Recent Advances, Opportunities, and Challenges in Developing Nucleic Acid Integrated Wearable Biosensors for Expanding the Capabilities of Wearable Technologies in Health Monitoring
Abstract
:1. Introduction
2. Fabrication Requirements
2.1. Microfluidics/Reagent Layer
2.2. Sensing Layer
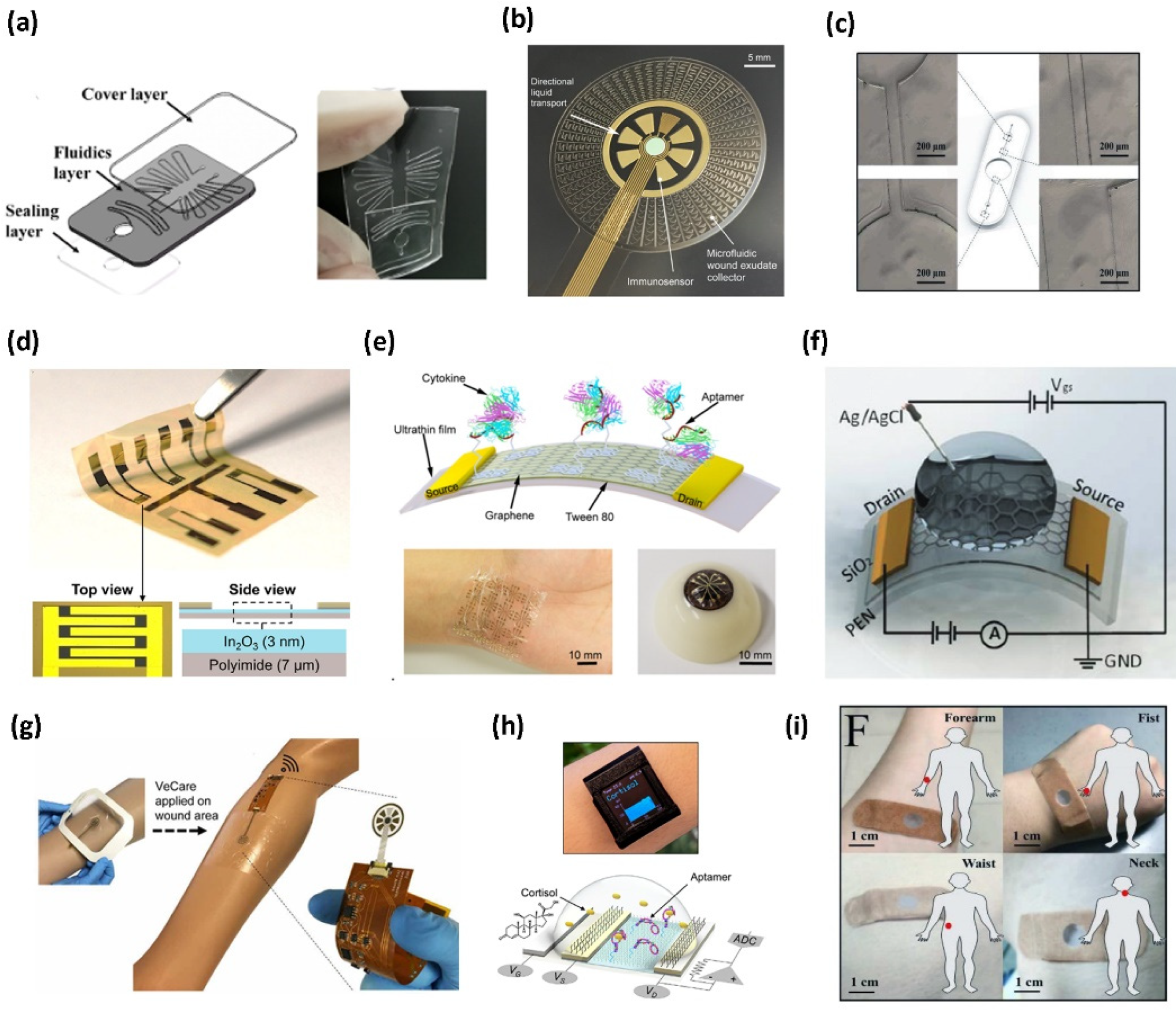
2.3. Readout/Packaging Layer
3. Biorecognition Requirements—Nucleic Acid-Based Assays
3.1. Synthetic Oligonucleotides
| Sensing Material | Target Analyte | Transduction Method | Limit of Detection | Reference |
|---|---|---|---|---|
| Synthetic nucleotide | ||||
| Ecoflex microfluidics | Nucleic acid fragments of Zika virus | Fluorescence | 10 copies μL−1 (1.66 × 10−2 fM) | [30] |
| PDMS | E. coli O157:H7 SARS-CoV-2 | Colorimetry | 500 pg/reaction (2.44 × 10−1 nM) 600 fg/reaction (3.03 × 10−1 pM) | [32] |
| PDMS | HIV-1 DNA | Fluorescence | 100 copies mL−1 (1.66 × 10−4 fM) | [34] |
| Graphene—based FET with PDMS microfluidics | miRNA-4484 | FET | 10 fM | [46] |
| Hydrogel microneedles modified with gold nanowires | Epstein−Barr virus cell-free DNA | Electrochemical | 3.7 × 102 copies μL−1 (6.1 × 10−1 fM) | [50] |
| CRISPR-Cas | ||||
| CRISPER-based Freeze-dried cell-free synthetic circuit | MecA gene HIV RNA Ebola virus RNA | Fluorescence Colorimetry Luminescence | 2.7 fM 10 μM 300 nM | [25] |
| Aptamers | ||||
| In2O3 FET on polyamide with tape-based microfluidics | Cortisol | FET | 1 pM | [27] |
| Gold modified electrodes with Graphene-gold nanoparticles with SU-8 microfluidics | IL-6 IL-8 TGF–β1 Staphylococcus aureus | Electrochemical | 10 ng mL−1 (4.76 × 10−1 nM) 10 ng mL−1 (1.18 nM) 50 pg mL−1 (1.13 pM) 1 ×108 CFU mL−1 | [29] |
| Graphene—Based FET on ultrathin Mylar | TNF-α IFN-γ | FET | 2.75 pM 2.89 pM | [41] |
| Graphen-Based FET on SiO2 Coated PEN | TNF-α | FET | 26 pM | [42] |
| Platinum-Graphene Extended gate electrode | Cortisol | FET | 0.2 nM | [47] |
| MeHA functionalized hydrogel microneedles | Glucose ATP L-tyrosinamide thrombin | Optical | 1.1 mM 0.1 mM 3.5 µM 25 nM | [59] |
| Graphen-Nafion composite film | IFN-γ | FET | 740 fM | [79] |
| ZnO on microporous hydrophilic membrane | Cortisol | Electrochemical | 0.11 µM | [80] |
| ZnO coated nano-porous polyamide | Cortisol | Electrochemical | 2.7 nM | [81] |
| PDMS@CNC/CNT | Cortisol | Electrochemical | 5 nM | [82] |
| In2O3 nanoribbons on PET | Serotonin Dopamine | FET | 10 fM | [83] |
| PEDOT-PAN nanofibers FET on PET | Cortisol | FET | 10 pM | [84] |
