3.1. Characterization of the Protein Extracts and Antibodies
The total protein content was quantified using the Bradford method. The protein content was 8.6 and 5.9 mg/mL for the almond and peanut extracts, respectively. Then, SDS-PAGE electrophoresis was performed to identify the allergenic proteins and evaluate the effect of the extraction procedure on the protein’s stability. The protein profiles achieved for the almond and peanut extracts were similar to the reported ones. As observed in
Figure S1, up to 25 different protein bands, ranging from 12 to 66 KDa, were identified in the almond extract, the 20–22 and 38–42 KDa ranges being the most abundant. These two proteins are known to form the amandin protein (the most abundant protein in almonds, 360 KDa) when bonded together [
24,
25]. Moreover, out of the 25 proteins present in almonds, eight of them have been reported as allergenic [
26]. Eight allergens were identified in the almond extract (
Table S1). Regarding the peanut extract, the three major allergenic proteins were also identified: Ara h 1, Ara h 2, and Ara h 3, which have a molecular weight of 65, 17, and 61 KDa, respectively [
27]. The latter is based on two subunits corresponding to the bands at 45 and 22 KDa [
28].
Once the presence and stability of the allergenic proteins in the food extracts were confirmed, polyclonal antibodies were raised by immunizing the rabbits with the respective extracts. The immunization process was monitored by evaluating the antibody titer with ELISA. A titer of 1/10
5 and 1/6.1·10
7 was achieved for the almond and peanut anti-sera, respectively, denoting a successful production of specific antibodies (
Figure S2). Furthermore, a western blot was performed to identify the proteins in the food extract that present reactivity against the raised antibodies. As shown in
Figure S3, the antibodies recognize the major allergenic and non-allergenic proteins. This fact is of interest in the food industry, considering the current heterogeneous and rapidly changing hypersensitivity dispersion.
3.2. Development of Individual Assays
Single non-competitive (immunosandwich) assays were developed to evaluate the sensitivity and selectivity of the assay using the purified antibodies. The LFIA followed the conventional double line (test and control lines). AuNPs of 20 nm sizes were selected as the signal transducer (
Figure S4) due to their simple, cost-effective synthesis and straightforward bioconjugation via direct adsorption of the antibodies to the nanoparticle’s surface. Calibration curves were performed by interrogating serial dilutions of known concentrations of the extracted proteins. The collected data for each calibration curve was fitted to a four-parameter logistic (sigmoidal) equation (
Figures S5 and S6), and the analytical parameters related to the LoD, LoQ, IC
50, dynamic range, and linear regression coefficient (r
2) were determined (
Table S2). Both calibration curves followed a dose-response relationship within the ng range, which is more than enough, considering that the eliciting dose predicted to provoke a reaction in 50% of individuals (ED
50) with an almond allergy- and peanut-derived proteins is 20 and 29 mg, respectively [
29].
3.3. Design of the LFµIA Layout
Once the developed LFIAs had an appropriate sensitivity when detecting the allergenic proteins of interest, the next step was to design the microarray layout. As previously discussed, the idea was to (1) integrate the almond, and peanut assays into the same strip and (2) provide the end user with more analytical information than the conventional LFIA. To this end, assay duplicates were included in the microarray to detect almond- and peanut-extracted proteins (row R1), together with four replicates of a negative control assay (row R2), four replicates of an internal calibration curve, and a positive control assay (rows R3–R6). Duplicates of a second test zone for almond and peanut (row R7) and four replicates of a hook control assay (rows R8–R9) were also included (
Figure 2).
At this stage, the main objective was to miniaturize the microarray to fit 36 spots within a reduced sensing area. According to previous experience and the existing published advice regarding the design and development of LFIA [
13], the first row was printed 15 mm from the conjugate pad, providing enough interaction time for the immunocomplex formation between the conjugated AuNPs and the target analyte. Next, the diameter of the spots was optimized by evaluating different dispensed volumes (25, 50, and 75 nL). As expected, the diameter of the spots enlarged as the volume increased. The average diameter of the spots was 0.30 ± 0.03 mm, 0.40 ± 0.05 mm, and 0.38 ± 0.08 mm when dispensing 25, 50, and 75 nL, respectively (
Figure S7A).
