Aptamer-Gated Mesoporous Silica Nanoparticles for N Protein Triggered Release of Remdesivir and Treatment of Novel Coronavirus (2019-nCoV)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Instrumentation
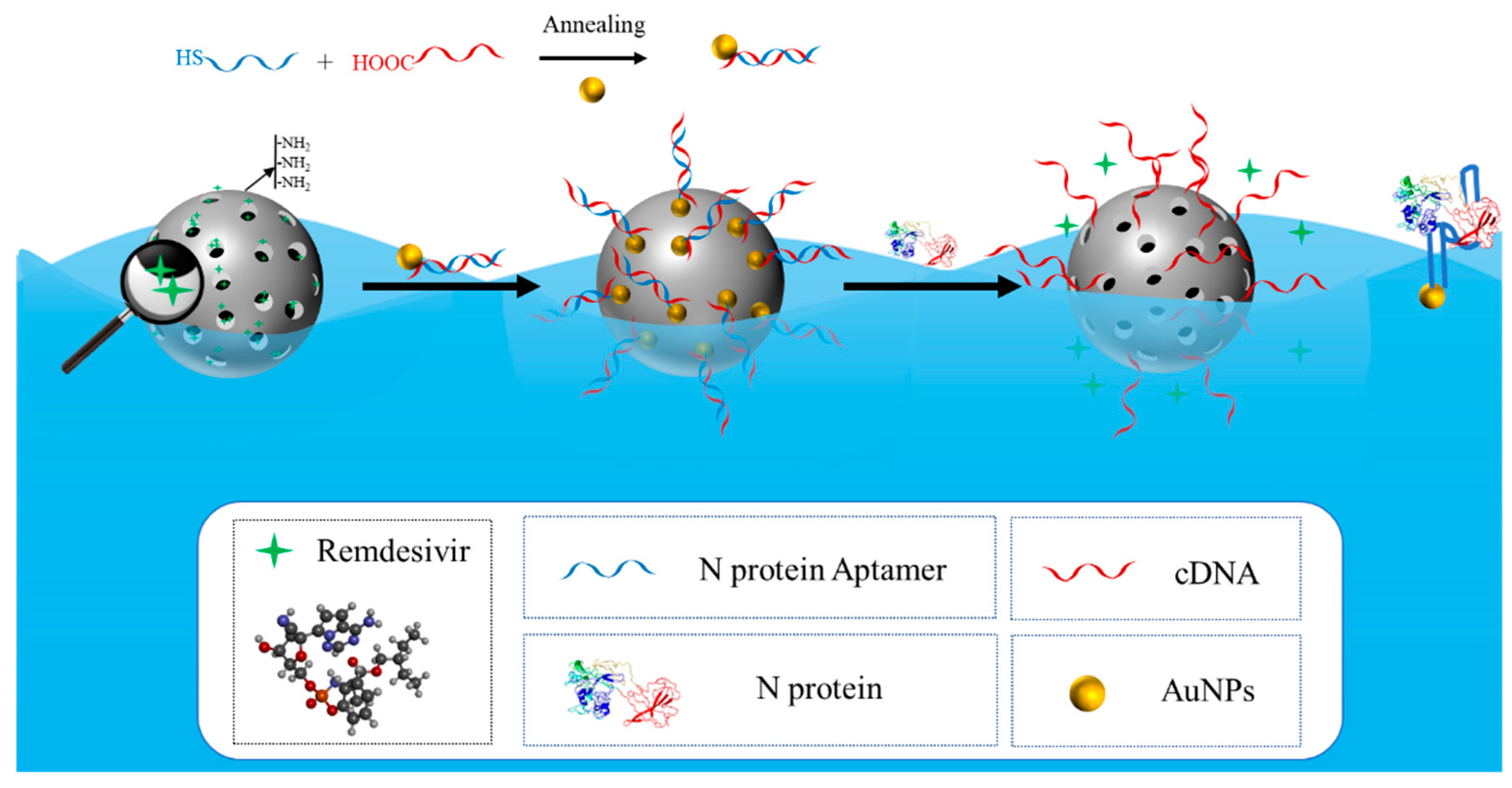
2.3. Binding of Aptamer to Gold Nanoparticles
2.4. Binding of Aptamers Modified with Gold Nanoparticles to MSN-NH2
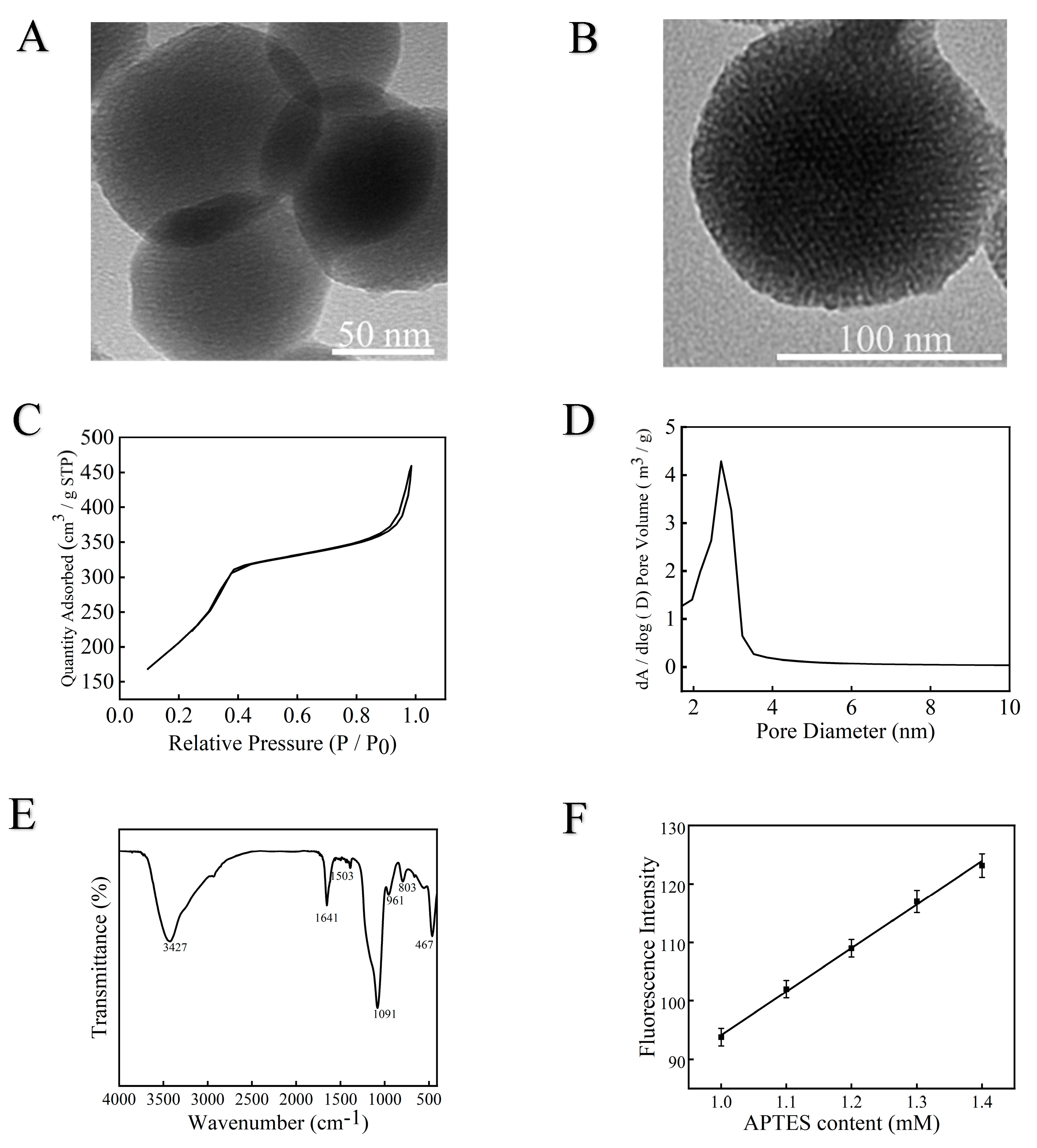
2.5. Characterization of Mesoporous Silica
2.6. N Protein Aptamer Selectivity
2.7. Analysis of RDV-Loaded Mesoporous Silica in Real Samples
3. Results and Discussion
3.1. Mesoporous Silica Characterization
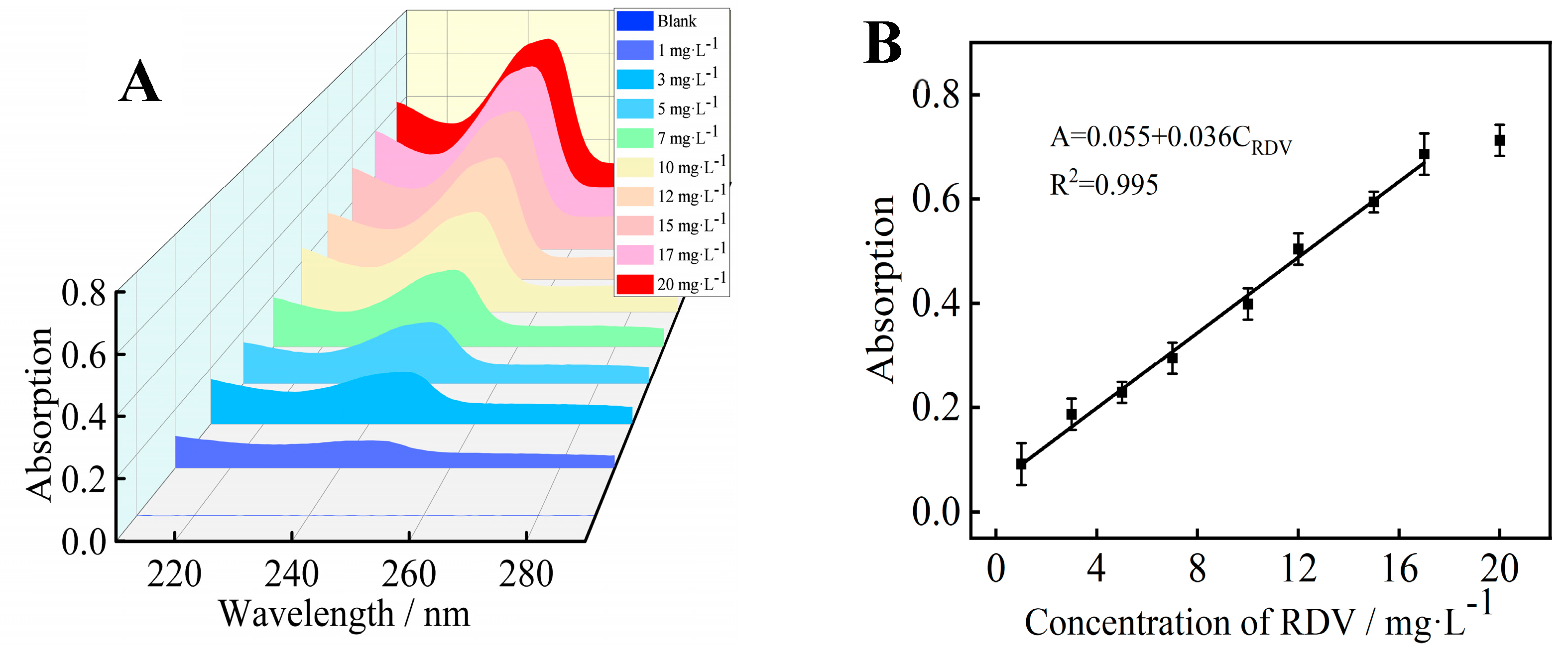
3.2. UV-vis Absorption Linear Range and Detection Limit of RDV
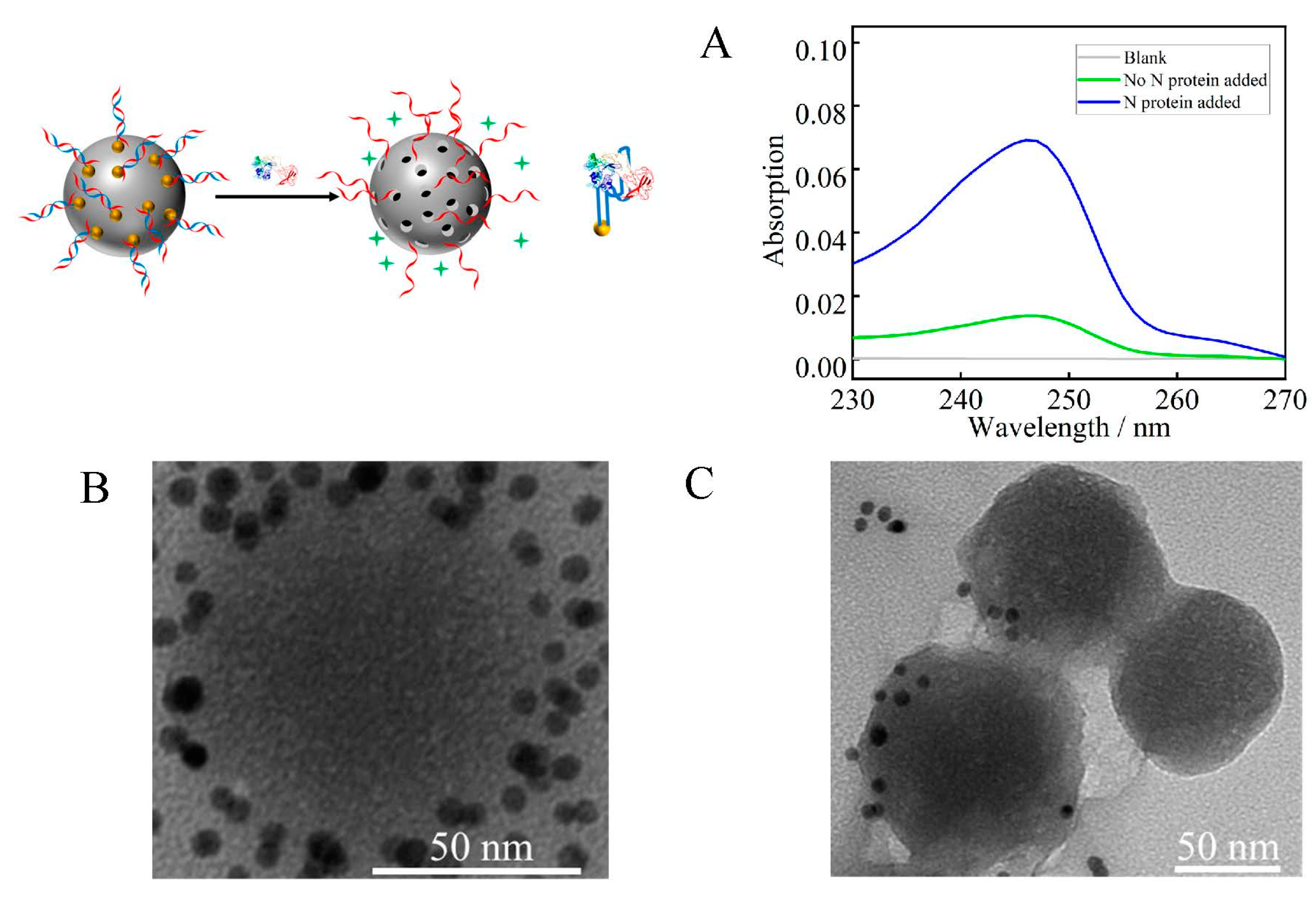
3.3. RDV Loading of Mesoporous Silica
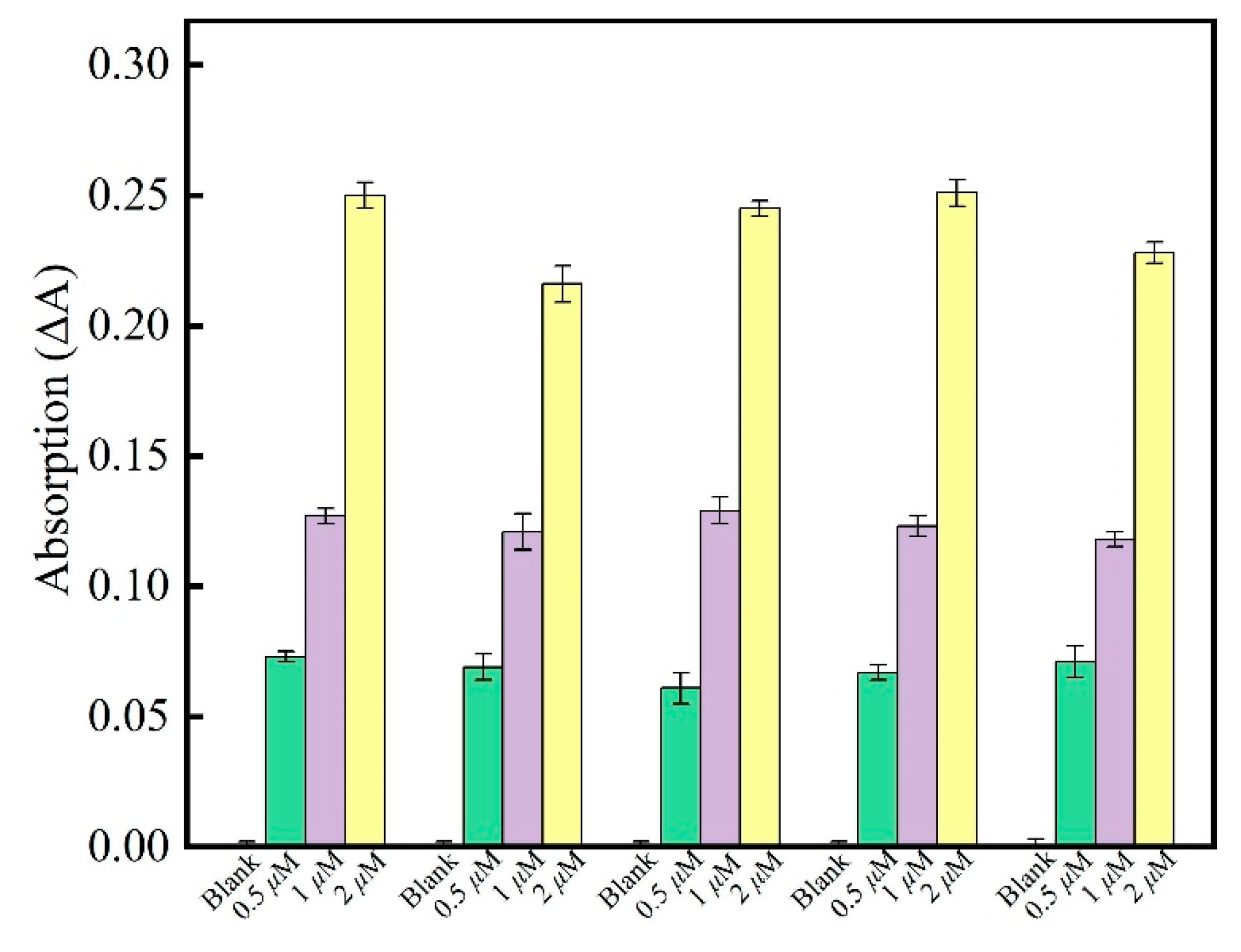
3.4. Aptamer Selectivity
3.5. Application Analysis in Human Blood
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hui, D.S.; Azhar, E.I.; Madani, T.A.; Ntoumi, F.; Kock, R.; Dar, O.; Ippolito, G.; McHugh, T.D.; Memish, Z.A.; Drosten, C.; et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020, 91, 264–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; De Clercq, E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat. Rev. Drug Discov. 2020, 19, 149–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Kang, H.; Liu, X.; Tong, Z. Combination of RT-qPCR testing and clinical features for diagnosis of COVID-19 facilitates management of SARS-CoV-2 outbreak. J. Med. Virol. 2020, 92, 538–539. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharyya, R.P.; Hanage, W.P. Challenges in inferring intrinsic severity of the SARSCoV-2 Omicron variant. N. Engl. J. Med. 2022, 386, e14. