Electrochemical Biosensors for Pathogen Detection: An Updated Review
Abstract
:1. Introduction
2. Pathogen Detection with Electrochemical Biosensors
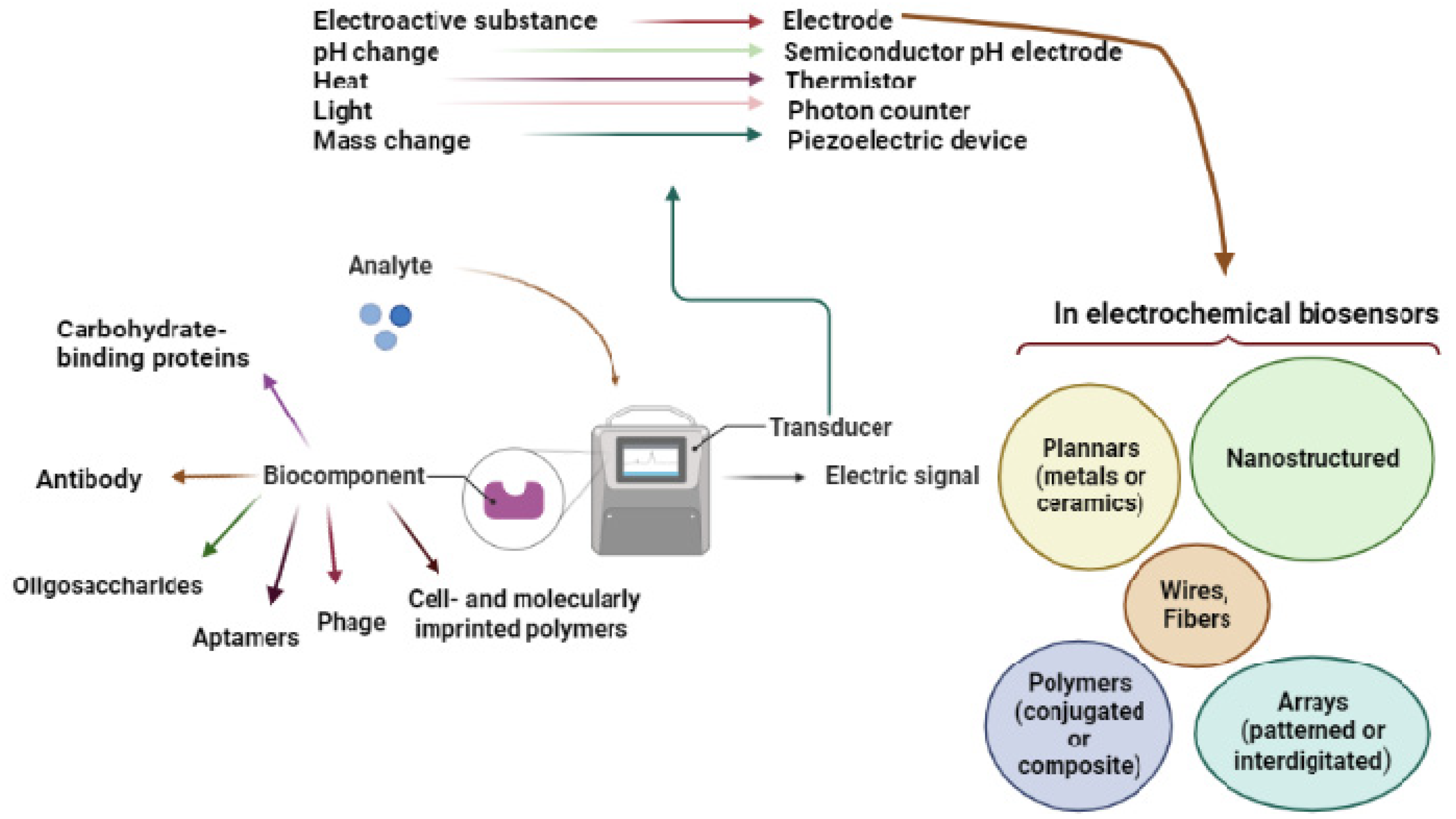
2.1. Transduction Elements
2.1.1. Metal Electrodes
2.1.2. Ceramic Electrodes
2.1.3. Polymer Electrodes
2.1.4. The shape and Design of the Electrodes
2.1.5. Electrode Nanostructuring
2.1.6. Complimentary Transduction Components
2.2. Biorecognition Elements
2.2.1. Antibodies and Antibody Fragments
2.2.2. Carbohydrate-Binding Proteins
2.2.3. Oligosaccharides
2.2.4. Oligonucleotides
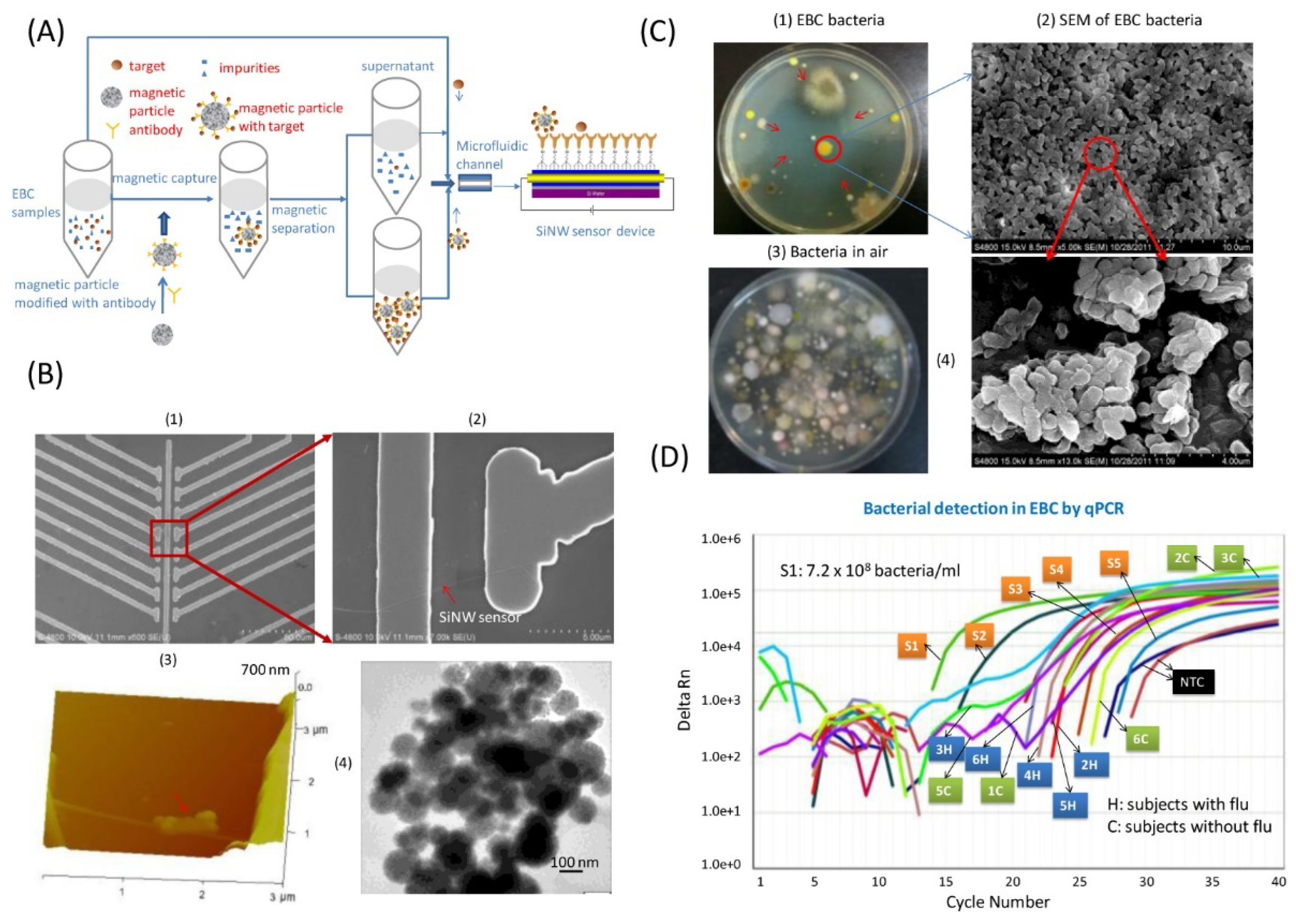
2.2.5. Phages
2.2.6. Cell and Molecularly Imprinted Polymers
2.2.7. Clustered Regularly Interspaced Short Palindromic Repeats/Associated Nuclease (CRISPR-Cas)
2.2.8. Antimicrobial Peptides
2.3. Biosensing and Surface Immobility
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dye, C. After 2015: Infectious diseases in a new era of health and development. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130426. [Google Scholar] [CrossRef] [Green Version]
- Khan, R.S.; Khurshid, Z.; Yahya Ibrahim Asiri, F. Advancing point-of-care (poc) testing using human saliva as liquid biopsy. Diagnostics 2017, 7, 39. [Google Scholar] [CrossRef]
- Moonla, C.; Lee, D.H.; Rokaya, D.; Rasitanon, N.; Kathayat, G.; Lee, W.-Y.; Kim, J.; Jeerapan, I. Review—Lab-in-a-mouth and advanced point-of-care sensing systems: Detecting bioinformation from the oral cavity and saliva. ECS Sens. Plus 2022, 1, 021603. [Google Scholar] [CrossRef]
- Nasrollahzadeh, M.; Sajjadi, M.; Soufi, G.J.; Iravani, S.; Varma, R.S. Nanomaterials and nanotechnology-associated innovations against viral infections with a focus on coronaviruses. Nanomaterials 2020, 10, 1072. [Google Scholar] [CrossRef]
- Hamid, H.; Khurshid, Z.; Adanir, N.; Zafar, M.S.; Zohaib, S. COVID-19 pandemic and role of human saliva as a testing biofluid in point-of-care technology. Eur. J. Dent. 2020, 14, S123–S129. [Google Scholar] [CrossRef]
- Yi, Z.; de Dieu Habimana, J.; Mukama, O.; Li, Z.; Odiwuor, N.; Jing, H.; Nie, C.; Hu, M.; Lin, Z.; Wei, H.; et al. Rational programming of cas12a for early-stage detection of COVID-19 by lateral flow assay and portable real-time fluorescence readout facilities. Biosensors 2022, 12, 11. [Google Scholar] [CrossRef]
- Simoska, O.; Stevenson, K.J. Electrochemical sensors for rapid diagnosis of pathogens in real time. Analyst 2019, 144, 6461–6478. [Google Scholar] [CrossRef]
- Dhar, B.C. Diagnostic assay and technology advancement for detecting SARS-CoV-2 infections causing the COVID-19 pandemic. Anal. Bioanal. Chem. 2022, 414, 2903–2934. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, J.; Song, X.; Xu, K.; Chen, H.; Zhao, C.; Li, J. Colorimetric immunoassay for listeria monocytogenes by using core gold nanoparticles, silver nanoclusters as oxidase mimetics, and aptamer-conjugated magnetic nanoparticles. Microchim. Acta 2018, 185, 360. [Google Scholar] [CrossRef] [PubMed]
- Cesewski, E.; Johnson, B.N. Electrochemical biosensors for pathogen detection. Biosens. Bioelectron. 2020, 159, 112214. [Google Scholar] [CrossRef] [PubMed]
- Cassedy, A.; Parle-McDermott, A.; O’Kennedy, R. Virus detection: A review of the current and emerging molecular and immunological methods. Front. Mol. Biosci. 2021, 76, 637559. [Google Scholar] [CrossRef] [PubMed]
- Alahi, M.E.E.; Mukhopadhyay, S.C. Detection methodologies for pathogen and toxins: A review. Sensors 2017, 17, 1885. [Google Scholar] [CrossRef] [Green Version]
- Lazcka, O.; Del Campo, F.J.; Munoz, F.X. Pathogen detection: A perspective of traditional methods and biosensors. Biosens. Bioelectron. 2007, 22, 1205–1217. [Google Scholar] [CrossRef]
- Zhao, G.; Xing, F.; Deng, S. A disposable amperometric enzyme immunosensor for rapid detection of vibrio parahaemolyticus in food based on agarose/nano-au membrane and screen-printed electrode. Electrochem. Commun. 2007, 9, 1263–1268. [Google Scholar] [CrossRef]
- Law, J.W.-F.; Ab Mutalib, N.-S.; Chan, K.-G.; Lee, L.-H. Rapid methods for the detection of foodborne bacterial pathogens: Principles, applications, advantages and limitations. Front. Microbiol. 2015, 5, 770. [Google Scholar] [CrossRef] [Green Version]
- Klein, D. Quantification using real-time pcr technology: Applications and limitations. Trends Mol. Med. 2002, 8, 257–260. [Google Scholar] [CrossRef]
- Malorny, B.; Tassios, P.T.; Rådström, P.; Cook, N.; Wagner, M.; Hoorfar, J. Standardization of diagnostic pcr for the detection of foodborne pathogens. Int. J. Food Microbiol. 2003, 83, 39–48. [Google Scholar] [CrossRef]
- Yang, S.; Rothman, R.E. Pcr-based diagnostics for infectious diseases: Uses, limitations, and future applications in acute-care settings. Lancet Infect. Dis. 2004, 4, 337–348. [Google Scholar] [CrossRef]
- Zeng, D.; Chen, Z.; Jiang, Y.; Xue, F.; Li, B. Advances and challenges in viability detection of foodborne pathogens. Front. Microbiol. 2016, 7, 1833. [Google Scholar] [CrossRef] [Green Version]
- Czajka, J.; Bsat, N.; Piani, M.; Russ, W.; Sultana, K.; Wiedmann, M.; Whitaker, R.; Batt, C. Differentiation of listeria monocytogenes and listeria innocua by 16s rrna genes and intraspecies discrimination of listeria monocytogenes strains by random amplified polymorphic DNA polymorphisms. Appl. Environ. Microbiol. 1993, 59, 304–308. [Google Scholar] [CrossRef]
- Hansen, B.M.; Hendriksen, N.B. Detection of enterotoxic bacillus cereus and bacillus thuringiensis strains by pcr analysis. Appl. Environ. Microbiol. 2001, 67, 185–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.-O.; Kim, S.-S. Bacterial pathogen detection by conventional culture-based and recent alternative (polymerase chain reaction, isothermal amplification, enzyme linked immunosorbent assay, bacteriophage amplification, and gold nanoparticle aggregation) methods in food samples: A review. J. Food Saf. 2021, 41, e12870. [Google Scholar]