Interestingly, it was also observed that the dispensing reproducibility significantly decreased when spotting 50 and 75 nL. The percentage relative standard deviation of the spots’ diameter was 8.9, 12.4, and 20.0% when dispensing 25, 50, and 75 nL, respectively. Analysis was also carried out on how the diameter of the spots affects the signal generated when performing LFIA. To this end, calibration curves were performed based on a direct immunoassay format by fixing the concentration of the secondary anti-rabbit IgG (0.12, 0.25, 0.50, 0.75, and 1.00 mg mL
−1 in PBS) in the nitrocellulose membrane of the LFIA strips. AuNPs conjugated with the purified antibodies were used as the biosensing element. As shown in
Figure S7B, the highest signal, analytical sensitivity, and regression coefficient were achieved when dispensing 25 nL of the capture bioreceptors. This result might be due to the high density of bioreceptors in the spots, which increases the number of AuNPs per mm
2. Moreover, the spots produced when dispensing 25 nL can be visualized by direct naked-eye inspection, since there is a proper separation distance of 0.28 ± 0.02 mm between other spots. Consequently, supported by the above results, 25 nL was selected as the optimal dispensing volume.
The microarray had dimensions of 5 × 2.5 mm and covered one-third of the detection pad in length and two-thirds in width. The arrangement of the various elements in the microarray was carefully designed. The test zone (spots) must be placed in row R1 to ensure that the differences in the generated signal are related to the concentration of the allergenic proteins rather than the differences in the number of available conjugated AuNPs. Thus, based on the selected non-competitive assay format, a positive sample containing proteins of both almond and peanut will trigger the generation of a high signal in the test zone. In contrast, a low signal will be related to a negative sample.
Similarly, the negative control assay must be placed as close as possible to the test zone, which in this case was in row R2. The reason is that the excess of conjugated AuNPs in this zone is still high. Hence, achieving a negligible signal in the negative control assay (fixed bovine serum albumin) proves that the conjugated AuNPs do not generate non-specific signals.
As far as the internal calibration assay is concerned, the signals come from the immobilized secondary anti-rabbit IgG (0.75, 0.50, 0.25, and 0.12 mg mL−1 as calibrators 1 to 4, respectively). Consequently, the signal generated will be high, regardless of the concentration of target analytes in the sample; this is why the internal calibration is also used as a positive control assay. The signal intensities will vary linearly, depending on the concentration of the secondary antibody immobilized on the nitrocellulose membrane. The analyte concentration in the sample will be quantified by comparing the signal intensity of the internal calibrators with the one in the test zone. Four replicates of each point were used to increase the internal signal calibration accuracy. The spots corresponding to the four concentrations were included in each row and exchanged in each replicate. The purpose of the latter is to check that there are no differences in the signal intensity when placing a spot on the left or the right of the microarray since this would indicate that the sample flow is not homogeneously distributed along the transverse axis of the LFIA strip.
Test zone 2 (row R7) is a replicate of test zone 1 (row R1) and aims to detect the almond and peanut proteins that cannot react at row 1, mainly due to the lack of interaction time or the saturation of the capture antibodies. Considering the lower amount of available AuNPs at this point of the LFIA strip, the second test zone could provide a shift in the dynamic range compared to the one achieved in the first test zone. In this way, more qualitative information for the end user is obtained. The spots related to the almond and peanut assay were also exchanged to check if the sample was flowing homogeneously. Finally, the hook effect control zone corresponds to rows 8–9. The hook effect is a false negative result produced by an extremely high concentration of the target analyte in the sample [
30]. Four replicates of almond and peanut proteins were included in the array layout to study this hook effect. Increasing target analyte concentrations in the sample will reduce the available detection antibodies conjugated to the AuNPs, which translates to progressively lower signal intensities in the hook effect control zone. In this sense, the lack of signal in the test zones will be attributed to the hook effect and not to the interrogation of a negative sample, provided that there is no signal in the hook effect control zone.
3.4. Development of Multiplexed Assays in LFµIA
The next step involved calibration curves to simultaneously detect almond- and peanut-extracted proteins. The signal intensity in the test zones increased with higher target analyte concentrations. Signal saturation was reached at 3000 ng mL
−1 before experiencing a decrease due to the hook effect. The signal intensities generated in the first test zone (row 1, indicated with an arrow in
Figure 3A) were normalized (min–max), fitted to a four-parameter logistic (sigmoidal) equation (
Figure 3B), and the analytical parameters related to the LoD, LoQ, IC
50, dynamic range and linear regression coefficient (r
2) were determined (
Table S3). The achieved sensitivity expressed as LoD was 185 and 229 µg/kg for the almond and peanut assay, respectively. Compared with the analytical parameters achieved with the individual calibration curves in the conventional LFIA format (
Section 3.2), the almond assay showed slightly lower sensitivity and a shift of the IC
50 and dynamic range to higher concentrations of a target analyte. Conversely, the peanut assay presented a similar LoD, LoQ, IC
50, and dynamic range. The calibration curve’s relative standard deviations (RSD) were between 5% and 25% within 100 to 3000 µg/mL for both analytes. Thus, it was concluded that integrating the almond and peanut assays within the same strip in a microarray format did not significantly vary the analytical performances.
