value | Affinity | Residue Interactions |
|---|---|---|---|---|
| Bromohexine | 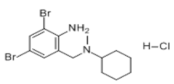 | 4.823 × 10−22 mol/L | −5.8 kcal/mol | HB: Tyr385, Asp382 HI: His401, His378, Ala348, Asp350, Thr347 |
| Shuanghuanglian oral liquid | — | 6.083 × 10−21 mol/L | — | — |
| Arbidol | 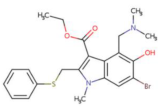 | 8.140 × 10−21 mol/L | −5.9 kcal/mol | HB: Asp206, Trp203, Lys562 HI: Tyr202, Asn397, Asn394, Gly205, Glu398, Tyr196 |
| Lopinavir |  | 6.325 × 10−20 mol/L | −8.9 kcal/mol | HB: Asn394 HI: Trp69, Ala99, Leu73, Leu391, Arg393, Phe390, Asp350, Phe40, Trp349, Ala348, His378, His401, Glu402 |
| Ribavirin | 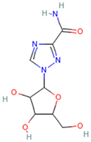 | 1.270 × 10−19 mol/L | −7.0 kcal/mol | HB: Ala396, Glu208, Trp566, Gln98, Asn210 HI: Pro565, Val209, Lys562, Leu95 |
| Chloroquine diphosphate | 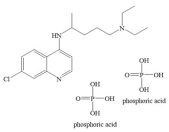 | 2.644 × 10−19 mol/L | −4.7 kcal/mol | HB: Leu391 HI: Phe390, Leu73, Lys74, Gln102 |
| Hydroxychloroquine |  | 3.825 × 10−15 mol/L | −6.5 kcal/mol | HB: Tyr196, Glu208, Gln102, Gln98, Asn210 HI: Val209, Lys562, Gly205, Leu95 |
| Sensor Type | Analyte | Electrode Type | Minimum Detection Limit | Measurement Method |
|---|---|---|---|---|
| electrochemical biosensor [29] | spike protein | screen-printed carbon electrode(SPCE) | 0.3 fg/mL | differential pulse voltammetry(DPV) |
| surface-enhanced Raman spectroscopy(SERS) sensor [30] | spike protein RBD | silicon nanowires | 9.3 × 10−12 mol/L | SERS spectroscopy |
| aptasensor [31] | spike protein RBD | screen-printed carbon electrode | 66 pg/mL | electrochemical impedance spectroscopy |
| electrochemical biosensor [32] | spike protein | glassy carbon electrode and SPCE | 1 ag/mL~10 fg/mL | voltammetry |
| electrochemical immunosensor [33] | spike protein | carbon electrodes (DEP) | 0.4 pg/mL for HCoV, 1.0 pg/mL for MERS-CoV | square wave voltammetry (SWV) |
| SERS-based biosensor [34] | SARS-CoV-2 virus in untreated saliva | silicon wafer | 6.07 pg/mL | SERS spectroscopy |
| electrochemical biosensor combined with recombinase polymerase amplification (RPA) [35] | SARS-CoV-2 | multi-microelectrode array | 0.972 fg/μL for RdRP gene, 3.925 fg/μL for N gene | DPV |
| a nucleic acid amplification-free electrochemical biosensor [1] | SARS-CoV-2 RNA | SPCE | 5.0 ag/μL for S gene, 6.8 ag/μL for Orf gene | square-wave voltammetry (SWV) |
| molecularly imprinted polymer-based electrochemical sensor [36] | SARS-CoV-2 nucleoprotein (ncovNP) | thin film electrode | 1.5 × 10−14 mol/L | DPV |
| electrochemical receptor sensors (this study) | spike protein | glassy carbon electrode | 3.3 × 10−20 mol/L | cyclic voltammetry |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, D.; Liu, D.; Wang, X.; Liu, Y.; Liu, Y.; Ren, R.; Pang, G. Kinetics of Drug Molecule Interactions with a Newly Developed Nano-Gold-Modified Spike Protein Electrochemical Receptor Sensor. Biosensors 2022, 12, 888. https://doi.org/10.3390/bios12100888
Lu D, Liu D, Wang X, Liu Y, Liu Y, Ren R, Pang G. Kinetics of Drug Molecule Interactions with a Newly Developed Nano-Gold-Modified Spike Protein Electrochemical Receptor Sensor. Biosensors. 2022; 12(10):888. https://doi.org/10.3390/bios12100888
Chicago/Turabian StyleLu, Dingqiang, Danyang Liu, Xinqian Wang, Yujiao Liu, Yixuan Liu, Ruijuan Ren, and Guangchang Pang. 2022. "Kinetics of Drug Molecule Interactions with a Newly Developed Nano-Gold-Modified Spike Protein Electrochemical Receptor Sensor" Biosensors 12, no. 10: 888. https://doi.org/10.3390/bios12100888
APA StyleLu, D., Liu, D., Wang, X., Liu, Y., Liu, Y., Ren, R., & Pang, G. (2022). Kinetics of Drug Molecule Interactions with a Newly Developed Nano-Gold-Modified Spike Protein Electrochemical Receptor Sensor. Biosensors, 12(10), 888. https://doi.org/10.3390/bios12100888