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Dellibovi-Ragheb, T.A.; Kerviel, A.; Pak, E.; Qiu, Q.; Fisher, M.; Takvorian, P.M.; Bleck, C.; Hsu, V.W.; Fehr, A.R.; et al. β-Coronaviruses Use Lysosomes for Egress Instead of the Biosynthetic Secretory Pathway. Cell 2020, 183, 1520–1535. [Google Scholar] [CrossRef]
- Zhang, G.; Li, B.; Yoo, D.; Qin, T.; Zhang, X.; Jia, Y.; Cui, S. Animal coronaviruses and SARS-CoV-2. Transbound. Emerg. Dis. 2021, 68, 1097–1110. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Karki, R.; Williams, E.P.; Yang, D.; Fitzpatrick, E.; Vogel, P.; Jonsson, C.B.; Kanneganti, T.-D. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat. Immunol. 2021, 22, 829–838. [Google Scholar] [CrossRef]
- Bernal, J.L.; Andrews, N.; Gower, C.; Gallagher, E.; Simmons, R.; Thelwall, S.; Stowe, J.; Tessier, E.; Groves, N.; Dabrera, G.; et al. Effectiveness of COVID-19 Vaccines against the B.1.617.2 (Delta) Variant. N. Engl. J. Med. 2021, 385, 585–594. [Google Scholar] [CrossRef]
- Tong, C.; Shi, W.; Zhang, A.; Shi, Z. Tracking and controlling the spatiotemporal spread of SARS-CoV-2 Omicron variant in South Africa. Trav. Med. Infect. Dis. 2022, 46, 102252–102260. [Google Scholar] [CrossRef]
- Ladhani, S.N.; Ireland, G.; Baawuah, F.; Beckmann, J.; Okike, I.O.; Ahmad, S.; Garstang, J.; Brent, A.J.; Brent, B.; Aiano, F.; et al. Emergence of SARS-CoV-2 Alpha (B.1.1.7) variant, infection rates, antibody seroconversion and seroprevalence rates in secondary school students and staff: Active prospective surveillance, December 2020 to March 2021, England. J. Infect. 