- Sakamoto, S.; Putalun, W.; Vimolmangkang, S.; Phoolcharoen, W.; Shoyama, Y.; Tanaka, H.; Morimoto, S. Enzyme-linked immunosorbent assay for the quantitative/qualitative analysis of plant secondary metabolites. J. Nat. Med. 2018, 72, 32–42. [Google Scholar] [CrossRef] [Green Version]
- Justino, C.I.; Duarte, A.C.; Rocha-Santos, T.A. Recent progress in biosensors for environmental monitoring: A review. Sensors 2017, 17, 2918. [Google Scholar] [CrossRef] [Green Version]
- Scognamiglio, V.; Rea, G.; Arduini, F.; Palleschi, G. Biosensors for Sustainable Food-New Opportunities and Technical Challenges; Elsevier: Amsterdam, The Netherlands, 2016. [Google Scholar]
- Sin, M.L.; Mach, K.E.; Wong, P.K.; Liao, J.C. Advances and challenges in biosensor-based diagnosis of infectious diseases. Expert Rev. Mol. Diagn. 2014, 14, 225–244. [Google Scholar] [CrossRef] [Green Version]
- Clark, K.D.; Zhang, C.; Anderson, J.L. Sample Preparation for Bioanalytical and Pharmaceutical Analysis; ACS Publications: Washington, DC, USA, 2016. [Google Scholar]
- Silverman, J.D.; Bloom, R.J.; Jiang, S.; Durand, H.K.; Mukherjee, S.; David, L.A. Measuring and mitigating pcr bias in microbiome data. PLoS Comput. Biol. 2021, 17, e1009113. [Google Scholar] [CrossRef]
- Thévenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical biosensors: Recommended definitions and classification. Biosens. Bioelectron. 2001, 16, 121–131. [Google Scholar] [CrossRef]
- Ferrari, A.G.-M.; Crapnell, R.D.; Banks, C.E. Electroanalytical overview: Electrochemical sensing platforms for food and drink safety. Biosensors 2021, 11, 291. [Google Scholar] [CrossRef] [PubMed]
- Daniels, J.S.; Pourmand, N. Label-free impedance biosensors: Opportunities and challenges. Electroanal. Int. J. Devoted Fundam. Pract. Asp. Electroanal. 2007, 19, 1239–1257. [Google Scholar] [CrossRef]
- Rapp, B.E.; Gruhl, F.J.; Länge, K. Biosensors with label-free detection designed for diagnostic applications. Anal. Bioanal. Chem. 2010, 398, 2403–2412. [Google Scholar] [CrossRef] [PubMed]
- Sang, S.; Wang, Y.; Feng, Q.; Wei, Y.; Ji, J.; Zhang, W. Progress of new label-free techniques for biosensors: A review. Crit. Rev. Biotechnol. 2016, 36, 465–481. [Google Scholar] [CrossRef] [PubMed]
- Vestergaard, M.; Kerman, K.; Tamiya, E. An overview of label-free electrochemical protein sensors. Sensors 2007, 7, 3442–3458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Resch-Genger, U.; Grabolle, M.; Cavaliere-Jaricot, S.; Nitschke, R.; Nann, T. Quantum dots versus organic dyes as fluorescent labels. Nat. Methods 2008, 5, 763–775. [Google Scholar] [CrossRef]
- Cooper, M.A. Label-Free Biosensors: Techniques and Applications; Cambridge University Press: Cambridge, UK, 2009. [Google Scholar]
- Syahir, A.; Usui, K.; Tomizaki, K.-y.; Kajikawa, K.; Mihara, H. Label and label-free detection techniques for protein microarrays. Microarrays 2015, 4, 228–244. [Google Scholar] [CrossRef] [Green Version]
- Yoo, S.M.; Lee, S.Y. Optical biosensors for the detection of pathogenic microorganisms. Trends Biotechnol. 2016, 34, 7–25. [Google Scholar] [CrossRef] [PubMed]
- Felix, F.S.; Angnes, L. Electrochemical immunosensors–a powerful tool for analytical applications. Biosens. Bioelectron. 2018, 102, 470–478. [Google Scholar] [CrossRef]
- Da Silva Neves, M.M.P.; González-García, M.B.; Hernandez-Santos, D.; Fanjul-Bolado, P. Future trends in the market for electrochemical biosensing. Curr. Opin. Electrochem. 2018, 10, 107–111. [Google Scholar] [CrossRef]
- Saucedo, N.M.; Srinives, S.; Mulchandani, A. Electrochemical biosensor for rapid detection of viable bacteria and antibiotic screening. J. Anal. Test. 2019, 3, 117–122. [Google Scholar] [CrossRef]
- Amiri, M.; Bezaatpour, A.; Jafari, H.; Boukherroub, R.; Szunerits, S. Electrochemical methodologies for the detection of pathogens. ACS Sens. 2018, 3, 1069–1086. [Google Scholar] [CrossRef]
- Duffy, G.; Moore, E. Electrochemical immunosensors for food analysis: A review of recent developments. Anal. Lett. 2017, 50, 1–32. [Google Scholar] [CrossRef]
- Furst, A.L.; Francis, M.B. Impedance-based detection of bacteria. Chem. Rev. 2018, 119, 700–726. [Google Scholar] [CrossRef] [PubMed]
- Mishra, G.K.; Sharma, V.; Mishra, R.K. Electrochemical aptasensors for food and environmental safeguarding: A review. Biosensors 2018, 8, 28. [Google Scholar] [CrossRef] [Green Version]
- Monzó, J.; Insua, I.; Fernandez-Trillo, F.; Rodriguez, P. Fundamentals, achievements and challenges in the electrochemical sensing of pathogens. Analyst 2015, 140, 7116–7128. [Google Scholar] [CrossRef] [PubMed]
- Chin, S.F.; Lim, L.S.; Pang, S.C.; Sum, M.S.H.; Perera, D. Carbon nanoparticle modified screen printed carbon electrode as a disposable electrochemical immunosensor strip for the detection of japanese encephalitis virus. Microchim. Acta 2017, 184, 491–497. [Google Scholar] [CrossRef]
- Bhardwaj, J.; Devarakonda, S.; Kumar, S.; Jang, J. Development of a paper-based electrochemical immunosensor using an antibody-single walled carbon nanotubes bio-conjugate modified electrode for label-free detection of foodborne pathogens. Sens. Actuators B Chem. 2017, 253, 115–123. [Google Scholar] [CrossRef]
- Mathelié-Guinlet, M.; Cohen-Bouhacina, T.; Gammoudi, I.; Martin, A.; Beven, L.; Delville, M.-H.; Grauby-Heywang, C. Silica nanoparticles-assisted electrochemical biosensor for the rapid, sensitive and specific detection of escherichia coli. Sens. Actuators B Chem. 2019, 292, 314–320. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, L.; Qiao, Z.; Lei, C.; Fu, Y.; Xie, Q.; Yao, S.; Li, Y.; Ying, Y. Electrochemical conversion of Fe3O4 magnetic nanoparticles to electroactive prussian blue analogues for self-sacrificial label biosensing of avian influenza virus h5n1. Anal. Chem. 2017, 89, 12145–12151. [Google Scholar] [CrossRef]
- Hai, W.; Goda, T.; Takeuchi, H.; Yamaoka, S.; Horiguchi, Y.; Matsumoto, A.; Miyahara, Y. Specific recognition of human influenza virus with pedot bearing sialic acid-terminated trisaccharides. ACS Appl. Mater. Interfaces 2017, 9, 14162–14170. [Google Scholar] [CrossRef]