The signal intensities related to the second test zone were then analyzed. As expected, these were much lower than their counterparts in the first test zone due to the fewer available conjugated AuNPs and target analytes. However, the study took advantage of this condition to develop a test zone with a right-shifted dynamic range. Following the ratiometric concept [
31], when combining the same concentration of immobilized capture bioreceptors with lower amounts of conjugated AuNPs, a much higher target analyte concentration will be required to achieve a dose-response relationship. This approach enabled an 86.8 and 47.2% higher IC
50 in the almond and peanut assays, respectively, of the second test zone compared with the first test zone (
Table S3). This shift in the dynamic range was beneficial since it overcame the hook effect observed when evaluating 3000 ng mL
−1 of the target analyte in the first test zone (
Figure S8). Moreover, the dynamic range of the second test zone for the almond and peanut assays also covered a 3.4- and 1.5-fold higher concentration of target analyte compared to the first test zone. To this end, using the first and second test zones provides an enhanced amount of information because they cover a wider dynamic range than a single test zone. Moreover, the degree of shift in the dynamic range of the second test zone can be modulated by the immobilization of higher or lower concentrations of capture antibodies.
3.5. Assay Selectivity
One of the most important aspects when developing multiplexed biosensing platforms is to assess the assay selectivity, as this parameter indicates the degree of cross-reactivity between the antibodies and target analytes of the different assays. A high selectivity ensures that the signal achieved in the test zone is related to detecting the desired target analyte. Therefore, calibration curves were performed for the individual detection of almond (
Figure 4A) and peanut (
Figure 4B) proteins using LFµIA, and the signal intensities were measured in the spots of the first test zone (
Figure 4 inset). As observed, a dose-response relationship was only achieved in the assays of each particular analyte. In contrast, the other assays show a low and constant signal independent of the concentration of proteins. The percentage cross-reactivity between the assays was quantified by dividing the signal intensity achieved when detecting the proteins of interest with the non-intended match pair by the one achieved with the intended match pair [
32]. The cross-reactivity of the assay was calculated by using 1000 ng mL
−1 of each protein extract since this value is close to the IC
50 in both assays. The peanut assay has a 10% cross-reactivity with the almond proteins, while the almond assay shows an 8% cross-reactivity with the peanut proteins. The obtained results determine that the developed multiplexed LFµIA is highly selective. The antibodies and target analytes related to almond and peanut assays practically do not cross-react.
The cross-reactivity of the developed LFµIA with other common allergenic proteins was also assessed, comparing the obtained signal response when analyzing other allergens with the expected response when analyzing the intended allergens. The cross-reactivity of the almond and peanut assays was lower than 23% when evaluating 1000 ng mL
−1 of shrimp, hazelnut, egg, milk, wheat, and fish allergens (
Figure S9 and
Table S4). Furthermore, the signal recovery (
) when analyzing 1000 ng mL
−1 of the almond and peanut prick test allergens was 110.3 ± 30.5% and 142.6 ± 13.8%, respectively. Such high percentage signal recoveries reveal strong analytical performances of LFµIA.
3.6. Assay Controls and Internal Calibration
After characterizing the analytical properties of the test zones, the next step was to analyze the information provided by the other elements of the microarray. Thus, the signal intensities in the negative and positive control assays were examined. This analysis confirmed that the values stayed constant (RSD < 4%), regardless of the extracted protein concentration added to the LFµIA strip. The high signal intensities in the positive control assay can be used to confirm that (1) the AuNPs are successfully conjugated to the purified antibodies and (2) the AuNPs conjugated solution flows appropriately along the nitrocellulose membrane (
Figure 5A). On the other hand, the intensity of the negative control was negligible and similar to the background signal observed in the nitrocellulose membrane, thus proving the absence of non-specific signals.