2021, 83, 573–580. [Google Scholar] [CrossRef]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021, 592, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Jaspe, R.C.; Loureiro, C.L.; Sulbaran, Y.; Moros, Z.C.; D’Angelo, P.; Rodríguez, L.; Zambrano, J.L.; Hidalgo, M.; Vizzi, E.; Alarcón, V.; et al. Introduction and rapid dissemination of SARS-CoV-2 Gamma Variant of Concern in Venezuela. Infect. Genet. Evol. 2021, 96, 105147. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Huang, J.; Zhang, L.; Chen, S.; Gao, J.; Jiao, H. The global transmission of new coronavirus variants. Environ. Res. 2022, 206, 112240. [Google Scholar] [CrossRef] [PubMed]
- Shuai, H.; Chan, J.F.; Hu, B.; Chai, Y.; Yuen, T.T.; Yin, F.; Huang, X.; Yoon, C.; Hu, J.C.; Liu, H.; et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature 2022, 603, 693–699. [Google Scholar] [CrossRef]
- Chen, J.; Wang, R.; Gilby, N.B.; Wei, G.-W. Omicron Variant (B.1.1.529): Infectivity, Vaccine Breakthrough, and Antibody Resistance. J. Chem. Inf. Model. 2022, 62, 412–422. [Google Scholar] [CrossRef]
- Khamees, A.; Bani-Issa, J.; Zoubi, M.S.A.; Qasem, T.; AbuAlArjah, M.I.; Alawadin, S.A.; Al-Shami, K.; Hussein, F.E.; Hussein, E.; Bashayreh, I.H.; et al. SARS-CoV-2 and Coronavirus Disease Mitigation: Treatment Options, Vaccinations and Variants. Pathogebs 2022, 11, 275. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, M. Omicron Makes A Feeble Attack on the Lungs. Nature 2022, 601, 177. [Google Scholar] [CrossRef]
- Madhi, S.A.; Kwatra, G.; Myers, J.E.; Jassat, W.; Dhar, N.; Mukendi, C.K.; Nana, A.J.; Blumberg, L.; Welch, R.; Ngorima-Mabhena, N.; et al. Population Immunity and COVID-19 Severity with Omicron Variant in South Africa. N. Engl. J. Med. 2022, 386, 1314–1326. [Google Scholar] [CrossRef]