- Chand, R.; Neethirajan, S. Microfluidic platform integrated with graphene-gold nano-composite aptasensor for one-step detection of norovirus. Biosens. Bioelectron. 2017, 98, 47–53. [Google Scholar] [CrossRef]
- Abbaspour, A.; Norouz-Sarvestani, F.; Noori, A.; Soltani, N. Aptamer-conjugated silver nanoparticles for electrochemical dual-aptamer-based sandwich detection of staphylococcus aureus. Biosens. Bioelectron. 2015, 68, 149–155. [Google Scholar] [CrossRef]
- Idil, N.; Hedström, M.; Denizli, A.; Mattiasson, B. Whole cell based microcontact imprinted capacitive biosensor for the detection of escherichia coli. Biosens. Bioelectron. 2017, 87, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Nahhas, A.F.; Nahhas, A.F.; Webster, T.J. Nanoscale pathogens treated with nanomaterial-like peptides: A platform technology appropriate for future pandemics. Nanomedicine 2021, 16, 1237–1254. [Google Scholar]
- Huang, C.; Zhao, J.; Lu, R.; Wang, J.; Nugen, S.R.; Chen, Y.; Wang, X. A phage-based magnetic relaxation switching biosensor using bioorthogonal reaction signal amplification for salmonella detection in foods. Food Chem. 2023, 400, 134035. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, H.; Li, C.; Ding, Y.; Wang, Y.; Zhu, W.; Wang, J.; Shao, Y.; Pan, H.; Wang, X. Eis biosensor based on a novel myoviridae bacteriophage sep37 for rapid and specific detection of salmonella in food matrixes. Food Res. Int. 2022, 158, 111479. [Google Scholar] [CrossRef]
- Radke, S.M.; Alocilja, E.C. A high density microelectrode array biosensor for detection of E. Coli O157: H7. Biosens. Bioelectron. 2005, 20, 1662–1667. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Goel, A.; Singh, L.; Rao, V. Immunological biosensor for detection of vibrio cholerae o1in environmental water samples. World J. Microbiol. Biotechnol. 2006, 22, 1155–1159. [Google Scholar] [CrossRef]
- Wang, R.; Ruan, C.; Kanayeva, D.; Lassiter, K.; Li, Y. Tio2 nanowire bundle microelectrode based impedance immunosensor for rapid and sensitive detection of listeria monocytogenes. Nano Lett. 2008, 8, 2625–2631. [Google Scholar] [CrossRef]
- Das, R.D.; RoyChaudhuri, C.; Maji, S.; Das, S.; Saha, H. Macroporous silicon based simple and efficient trapping platform for electrical detection of salmonella typhimurium pathogens. Biosens. Bioelectron. 2009, 24, 3215–3222. [Google Scholar] [CrossRef]
- Nguyen, B.T.; Koh, G.; Lim, H.S.; Chua, A.J.; Ng, M.M.; Toh, C.-S. Membrane-based electrochemical nanobiosensor for the detection of virus. Anal. Chem. 2009, 81, 7226–7234. [Google Scholar] [CrossRef]
- Pal, S.; Alocilja, E.C. Electrically active polyaniline coated magnetic (eapm) nanoparticle as novel transducer in biosensor for detection of bacillus anthracis spores in food samples. Biosens. Bioelectron. 2009, 24, 1437–1444. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Yang, G.; Meng, W.; Wu, L.; Zhu, A. An electrochemical impedimetric immunosensor for label-free detection of campylobacter jejuni in diarrhea patients’ stool based on o-carboxymethylchitosan surface modified Fe3O4 nanoparticles. Biosens. Bioelectron. 2010, 25, 1204–1211. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Nartker, S.; Miller, H.; Hochhalter, D.; Wiederoder, M.; Wiederoder, S.; Setterington, E.; Drzal, L.T.; Alocilja, E.C. Surface functionalization of electrospun nanofibers for detecting E. Coli O157: H7 and bvdv cells in a direct-charge transfer biosensor. Biosens. Bioelectron. 2010, 26, 1612–1617. [Google Scholar] [CrossRef] [PubMed]
- Mannoor, M.S.; Tao, H.; Clayton, J.D.; Sengupta, A.; Kaplan, D.L.; Naik, R.R.; Verma, N.; Omenetto, F.G.; McAlpine, M.C. Graphene-based wireless bacteria detection on tooth enamel. Nat. Commun. 2012, 3, 763. [Google Scholar] [CrossRef] [PubMed]
- Tolba, M.; Ahmed, M.U.; Tlili, C.; Eichenseher, F.; Loessner, M.J.; Zourob, M. A bacteriophage endolysin-based electrochemical impedance biosensor for the rapid detection of listeria cells. Analyst 2012, 137, 5749–5756. [Google Scholar] [CrossRef] [PubMed]
- Samardzic, R.; Sussitz, H.F.; Jongkon, N.; Lieberzeit, P.A. Quartz crystal microbalance in-line sensing of Escherichia coli in a bioreactor using molecularly imprinted polymers. Sens. Lett. 2014, 12, 1152–1155. [Google Scholar] [CrossRef]
- Chen, G.-Z.; Yin, Z.-Z.; Lou, J.-F. Electrochemical immunoassay of Escherichia coli O157: H7 using Ag@SiO2 nanoparticles as labels. J. Anal. Methods Chem. 2014, 2014, 247034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dastider, S.G.; Barizuddin, S.; Yuksek, N.S.; Dweik, M.; Almasri, M.F. Efficient and rapid detection of salmonella using microfluidic impedance based sensing. J. Sens. 2015, 2015, 293461. [Google Scholar]
- Andrade, C.A.; Nascimento, J.M.; Oliveira, I.S.; de Oliveira, C.V.; de Melo, C.P.; Franco, O.L.; Oliveira, M.D. Nanostructured sensor based on carbon nanotubes and clavanin a for bacterial detection. Colloids Surf. B Biointerfaces 2015, 135, 833–839. [Google Scholar] [CrossRef]
- Luna, D.M.; Avelino, K.Y.; Cordeiro, M.T.; Andrade, C.A.; Oliveira, M.D. Electrochemical immunosensor for dengue virus serotypes based on 4-mercaptobenzoic acid modified gold nanoparticles on self-assembled cysteine monolayers. Sens. Actuators B Chem. 2015, 220, 565–572. [Google Scholar] [CrossRef]
- Kitajima, M.; Wang, N.; Tay, M.Q.; Miao, J.; Whittle, A.J. Development of a mems-based electrochemical aptasensor for norovirus detection. Micro Nano Lett. 2016, 11, 582–585. [Google Scholar] [CrossRef]
- Attar, A.; Mandli, J.; Ennaji, M.M.; Amine, A. Label-free electrochemical impedance detection of rotavirus based on immobilized antibodies on gold sononanoparticles. Electroanalysis 2016, 28, 1839–1846. [Google Scholar] [CrossRef]