The subsequent step involved analyzing the signal intensities generated in the hook effect control and fitting the data into a linear regression curve. As observed in
Figure 5B, the signal intensity is inversely proportional to the concentration of the target analyte (O.I. = −0.044 ln [protein extract (ng mL
−1)] + 0.57 R
2 = 0.98). The hook effect control assays are direct immunoassays between the almond- and peanut-extracted proteins and the conjugated AuNPs. Thus, higher target analyte concentrations in the sample block a higher number of binding sites of the antibodies conjugated to the AuNPs, which hinders the biorecognition of the conjugated AuNPs by the immobilized proteins of the hook effect control zone. For that reason, the hook effect control assay is useful in situations like that observed in
Figure 3B, where similar signal intensities are achieved when evaluating 1000 and 10,000 ng mL
−1 (
Figure 5B strips ii and iv). The differences in signal intensity (0.3 and 0.15, respectively) observed in the hook effect control assays of these two strips can be successfully used to distinguish both concentrations, thus reducing the occurrence of false results.
The signal intensities of the internal calibration spots were also measured alongside a comparison with those from the first test zone (external calibration) (
Figure 5C). As shown in
Figure 5D, the internal curve is similar to the external calibration (
Table S5), revealing an accurate assay quantification in a single step.
3.7. Real Sample Analysis
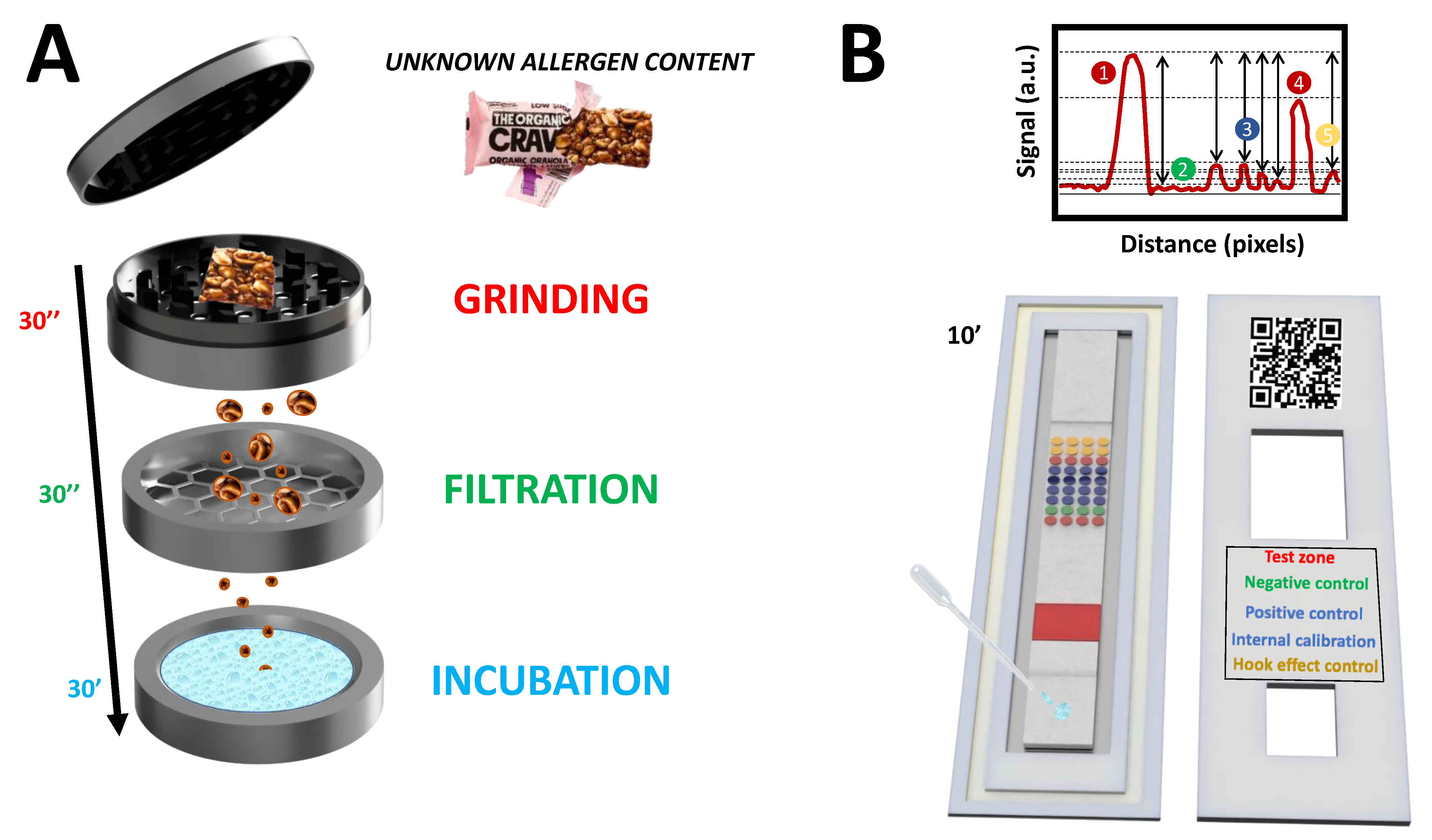
The LFµIA was evaluated by testing a snack bar with a declared content of peanuts (34%) and possible traces of almonds. This testing allowed for (1) determining the percentage recovery when detecting peanuts and (2) checking whether the proposed biosensing platform had enough sensitivity to quantify traces of almonds. The protein extraction was performed using a fast, power-free method that can be easily performed on-site. A grinding device with dimensions of 4.8 ×4.5 cm was used, containing all the required elements for extracting the allergenic proteins from the sample. As shown in
Figure S10, the method consisted of the manual grinding of the snack bar, followed by the filtering of the shreds, the addition and incubation of the extraction buffer, and the collection of the solution containing the extracted proteins. Compared with a standard commercial extraction approach, which requires bulky external instrumentation (water bath, rotatory station, and centrifuge) and at least seven different steps [
23], the proposed method is 3.6-fold faster. It enables the extraction of 76.7% of the proteins (according to the Bradford method).
The next step was to analyze the extracted proteins with the developed LFµIA. As is shown in