- Moore, P.L.; Baden, L.R. Omicron-Decoupling Infection from Severe Disease. N. Engl. J. Med. 2022, 386, 1361–1362. [Google Scholar] [CrossRef]
- Peiris, J.S.M.; Guan, Y.; Yuen, K.Y. Severe acute respiratory syndrome. Nat. Med. 2004, 10, S88–S97. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.; Wang, X.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.; Zhu, Y.; Li, B.; Huang, C.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhao, X.; Niu, J.; Li, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Hightower, L.E.; Santoro, M.G. Coronaviruses and stress: From cellular to global. Cell Stress Chaperones 2020, 25, 701–705. [Google Scholar] [CrossRef]
- Martinez, M.A. Compounds with Therapeutic Potential against Novel Respiratory 2019 Coronavirus. Antimicrob. Agents Chemother. 2020, 64, e00399-20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, C.J.; Tchesnokov, E.P.; Feng, J.Y.; Porter, D.P.; Götte, M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020, 295, 4773–4779. [Google Scholar] [CrossRef] [Green Version]
- Gordon, C.J.; Tchesnokov, E.P.; Woolner, E.; Perry, J.K.; Feng, J.Y.; Porter, D.P.; Götte, M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020, 295, 6785–6797. [Google Scholar] [CrossRef] [Green Version]
- Hong, Y.-N.; Xu, J.; Sasa, G.B.K.; Zhou, K.-X.; Ding, X.-F. Remdesivir as a broad-spectrum antiviral drug against COVID-19. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 541–548. [Google Scholar]