- Golabi, M.; Kuralay, F.; Jager, E.W.; Beni, V.; Turner, A.P. Electrochemical bacterial detection using poly (3-aminophenylboronic acid)-based imprinted polymer. Biosens. Bioelectron. 2017, 93, 87–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hai, W.; Goda, T.; Takeuchi, H.; Yamaoka, S.; Horiguchi, Y.; Matsumoto, A.; Miyahara, Y. Human influenza virus detection using sialyllactose-functionalized organic electrochemical transistors. Sens. Actuators B Chem. 2018, 260, 635–641. [Google Scholar] [CrossRef]
- Jantra, J.; Kanatharana, P.; Asawatreratanakul, P.; Hedström, M.; Mattiasson, B.; Thavarungkul, P. Real-time label-free affinity biosensors for enumeration of total bacteria based on immobilized concanavalin a. J. Environ. Sci. Health Part A 2011, 46, 1450–1460. [Google Scholar] [CrossRef]
- Hou, T.; Zeng, W.; Yang, M.; Chen, W.; Ren, L.; Ai, J.; Wu, J.; Liao, Y.; Gou, X.; Li, Y. Development and evaluation of a rapid crispr-based diagnostic for COVID-19. PLoS Pathog. 2020, 16, e1008705. [Google Scholar] [CrossRef]
- Bacchu, M.S.; Ali, M.R.; Das, S.; Akter, S.; Sakamoto, H.; Suye, S.I.; Rahman, M.M.; Campbell, K.; Khan, M.Z.H. A DNA functionalized advanced electrochemical biosensor for identification of the foodborne pathogen salmonella enterica serovar typhi in real samples. Anal. Chim. Acta 2022, 1192, 339332. [Google Scholar] [CrossRef]
- Fortunati, S.; Giliberti, C.; Giannetto, M.; Bolchi, A.; Ferrari, D.; Donofrio, G.; Bianchi, V.; Boni, A.; De Munari, I.; Careri, M. Rapid quantification of SARS-CoV-2 spike protein enhanced with a machine learning technique integrated in a smart and portable immunosensor. Biosensors 2022, 12, 426. [Google Scholar] [CrossRef]
- Yamanaka, K.; Vestergaard, M.d.C.; Tamiya, E. Printable electrochemical biosensors: A focus on screen-printed electrodes and their application. Sensors 2016, 16, 1761. [Google Scholar] [CrossRef] [Green Version]
- Arduini, F.; Micheli, L.; Moscone, D.; Palleschi, G.; Piermarini, S.; Ricci, F.; Volpe, G. Electrochemical biosensors based on nanomodified screen-printed electrodes: Recent applications in clinical analysis. TrAC Trends Anal. Chem. 2016, 79, 114–126. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Wang, J.; Cinti, S. An overview on recent progress in screen-printed electroanalytical (bio) sensors. ECS Sens. Plus 2022, 1, 023401. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Z.; Wu, G.; Xu, C.; Wu, J.; Zhang, X.; Liu, J. Applications of electrochemical biosensors based on functional antibody modified screen printed electrodes: A review. Anal. Methods 2021, 14, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Kaya, H.O.; Cetin, A.E.; Azimzadeh, M.; Topkaya, S.N. Pathogen detection with electrochemical biosensors: Advantages, challenges and future perspectives. J. Electroanal. Chem. 2021, 882, 114989. [Google Scholar] [CrossRef] [PubMed]
- Dyussembayev, K.; Sambasivam, P.; Bar, I.; Brownlie, J.C.; Shiddiky, M.J.A.; Ford, R. Biosensor technologies for early detection and quantification of plant pathogens. Front. Chem. 2021, 9, 636245. [Google Scholar] [CrossRef] [PubMed]
- Topkaya, S.N.; Azimzadeh, M.; Ozsoz, M. Electrochemical biosensors for cancer biomarkers detection: Recent advances and challenges. Electroanalysis 2016, 28, 1402–1419. [Google Scholar] [CrossRef]
- Srimaneepong, V.; Rokaya, D.; Thunyakitpisal, P.; Qin, J.; Saengkiettiyut, K. Corrosion resistance of graphene oxide/silver coatings on ni–ti alloy and expression of il-6 and il-8 in human oral fibroblasts. Sci. Rep. 2020, 10, 3247. [Google Scholar] [CrossRef] [Green Version]
- Faulkner, L.R.; Bard, A.J. Electrochemical Methods: Fundamentals and Applications; John Wiley and Sons: Hoboken, NJ, USA, 2002. [Google Scholar]
- Hierlemann, A.; Brand, O.; Hagleitner, C.; Baltes, H. Microfabrication techniques for chemical/biosensors. Proc. IEEE 2003, 91, 839–863. [Google Scholar] [CrossRef] [Green Version]
- Taleat, Z.; Khoshroo, A.; Mazloum-Ardakani, M. Screen-printed electrodes for biosensing: A review (2008–2013). Microchim. Acta 2014, 181, 865–891. [Google Scholar] [CrossRef]
- Bhat, K.S.; Ahmad, R.; Yoo, J.-Y.; Hahn, Y.-B. Fully nozzle-jet printed non-enzymatic electrode for biosensing application. J. Colloid Interface Sci. 2018, 512, 480–488. [Google Scholar] [CrossRef]
- Medina-Sánchez, M.; Martínez-Domingo, C.; Ramon, E.; Merkoçi, A. An inkjet-printed field-effect transistor for label-free biosensing. Adv. Funct. Mater. 2014, 24, 6291–6302. [Google Scholar] [CrossRef]
- Pavinatto, F.J.; Paschoal, C.W.; Arias, A.C. Printed and flexible biosensor for antioxidants using interdigitated ink-jetted electrodes and gravure-deposited active layer. Biosens. Bioelectron. 2015, 67, 553–559. [Google Scholar] [CrossRef]
- Foo, C.; Lim, H.; Mahdi, M. Three-dimensional printed electrode and its novel applications in electronic devices. Sci. Rep. 2018, 8, 7399. [Google Scholar] [CrossRef] [PubMed]
- Loo, A.H.; Chua, C.K.; Pumera, M. DNA biosensing with 3d printing technology. Analyst 2017, 142, 279–283. [Google Scholar] [CrossRef]
- Ambrosi, A.; Moo, J.G.S.; Pumera, M. Helical 3d-printed metal electrodes as custom-shaped 3d platform for electrochemical devices. Adv. Funct. Mater. 2016, 26, 698–703. [Google Scholar] [CrossRef]
- Dos Santos, M.B.; Azevedo, S.; Agusil, J.; Prieto-Simón, B.; Sporer, C.; Torrents, E.; Juárez, A.; Teixeira, V.; Samitier, J. Label-free ito-based immunosensor for the detection of very low concentrations of pathogenic bacteria. Bioelectrochemistry 2015, 101, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Aydın, E.B.; Sezgintürk, M.K. Indium tin oxide (ito): A promising material in biosensing technology. TrAC Trends Anal. Chem. 2017, 97, 309–315. [Google Scholar] [CrossRef]
- Yang, L.; Li, Y. Afm and impedance spectroscopy characterization of the immobilization of antibodies on indium–tin oxide electrode through self-assembled monolayer of epoxysilane and their capture of Escherichia coli O157: H7. Biosens. Bioelectron. 2005, 20, 1407–1416. [Google Scholar] [CrossRef]