Figure S11i, low signal intensity in the almond assay was detected, alongside a significantly high signal intensity in the peanut assay. By comparing the achieved signals with the ones of the internal signal calibration curve, a concentration of almond proteins of 212 ± 45 ng mL
−1 (RSD 21%) was determined, a value that was slightly lower than the LoQ (273.9 ng mL
−1) but higher than the LoD (184.8 ng mL
−1). Thus, the LFµIA could not provide a reliable value of the almond content, but the assay showed enough analytical sensitivity to detect traces of almonds. The detected quantity of peanut protein in the sample was 516 ng mL
−1. However, the hook effect control assay showed a negligible signal intensity in the peanut assay, indicative of the hook effect due to the presence of peanut proteins at a concentration higher than 3000 ng mL
−1. The analysis of the peanut protein content with the Bradford method confirmed a concentration of peanut proteins of 23,000 ng mL
−1 (considering that the snack bar declared a 34% peanut content and that 26% of the peanut content is related to proteins). Therefore, it was proved that the developed LFµIA provided helpful information for identifying the hook effect. It is worth mentioning that, without the hook effect control assay, the performed assay would have returned a false negative result by quantifying a 45-fold lower concentration of peanut in the sample.
Then, the extracted sample was diluted to adjust the peanut protein content to 2300 ng mL
−1, which is within the operating range of the calibration curve. This dilution enabled the proper quantification of the peanut protein content with the internal signal calibration of the LFµIA (
Figure S11ii). The LFµIA results revealed a concentration of peanut protein of 1343 ng mL
−1, corresponding to a recovery of 58 ± 8%. Considering that the Bradford method calculated the peanut protein concentration, this result can be regarded as appropriate. Finally, as expected, a decrease was observed in the signal intensity of the peanut assay, alongside a signal intensity increase in the hook effect control assay, when diluting the sample up to a peanut protein content of 230 ng mL
−1 (
Figure S11iii). These data revealed that this sample had a lower peanut protein concentration than the previously analyzed one. In quantitative terms, the peanut proteins were detected at a concentration of 451 ng mL
−1, resulting in a 196% recovery. However, it is worth mentioning that the peanut protein content taken as reference (230 ng mL
−1) was out of the dynamic range of the calibration curve (300–3000 ng mL
−1) and lower than the LoQ (264.7 ng mL
−1). The data revealed that the developed LFµIA could provide accurate qualitative information on the almond and peanut protein content and semiquantitative information when the protein concentration was within the dynamic range. Moreover, it is fundamental to highlight the appropriateness of the positive and negative control assays integrated into the LFµIA, since the signals remain constant when analyzing such a complex matrix.