- Saha, A.; Sharma, A.R.; Bhattacharya, M.; Sharma, G.; Lee, S.-S.; Chakraborty, C. Probable molecular mechanism of Remdesivir for the Treatment of COVID-19: Need to Know More. Arch. Med. Res. 2020, 51, 585–586. [Google Scholar] [CrossRef]
- Tchesnokov, E.P.; Feng, J.Y.; Porter, D.P.; Götte, M. Mechanism of Inhibition of Ebola Virus RNA-Dependent RNA Polymerase by Remdesivir. Viruses 2019, 11, 326. [Google Scholar] [CrossRef] [Green Version]
- Grein, J.; Ohmagari, N.; Shin, D.; Diaz, G.; Asperges, E.; Castagna, A.; Feldt, T.; Green, G.; Green, M.L.; Lescure, F.-X.; et al. Compassionate Use of Remdesivir for Patients with Severe COVID-19. N. Engl. J. Med. 2020, 382, 2327–2336. [Google Scholar] [CrossRef] [PubMed]
- Yan, V.C.; Muller, F.L. Advantages of the Parent Nucleoside GS-441524 over Remdesivir for COVID-19 Treatment. ACS Med. Chem. Lett. 2020, 11, 1361–1366. [Google Scholar] [CrossRef] [PubMed]
- Ganewatta, M.S.; Tang, C.; Wang, Z. Chemical syntheses of bioinspired and biomimetic polymers toward biobased materials. Nat. Rev. Chem. 2021, 5, 753–772. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Ganewatta, M.S.; Tang, C. Sustainable polymers from biomass: Bridging chemistry with materials and processing. Prog. Polym. Sci. 2020, 101, 101197–101210. [Google Scholar] [CrossRef]
- Zhu, A.; Jiao, T.; Ali, S.; Xu, Y.; Ouyang, Q.; Chen, Q. SERS Sensors Based on Aptamer-Gated Mesoporous Silica Nanoparticles for Quantitative Detection of Staphylococcus aureus with Signal Molecular Release. Anal. Chem. 2021, 93, 9788–9796. [Google Scholar] [CrossRef] [PubMed]
- Climent, E.; Martínez-Máñez, R.; Sancenón, F.; Marcos, M.D.; Soto, J.; Maquieira, A.; Amorós, P. Controlled Delivery Using Oligonucleotide-Capped Mesoporous Silica Nanoparticles. Angew. Chem. Int. Ed. 2010, 49, 7281–7283. [Google Scholar] [CrossRef]