- Wenzel, T.; Härtter, D.; Bombelli, P.; Howe, C.J.; Steiner, U. Porous translucent electrodes enhance current generation from photosynthetic biofilms. Nat. Commun. 2018, 9, 1299. [Google Scholar] [CrossRef]
- Guimard, N.K.; Gomez, N.; Schmidt, C.E. Conducting polymers in biomedical engineering. Prog. Polym. Sci. 2007, 32, 876–921. [Google Scholar] [CrossRef]
- Arshak, K.; Velusamy, V.; Korostynska, O.; Oliwa-Stasiak, K.; Adley, C. Conducting polymers and their applications to biosensors: Emphasizing on foodborne pathogen detection. IEEE Sens. J. 2009, 9, 1942–1951. [Google Scholar] [CrossRef]
- Kaur, G.; Adhikari, R.; Cass, P.; Bown, M.; Gunatillake, P. Electrically conductive polymers and composites for biomedical applications. RSC Adv. 2015, 5, 37553–37567. [Google Scholar] [CrossRef]
- Terán-Alcocer, Á.; Bravo-Plascencia, F.; Cevallos-Morillo, C.; Palma-Cando, A. Electrochemical sensors based on conducting polymers for the aqueous detection of biologically relevant molecules. Nanomaterials 2021, 11, 252. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Morrin, A.; Li, M.; Liu, N.; Luo, X. Nanomaterial-doped conducting polymers for electrochemical sensors and biosensors. J. Mater. Chem. B 2018, 6, 4173–4190. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.A.; Kumar, P.; Park, D.-S.; Shim, Y.-B. Electrochemical sensors based on organic conjugated polymers. Sensors 2008, 8, 118–141. [Google Scholar] [CrossRef] [PubMed]
- Soylemez, S.; Kaya, H.Z.; Udum, Y.A.; Toppare, L. A multipurpose conjugated polymer: Electrochromic device and biosensor construction for glucose detection. Org. Electron. 2019, 65, 327–333. [Google Scholar] [CrossRef]
- Pavase, T.R.; Lin, H.; Shaikh, Q.-u.-a.; Hussain, S.; Li, Z.; Ahmed, I.; Lv, L.; Sun, L.; Shah, S.B.H.; Kalhoro, M.T. Recent advances of conjugated polymer (cp) nanocomposite-based chemical sensors and their applications in food spoilage detection: A comprehensive review. Sens. Actuators B Chem. 2018, 273, 1113–1138. [Google Scholar] [CrossRef]
- Dong, J.; Zhao, H.; Xu, M.; Ma, Q.; Ai, S. A label-free electrochemical impedance immunosensor based on aunps/pamam-mwcnt-chi nanocomposite modified glassy carbon electrode for detection of salmonella typhimurium in milk. Food Chem. 2013, 141, 1980–1986. [Google Scholar] [CrossRef]
- Lee, D.; Chander, Y.; Goyal, S.M.; Cui, T. Carbon nanotube electric immunoassay for the detection of swine influenza virus h1n1. Biosens. Bioelectron. 2011, 26, 3482–3487. [Google Scholar] [CrossRef]
- Li, Y.; Cheng, P.; Gong, J.; Fang, L.; Deng, J.; Liang, W.; Zheng, J. Amperometric immunosensor for the detection of Escherichia Coli O157: H7 in food specimens. Anal. Biochem. 2012, 421, 227–233. [Google Scholar] [CrossRef]
- Viswanathan, S.; Rani, C.; Ho, J.-a.A. Electrochemical immunosensor for multiplexed detection of food-borne pathogens using nanocrystal bioconjugates and mwcnt screen-printed electrode. Talanta 2012, 94, 315–319. [Google Scholar] [CrossRef] [Green Version]
- Tamara, F.R.; Lin, C.; Mi, F.-L.; Ho, Y.-C. Antibacterial effects of chitosan/cationic peptide nanoparticles. Nanomaterials 2018, 8, 88. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Obodo, D.; Yadavalli, V.K. The design, fabrication, and applications of flexible biosensing devices. Biosens. Bioelectron. 2019, 124, 96–114. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.L.; Tamargo, I.A.; Kim, H.; Johnson, B.N.; Gupta, M.K.; Koh, T.-W.; Chin, H.-A.; Steingart, D.A.; Rand, B.P.; McAlpine, M.C. 3d printed quantum dot light-emitting diodes. Nano Lett. 2014, 14, 7017–7023. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; He, P.; Hussain, S.; Lu, H.; Zhou, X.; Lv, F.; Liu, L.; Dai, Z.; Wang, S. Conjugated polymer-based photoelectrochemical cytosensor with turn-on enable signal for sensitive cell detection. ACS Appl. Mater. Interfaces 2018, 10, 6618–6623. [Google Scholar] [CrossRef]
- Kim, J.; Campbell, A.S.; de Ávila, B.E.-F.; Wang, J. Wearable biosensors for healthcare monitoring. Nat. Biotechnol. 2019, 37, 389–406. [Google Scholar] [CrossRef]
- Mohankumar, P.; Ajayan, J.; Mohanraj, T.; Yasodharan, R. Recent developments in biosensors for healthcare and biomedical applications: A review. Measurement 2021, 167, 108293. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, J.; Joo, H.; Raj, M.S.; Ghaffari, R.; Kim, D.H. Wearable sensing systems with mechanically soft assemblies of nanoscale materials. Adv. Mater. Technol. 2017, 2, 1700053. [Google Scholar] [CrossRef]
- Cesewski, E.; Haring, A.P.; Tong, Y.; Singh, M.; Thakur, R.; Laheri, S.; Read, K.A.; Powell, M.D.; Oestreich, K.J.; Johnson, B.N. Additive manufacturing of three-dimensional (3d) microfluidic-based microelectromechanical systems (mems) for acoustofluidic applications. Lab A Chip 2018, 18, 2087–2098. [Google Scholar] [CrossRef]
- Xu, Y.; Wu, X.; Guo, X.; Kong, B.; Zhang, M.; Qian, X.; Mi, S.; Sun, W. The boom in 3d-printed sensor technology. Sensors 2017, 17, 1166. [Google Scholar] [CrossRef]
- Varshney, M.; Li, Y. Interdigitated array microelectrodes based impedance biosensors for detection of bacterial cells. Biosens. Bioelectron. 2009, 24, 2951–2960. [Google Scholar] [CrossRef]
- Yang, L.; Li, Y. Detection of viable salmonella using microelectrode-based capacitance measurement coupled with immunomagnetic separation. J. Microbiol. Methods 2006, 64, 9–16. [Google Scholar] [CrossRef]
- Yang, H.; Rahman, M.T.; Du, D.; Panat, R.; Lin, Y. 3-d printed adjustable microelectrode arrays for electrochemical sensing and biosensing. Sens. Actuators B Chem. 2016, 230, 600–606. [Google Scholar] [CrossRef] [Green Version]
- Teleanu, D.M.; Negut, I.; Grumezescu, V.; Grumezescu, A.M.; Teleanu, R.I. Nanomaterials for drug delivery to the central nervous system. Nanomaterials 2019, 9, 371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, A.; Akin, D.; Bashir, R. Single virus particle mass detection using microresonators with nanoscale thickness. Appl. Phys. Lett. 2004, 84, 1976–1978. [Google Scholar] [CrossRef]