- Zhu, X.; Tang, F.; Yang, J.; Gao, Y. Determination of Acrolein-Derived 3-Hydroxypropylmercapturic Acid in Human Urine Using Solid-phase Extraction Combined with Molecularly Imprinted Mesoporous Silica and LC-MS/MS Detection. J. Chin. Biochem. Soc. 2014, 61, 227–232. [Google Scholar] [CrossRef]
- Zhu, C.-L.; Lu, C.-H.; Song, X.-Y.; Yang, H.-H.; Wang, X.-R. Bioresponsive Controlled Release Using Mesoporous Silica Nanoparticles Capped with Aptamer-Based Molecular Gate. J. Am. Chem. Soc. 2011, 133, 1278–1281. [Google Scholar] [CrossRef]
- Li, C.; Yang, J.; Xu, R.; Wang, H.; Zhang, Y.; Wei, Q. Progress and Prospects of Electrochemiluminescence Biosensors Based on Porous Nanomaterials. Biosensors 2022, 12, 508. [Google Scholar] [CrossRef]
- Zhu, M.; Tang, J.; Shi, T.; Ma, X.; Wang, Y.; Wu, X.; Li, H.; Hua, R. Uptake, translocation and metabolism of imidacloprid loaded within fluorescent mesoporous silica nanoparticles in tomato (Solanum lycopersicum). Ecotoxicol. Environ. Saf. 2022, 232, 113243–113251. [Google Scholar] [CrossRef]
- Zhang, L.; Fang, X.; Liu, X.; Ou, H.; Zhang, H.; Wang, J.; Li, Q.; Cheng, H.; Zhang, W.; Luo, Z. Discovery of sandwich type COVID-19 nucleocapsid protein DNA aptamers. Chem. Commun. 2020, 56, 10235–10238. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Jiao, X.; Liang, Z.; Zhao, H.; Zhao, Y.; Xie, J.; Jiang, Y.; Yu, X.; Fang, X.; Dai, X. Lateral flow immunoassay coupled with copper enhancement for rapid and sensitive SARS-CoV-2 nucleocapsid protein detection. Biosensors 2021, 12, 13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, X.; Feng, P.; Han, Q.; Liu, W.; Lu, Y.; Song, C.; Li, F. Photodriven regeneration of G-quadruplex aptasensor for sensitively detecting thrombin. Anal. Chem. 2020, 92, 7419–7424. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Zhang, X.; Zhang, X.; Zahra, Q.; Huang, Z.; Chen, Y.; Song, C.; Song, M.; Jiang, H.; Luo, Z.; et al. An electrochemical aptasensor with N protein binding aptamer-complementary oligonucleotides as probe for ultra-sensitive detection of COVID-19. Biosens. Bioelectron. 2022, 213, 114436–114441. [Google Scholar] [CrossRef]
- Cheng, S.; Tu, Z.; Zheng, S.; Cheng, X.; Han, H.; Wang, C.; Xiao, R.; Gu, B. An efficient SERS platform for the ultrasensitive detection of Staphylococcus aureus and Listeria monocytogenes via wheat germ agglutinin-modified magnetic SERS substrate and streptavidin/aptamer co-functionalized SERS tags. Anal. Chim. Acta 2021, 1187, 339155–339165. [Google Scholar] [CrossRef]
- Rostamizadeh, S.; Nojavan, M. An Environmentally Benign Multicomponent Synthesis of Some Novel 2-Methylthio Pyrimidine Derivatives Using MCM-41-NH2 as Nanoreactor and Nanocatalyst. J. Heterocycl. Chem. 2014, 51, 418–422. [Google Scholar] [CrossRef]
- Lu, T.; Chen, F. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef]
- Ding, H.; Zhang, Y.; Wang, S.; Xu, J.; Xu, S.; Li, G. Fe3O4@SiO2 core/shell nanoparticles: The silica coating regulations with a single core for different core sizes and shell thicknesses. Chem. Mater. 2012, 24, 4572–4580. [Google Scholar] [CrossRef]