- Wei, D.; Bailey, M.J.; Andrew, P.; Ryhänen, T. Electrochemical biosensors at the nanoscale. Lab A Chip 2009, 9, 2123–2131. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.A.; Kwon, J.; Kim, D.; Yang, S. A rapid, sensitive and selective electrochemical biosensor with concanavalin a for the preemptive detection of norovirus. Biosens. Bioelectron. 2015, 64, 338–344. [Google Scholar] [CrossRef]
- Patris, S.; Vandeput, M.; Kauffmann, J.-M. Antibodies as target for affinity biosensors. TrAC Trends Anal. Chem. 2016, 79, 239–246. [Google Scholar] [CrossRef]
- Yogeswaran, U.; Chen, S.-M. A review on the electrochemical sensors and biosensors composed of nanowires as sensing material. Sensors 2008, 8, 290–313. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Odom, T.W.; Lieber, C.M. Chemistry and physics in one dimension: Synthesis and properties of nanowires and nanotubes. Acc. Chem. Res. 1999, 32, 435–445. [Google Scholar] [CrossRef]
- Wanekaya, A.K.; Chen, W.; Myung, N.V.; Mulchandani, A. Nanowire-based electrochemical biosensors. Electroanal. Int. J. Devoted Fundam. Pract. Asp. Electroanal. 2006, 18, 533–550. [Google Scholar]
- Shen, F.; Wang, J.; Xu, Z.; Wu, Y.; Chen, Q.; Li, X.; Jie, X.; Li, L.; Yao, M.; Guo, X. Rapid flu diagnosis using silicon nanowire sensor. Nano Lett. 2012, 12, 3722–3730. [Google Scholar] [CrossRef]
- Chartuprayoon, N.; Rheem, Y.; Ng, J.C.; Nam, J.; Chen, W.; Myung, N.V. Polypyrrole nanoribbon based chemiresistive immunosensors for viral plant pathogen detection. Anal. Methods 2013, 5, 3497–3502. [Google Scholar] [CrossRef]
- Soleymani, L.; Fang, Z.; Sargent, E.H.; Kelley, S.O. Programming the detection limits of biosensors through controlled nanostructuring. Nat. Nanotechnol. 2009, 4, 844–848. [Google Scholar] [CrossRef]
- Ludwig, K.A.; Uram, J.D.; Yang, J.; Martin, D.C.; Kipke, D.R. Chronic neural recordings using silicon microelectrode arrays electrochemically deposited with a poly (3, 4-ethylenedioxythiophene)(pedot) film. J. Neural Eng. 2006, 3, 59. [Google Scholar] [CrossRef] [Green Version]
- Alkire, R.C.; Gogotsi, Y.; Simon, P. Nanostructured Materials in Electrochemistry; John Wiley & Sons: Hoboken, NJ, USA, 2008. [Google Scholar]
- Wang, Y.; Ping, J.; Ye, Z.; Wu, J.; Ying, Y. Impedimetric immunosensor based on gold nanoparticles modified graphene paper for label-free detection of Escherichia coli O157: H7. Biosens. Bioelectron. 2013, 49, 492–498. [Google Scholar] [CrossRef]
- De Luna, P.; Mahshid, S.S.; Das, J.; Luan, B.; Sargent, E.H.; Kelley, S.O.; Zhou, R. High-curvature nanostructuring enhances probe display for biomolecular detection. Nano Lett. 2017, 17, 1289–1295. [Google Scholar] [CrossRef] [PubMed]
- Mahshid, S.S.; Vallée-Bélisle, A.; Kelley, S.O. Biomolecular steric hindrance effects are enhanced on nanostructured microelectrodes. Anal. Chem. 2017, 89, 9751–9757. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Goryll, M.; Sin, L.Y.M.; Wong, P.K.; Chae, J. Microfluidic-based biosensors toward point-of-care detection of nucleic acids and proteins. Microfluid. Nanofluid. 2011, 10, 231–247. [Google Scholar] [CrossRef] [PubMed]
- Bearinger, J.; Vörös, J.; Hubbell, J.; Textor, M. Electrochemical optical waveguide lightmode spectroscopy (ec-owls): A pilot study using evanescent-field optical sensing under voltage control to monitor polycationic polymer adsorption onto indium tin oxide (ito)-coated waveguide chips. Biotechnol. Bioeng. 2003, 82, 465–473. [Google Scholar] [CrossRef]
- Németh, E.; Adányi, N.; Halász, A.; Váradi, M.; Szendrő, I. Real-time study of the effect of different stress factors on lactic acid bacteria by electrochemical optical waveguide lightmode spectroscopy. Biomol. Eng. 2007, 24, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Li, C.M.; Dong, H. Poly (pyrrole-co-pyrrole propylic acid) film and its application in label-free surface plasmon resonance immunosensors. Anal. Chim. Acta 2008, 630, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Juan-Colás, J.; Johnson, S.; Krauss, T.F. Dual-mode electro-optical techniques for biosensing applications: A review. Sensors 2017, 17, 2047. [Google Scholar] [CrossRef] [Green Version]
- Johnson, B.N.; Mutharasan, R. Electrochemical piezoelectric-excited millimeter-sized cantilever (epemc) for simultaneous dual transduction biosensing. Analyst 2013, 138, 6365–6371. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.N.; Mutharasan, R. Reduction of nonspecific protein adsorption on cantilever biosensors caused by transverse resonant mode vibration. Analyst 2014, 139, 1112–1120. [Google Scholar] [CrossRef] [PubMed]
- Yeh, P.-Y.J.; Kizhakkedathu, J.N.; Madden, J.D.; Chiao, M. Electric field and vibration-assisted nanomolecule desorption and anti-biofouling for biosensor applications. Colloids Surf. B Biointerfaces 2007, 59, 67–73. [Google Scholar] [CrossRef]
- Hou, Y.-H.; Wang, J.-J.; Jiang, Y.-Z.; Lv, C.; Xia, L.; Hong, S.-L.; Lin, M.; Lin, Y.; Zhang, Z.-L.; Pang, D.-W. A colorimetric and electrochemical immunosensor for point-of-care detection of enterovirus 71. Biosens. Bioelectron. 2018, 99, 186–192. [Google Scholar] [CrossRef]
- Giouroudi, I.; Kokkinis, G. Recent advances in magnetic microfluidic biosensors. Nanomaterials 2017, 7, 171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mungroo, N.; Neethirajan, S. Optical biosensors for the detection of food borne pathogens. Google Sch. 2016, 179–206. [Google Scholar] [CrossRef]
- Pires, N.M.M.; Dong, T.; Hanke, U.; Hoivik, N. Recent developments in optical detection technologies in lab-on-a-chip devices for biosensing applications. Sensors 2014, 14, 15458–15479. [Google Scholar] [CrossRef] [Green Version]
- Sande, M.G.; Rodrigues, J.L.; Ferreira, D.; Silva, C.J.; Rodrigues, L.R. Novel biorecognition elements against pathogens in the design of state-of-the-art diagnostics. Biosensors 2021, 11, 418. [Google Scholar] [CrossRef]