- Kim, H.; Kim, T.; Roev, V.; Lee, H.; Kwon, H.; Lee, H.; Kwon, S.; Im, D. Enhanced electrochemical stability of Quasi−Solid−State electrolyte containing SiO2 nanoparticles for Li-O2 battery applications. ACS Appl. Mater. Interfaces 2016, 8, 1344–1350. [Google Scholar] [CrossRef]
- Marler, B.; Oberhagemann, U.; Vortmann, S.; Gies, H. Influence of the sorbate type on the XRD peak intensities ofloaded MCM−411. Microporous Mater. 1996, 6, 375–383. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Zhang, X.; Xu, A.; Yu, M.; Xu, Y.; Xu, Y.; Wang, C.; Yang, G.; Song, C.; Wu, X.; et al. Aptamer-Gated Mesoporous Silica Nanoparticles for N Protein Triggered Release of Remdesivir and Treatment of Novel Coronavirus (2019-nCoV). Biosensors 2022, 12, 950. https://doi.org/10.3390/bios12110950
Zhang X, Zhang X, Xu A, Yu M, Xu Y, Xu Y, Wang C, Yang G, Song C, Wu X, et al. Aptamer-Gated Mesoporous Silica Nanoparticles for N Protein Triggered Release of Remdesivir and Treatment of Novel Coronavirus (2019-nCoV). Biosensors. 2022; 12(11):950. https://doi.org/10.3390/bios12110950
Chicago/Turabian StyleZhang, Xiaohui, Xin Zhang, Aoqiong Xu, Mengdi Yu, Yu Xu, Ying Xu, Chao Wang, Gege Yang, Chunxia Song, Xiangwei Wu, and et al. 2022. "Aptamer-Gated Mesoporous Silica Nanoparticles for N Protein Triggered Release of Remdesivir and Treatment of Novel Coronavirus (2019-nCoV)" Biosensors 12, no. 11: 950. https://doi.org/10.3390/bios12110950
APA StyleZhang, X., Zhang, X., Xu, A., Yu, M., Xu, Y., Xu, Y., Wang, C., Yang, G., Song, C., Wu, X., & Lu, Y. (2022). Aptamer-Gated Mesoporous Silica Nanoparticles for N Protein Triggered Release of Remdesivir and Treatment of Novel Coronavirus (2019-nCoV). Biosensors, 12(11), 950. https://doi.org/10.3390/bios12110950