- Newell, D.G.; Koopmans, M.; Verhoef, L.; Duizer, E.; Aidara-Kane, A.; Sprong, H.; Opsteegh, M.; Langelaar, M.; Threfall, J.; Scheutz, F. Food-borne diseases—The challenges of 20 years ago still persist while new ones continue to emerge. Int. J. Food Microbiol. 2010, 139, S3–S15. [Google Scholar] [CrossRef] [PubMed]
- Leonard, P.; Hearty, S.; Brennan, J.; Dunne, L.; Quinn, J.; Chakraborty, T.; O’Kennedy, R. Advances in biosensors for detection of pathogens in food and water. Enzym. Microb. Technol. 2003, 32, 3–13. [Google Scholar] [CrossRef]
- Birch, J.R.; Racher, A.J. Antibody production. Adv. Drug Deliv. Rev. 2006, 58, 671–685. [Google Scholar] [CrossRef] [PubMed]
- Byrne, B.; Stack, E.; Gilmartin, N.; O’Kennedy, R. Antibody-based sensors: Principles, problems and potential for detection of pathogens and associated toxins. Sensors 2009, 9, 4407–4445. [Google Scholar] [CrossRef] [Green Version]
- Sharma, H.; Mutharasan, R. Half antibody fragments improve biosensor sensitivity without loss of selectivity. Anal. Chem. 2013, 85, 2472–2477. [Google Scholar] [CrossRef] [PubMed]
- Pavan, S.; Berti, F. Short peptides as biosensor transducers. Anal. Bioanal. Chem. 2012, 402, 3055–3070. [Google Scholar] [CrossRef] [PubMed]
- Etayash, H.; Jiang, K.; Azmi, S.; Thundat, T.; Kaur, K. Real-time detection of breast cancer cells using peptide functionalized microcantilever arrays. Sci. Rep. 2015, 5, 13967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dester, E.; Alocilja, E. Current methods for extraction and concentration of foodborne bacteria with glycan-coated magnetic nanoparticles: A review. Biosensors 2022, 12, 112. [Google Scholar] [CrossRef]
- Zeng, X.; Andrade, C.A.; Oliveira, M.D.; Sun, X.-L. Carbohydrate–protein interactions and their biosensing applications. Anal. Bioanal. Chem. 2012, 402, 3161–3176. [Google Scholar] [CrossRef]
- Lakhin, A.; Tarantul, V.; Gening, L. Aptamers: Problems, solutions and prospects. Acta Nat. 2013, 5, 34–43. [Google Scholar] [CrossRef] [Green Version]
- Reverdatto, S.; Burz, D.S.; Shekhtman, A. Peptide aptamers: Development and applications. Curr. Top. Med. Chem. 2015, 15, 1082. [Google Scholar] [CrossRef] [Green Version]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. Selex—A (r) evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007, 24, 381–403. [Google Scholar] [CrossRef] [PubMed]
- Giamberardino, A.; Labib, M.; Hassan, E.M.; Tetro, J.A.; Springthorpe, S.; Sattar, S.A.; Berezovski, M.V.; DeRosa, M.C. Ultrasensitive norovirus detection using DNA aptasensor technology. PLoS ONE 2013, 8, e79087. [Google Scholar] [CrossRef] [Green Version]
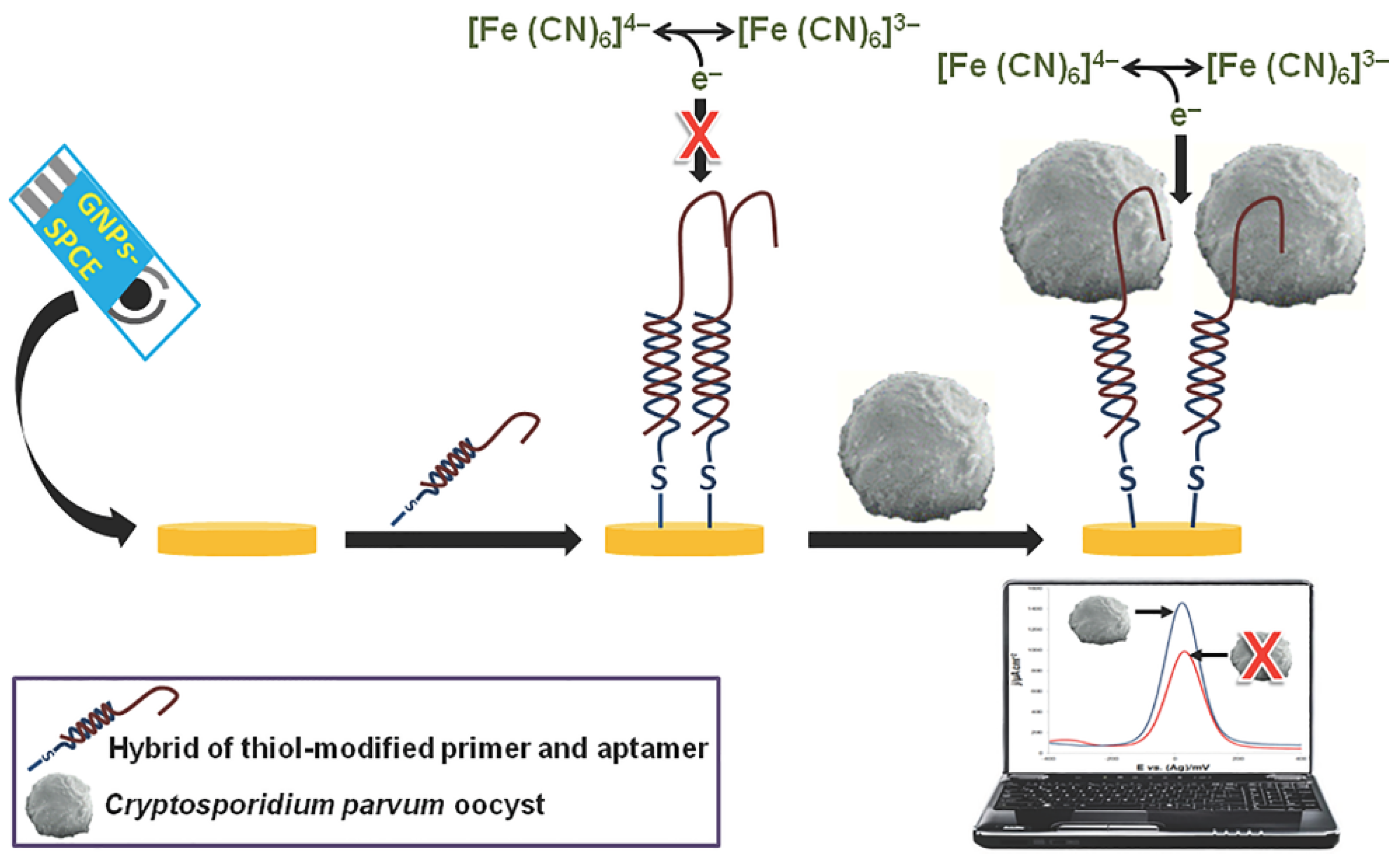
- Iqbal, A.; Labib, M.; Muharemagic, D.; Sattar, S.; Dixon, B.R.; Berezovski, M.V. Detection of cryptosporidium parvum oocysts on fresh produce using DNA aptamers. PLoS ONE 2015, 10, e0137455. [Google Scholar] [CrossRef]
- Azizian, R.; Nasser, A.; Askari, H.; Taheri Kalani, M.; Sadeghifard, N.; Pakzad, I.; Amini, R.; Mozaffari Nejad, A.S.; Azizi Jalilian, F. Sewage as a rich source of phage study against pseudomonas aeruginosa pao. Biol. J. Int. Assoc. Biol. Stand. 2015, 43, 238–241. [Google Scholar] [CrossRef]
- Nasser, A.; Azizian, R.; Tabasi, M.; Khezerloo, J.K.; Heravi, F.S.; Kalani, M.T.; Sadeghifard, N.; Amini, R.; Pakzad, I.; Radmanesh, A.; et al. Specification of bacteriophage isolated against clinical methicillin-resistant staphylococcus aureus. Osong Public Health Res. Perspect. 2019, 10, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Aliakbar Ahovan, Z.; Hashemi, A.; De Plano, L.M.; Gholipourmalekabadi, M.; Seifalian, A. Bacteriophage based biosensors: Trends, outcomes and challenges. Nanomaterials 2020, 10, 501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kutter, E.; Sulakvelidze, A. Bacteriophages: Biology and Applications; Crc press: Boca Raton, FL, USA, 2004. [Google Scholar]
- Kim, S.-K.; Oh, Y.-H.; Ko, D.-H.; Sung, H.; Oh, H.-B.; Hwang, S.-H. Nanoparticle-based visual detection of amplified DNA for diagnosis of hepatitis c virus. Biosensors 2022, 12, 744. [Google Scholar] [CrossRef]
- Shabani, A.; Zourob, M.; Allain, B.; Marquette, C.A.; Lawrence, M.F.; Mandeville, R. Bacteriophage-modified microarrays for the direct impedimetric detection of bacteria. Anal. Chem. 2008, 80, 9475–9482. [Google Scholar] [CrossRef]
- Mejri, M.; Baccar, H.; Baldrich, E.; Del Campo, F.; Helali, S.; Ktari, T.; Simonian, A.; Aouni, M.; Abdelghani, A. Impedance biosensing using phages for bacteria detection: Generation of dual signals as the clue for in-chip assay confirmation. Biosens. Bioelectron. 2010, 26, 1261–1267. [Google Scholar] [CrossRef] [Green Version]
- Pan, J.; Chen, W.; Ma, Y.; Pan, G. Molecularly imprinted polymers as receptor mimics for selective cell recognition. Chem. Soc. Rev. 2018, 47, 5574–5587. [Google Scholar] [CrossRef]
- Chen, L.; Wang, X.; Lu, W.; Wu, X.; Li, J. Molecular imprinting: Perspectives and applications. Chem. Soc. Rev. 2016, 45, 2137–2211. [Google Scholar] [CrossRef] [PubMed]
- Jafari, H.; Amiri, M.; Abdi, E.; Navid, S.L.; Bouckaert, J.; Jijie, R.; Boukherroub, R.; Szunerits, S. Entrapment of uropathogenic E. Coli cells into ultra-thin sol-gel matrices on gold thin films: A low cost alternative for impedimetric bacteria sensing. Biosens. Bioelectron. 2019, 124, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Yáñez-Sedeño, P.; Campuzano, S.; Pingarrón, J.M. Electrochemical sensors based on magnetic molecularly imprinted polymers: A review. Anal. Chim. Acta 2017, 960, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Cheong, W.J.; Yang, S.H.; Ali, F. Molecular imprinted polymers for separation science: A review of reviews. J. Sep. Sci. 2013, 36, 609–628. [Google Scholar] [CrossRef] [PubMed]
- MohammadSadeghi, A.; Karimzadeh, I.; Lankarani, K.B.; Banakar, M. Pharmacotherapy for reducing saliva and droplet production in airborne procedures may help to decrease the COVID-19 transmission: A hypothesis. Med. Hypotheses 2020, 144, 109874. [Google Scholar] [CrossRef]
- Zou, Y.; Xiang, C.; Sun, L.-X.; Xu, F. Glucose biosensor based on electrodeposition of platinum nanoparticles onto carbon nanotubes and immobilizing enzyme with chitosan-SiO2 sol–gel. Biosens. Bioelectron. 2008, 23, 1010–1016. [Google Scholar] [CrossRef] [PubMed]
- Smutok, O.; Ngounou, B.; Pavlishko, H.; Gayda, G.; Gonchar, M.; Schuhmann, W. A reagentless bienzyme amperometric biosensor based on alcohol oxidase/peroxidase and an os-complex modified electrodeposition paint. Sens. Actuators B Chem. 2006, 113, 590–598. [Google Scholar] [CrossRef]
- Shi, J.; Porterfield, D.M. Surface modification approaches for electrochemical biosensors. In Biosensors-Emerging Materials and Applications, 1st ed.; Serra, P.A., Ed.; IntechOpen: London, UK, 2011; pp. 209–226. [Google Scholar] [CrossRef]


| Recognition Element | Advantages | Disadvantages | Reference(s) |
|---|---|---|---|
| Antibodies | High affinity High selectivity | Selectivity may be affected by antibody labeling Alteration of binding affinity to antigen Low temperature High cost | [47,48,49] |
| Carbohydrate binding proteins | Binding ligands unique to the Target organism Low cost High-yield automated synthesis | Pathogens detection abilities using electrochemical biosensors lack sufficient data | [41,50] |
| Oligosaccharides | Pathogens have receptors for Carbohydrate-specific trisaccharides Utilized with electrochemical biosensors | Limited selectivity Low affinity Carbohydrate–protein interaction | [41,51] |
| Oligonucleotides | Utilized with electrochemical biosensors Strong binding affinity and selectivity Low cost Feasible to extract and amplify particular binding sequences | Possibility of cross-reactions Lack of repeatability when using different procedures Degradation of aptamers | [52,53] |
| Cell-and molecular-imprinted polymers | Use morphology particular to target | Poor selectivity | [54,55] |
| Phages | Utilized with electrochemical biosensors Effective biorecognition component in water monitoring | High cost | [56,57] |
| Bacteria/Virus | Method and Materials | Biorecognition Element | * LOD/LOQ | Year | Reference(s) |
|---|---|---|---|---|---|
| E. coli | Electrochemical impedance spectroscopy | Polyclonal anti-E. coli | 104 CFU/mL | 2005 | [58] |
| V. cholerae | Carbon electrode | Polyclonal anti-V. cholerae | 8 CFU/mL | 2006 | [59] |
| L. monocytogenes | Electrode nanostructuring | Monoclonal anti-L. monocytogenes | 4.7 × 102 CFU/mL | 2008 | [60] |
| S. typhimurium | Ceramic electrodes | Anti-S. typhimurium | 103 CFU/mL | 2009 | [61] |
| West Nile virus (WNV) | Anodic stripping voltammetry | Monoclonal anti-WNV | 0.02 viruses/mL | 2009 | [62] |
| B. anthracis | Ag electrode (Conductometry) | Monoclonal and polyclonal anti-B. anthracis | 420 spores/mL | 2009 | [63] |
| Campylobacter jejuni | Nanoparticles on carbon electrode | Monoclonal anti-Flagellin A | 103 CFU/mL | 2010 | [64] |
| Bovine viral diarrhea virus (BVDV) | Nanofiber array electrode (Conductometry) | Monoclonal and polyclonal anti-BVDV | 103 CCID **/mL | 2010 | [65] |
| Helicobacter pylori | Graphene interdigitated microelectrode array (Conductometry) | Odoranin-HP peptide | 100 cells | 2012 | [66] |
| L. innocua | Phage | L. innocua-specific bacteriophage | 1.1 × 104–105 CFU/mL | 2012 | [67] |
| E. coli | Cell- and molecularly imprinted polymers | Anti-E. coli | 1.6 × 108 Cells/mL | 2014 | [68] |
| E. coli | Composite on carbon electrode | Anti-E. coli | 13 CFU/mL | 2014 | [69] |
| S. typhimurium | Electrochemical Impedance Spectroscopy (EIS) | Monoclonal anti-S. typhimurium | 3 × 103 CFU/mL | 2015 | [70] |
| Enterococcus faecalis | Carbon-based electrodes on Au electrode | Clavanin A peptide | 103 CFU/mL | 2015 | [71] |
| Dengue virus | AuNPs on Au electrode | Anti-DENV | --------------- | 2015 | [72] |
| Norovirus | Au microelectrode (square wave voltammetry) | Anti-norovirus aptamer | 10 PFU ***/mL | 2016 | [73] |
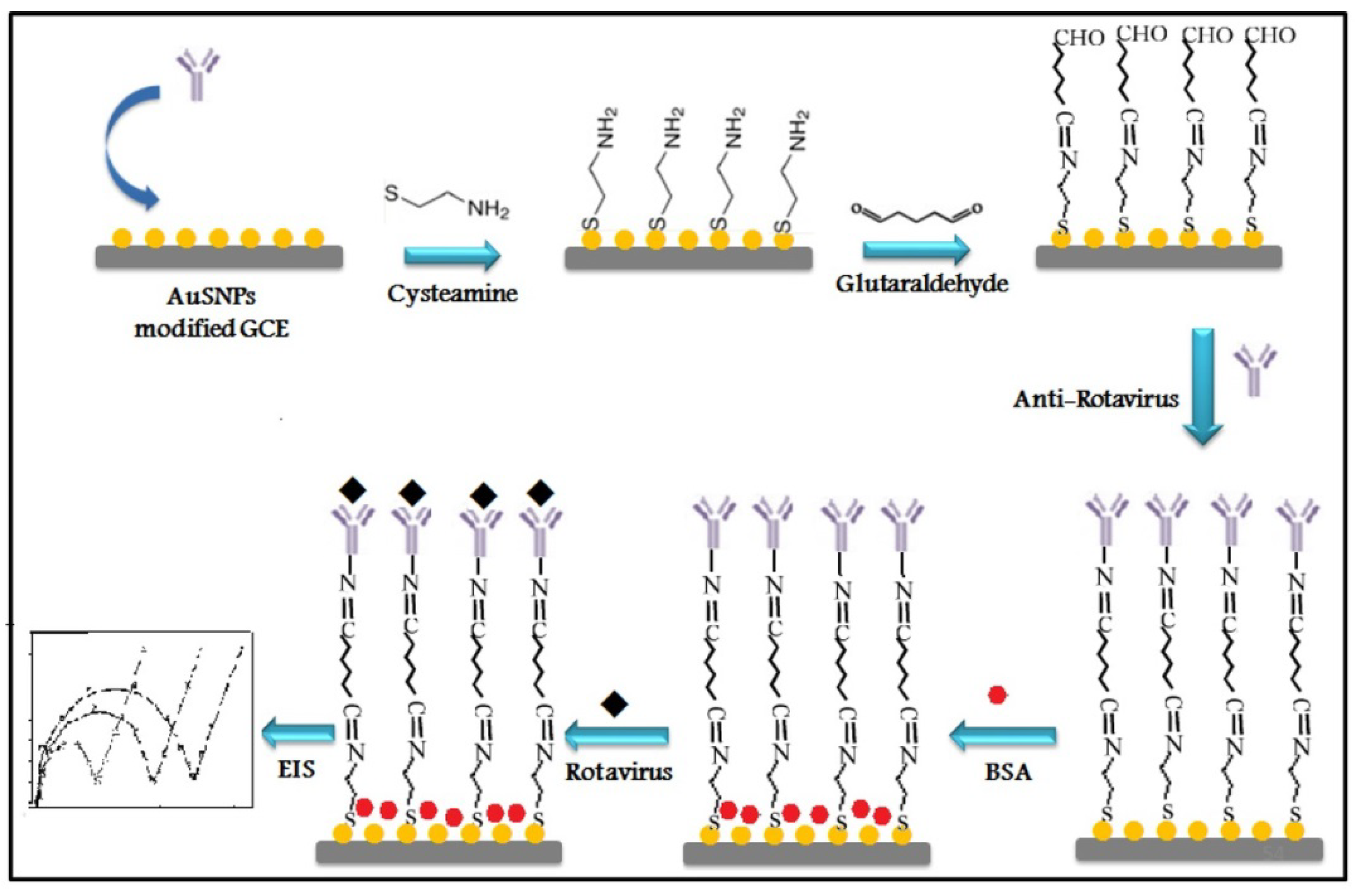
| Rotavirus | Electrochemical Impedance Spectroscopy (EIS) and nano structuring | Anti-rotavirus | 2.3 PFU/mL R2 ****: 0.993 | 2016 | [74] |
| S. epidermidis | Au microelectrode (Electrochemical Impedance Spectroscopy) | S. epidermidis-imprinted polymer film | 103 CFU/mL | 2017 | [75] |
| Influenza A virus (H1N1) | Oligosaccharides (PEDOT:PSS) | Hemagglutinin-specific trisaccharide ligand | 0.13 HAU ***** | 2017 | [51] |
| E. coli and human influenza A virus | Polymer electrode | Hemagglutinin-specific trisaccharide ligand | 0.025 HAU | 2018 | [76] |
| E. coli | Carbohydrate binding proteins | Anti-E. coli | 12 CFU/ml ----------------- 6.0 × 103–9.2 × 107 CFU/mL | 2011 ------ 2019 | [41,77] |
| SARS-CoV-2 | CRISPER-Cas | --------------- | Fold change: 10 | 2020 | [78] |
| S. typhimurium | DNA functionalized | Amine labeled S. Typhi | 6.8 × 10−25 molL−1 | 2022 | [79] |
| SARS-CoV-2 | Electrochemical immunosensor | SARS-CoV-2 spike protein | 12 ng/mL–40 ng/mL | 2022 | [80] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Banakar, M.; Hamidi, M.; Khurshid, Z.; Zafar, M.S.; Sapkota, J.; Azizian, R.; Rokaya, D. Electrochemical Biosensors for Pathogen Detection: An Updated Review. Biosensors 2022, 12, 927. https://doi.org/10.3390/bios12110927
Banakar M, Hamidi M, Khurshid Z, Zafar MS, Sapkota J, Azizian R, Rokaya D. Electrochemical Biosensors for Pathogen Detection: An Updated Review. Biosensors. 2022; 12(11):927. https://doi.org/10.3390/bios12110927
Chicago/Turabian StyleBanakar, Morteza, Masoud Hamidi, Zohaib Khurshid, Muhammad Sohail Zafar, Janak Sapkota, Reza Azizian, and Dinesh Rokaya. 2022. "Electrochemical Biosensors for Pathogen Detection: An Updated Review" Biosensors 12, no. 11: 927. https://doi.org/10.3390/bios12110927
APA StyleBanakar, M., Hamidi, M., Khurshid, Z., Zafar, M. S., Sapkota, J., Azizian, R., & Rokaya, D. (2022). Electrochemical Biosensors for Pathogen Detection: An Updated Review. Biosensors, 12(11), 927. https://doi.org/10.3390/bios12110927