3.2. Aptamers
3.3. CRISPR-Cas
4. Transduction Mechanism Requirements
4.1. Optical
4.2. Electrical
4.3. Electrochemical
5. Summary and Future Outlook
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wood, C.S.; Thomas, M.R.; Budd, J.; Mashamba-Thompson, T.P.; Herbst, K.; Pillay, D.; Peeling, R.W.; Johnson, A.M.; McKendry, R.A.; Stevens, M.M. Taking Connected Mobile-Health Diagnostics of Infectious Diseases to the Field. Nature 2019, 566, 467–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, J.-W.; Yeo, W.-H.; Akhtar, A.; Norton, J.J.S.; Kwack, Y.-J.; Li, S.; Jung, S.-Y.; Su, Y.; Lee, W.; Xia, J.; et al. Materials and Optimized Designs for Human-Machine Interfaces Via Epidermal Electronics. Adv. Mater. 2013, 25, 6839–6846. [Google Scholar] [CrossRef] [PubMed]
- Pusta, A.; Tertiș, M.; Cristea, C.; Mirel, S. Wearable Sensors for the Detection of Biomarkers for Wound Infection. Biosensors 2021, 12, 1. [Google Scholar] [CrossRef]
- Davies, E.H.; Johnston, J.; Toro, C.; Tifft, C.J. A Feasibility Study of MHealth and Wearable Technology in Late Onset GM2 Gangliosidosis (Tay-Sachs and Sandhoff Disease). Orphanet J. Rare Dis. 2020, 15, 199. [Google Scholar] [CrossRef] [PubMed]
- Cappon, G.; Acciaroli, G.; Vettoretti, M.; Facchinetti, A.; Sparacino, G. Wearable Continuous Glucose Monitoring Sensors: A Revolution in Diabetes Treatment. Electronics 2017, 6, 65. [Google Scholar] [CrossRef] [Green Version]
- Smith, E.N.; Santoro, E.; Moraveji, N.; Susi, M.; Crum, A.J. Integrating Wearables in Stress Management Interventions: Promising Evidence from a Randomized Trial. Int. J. Stress Manag. 2020, 27, 172–182. [Google Scholar] [CrossRef]
- Kim, J.; Campbell, A.S.; de Ávila, B.E.-F.; Wang, J. Wearable Biosensors for Healthcare Monitoring. Nat. Biotechnol. 2019, 37, 389–406. [Google Scholar] [CrossRef]
- Bhide, A.; Ganguly, A.; Parupudi, T.; Ramasamy, M.; Muthukumar, S.; Prasad, S. Next-Generation Continuous Metabolite Sensing toward Emerging Sensor Needs. ACS Omega 2021, 6, 6031–6040. [Google Scholar] [CrossRef]
- Mitra, P.; Sharma, P. POCT in Developing Countries. EJIFCC 2021, 32, 195–199. [Google Scholar]
- Slade Shantz, J.A.; Veillette, C.J.H. The Application of Wearable Technology in Surgery: Ensuring the Positive Impact of the Wearable Revolution on Surgical Patients. Front. Surg. 2014, 1, 39. [Google Scholar] [CrossRef] [Green Version]
- Weizman, Y.; Tan, A.M.; Fuss, F.K. Use of Wearable Technology to Enhance Response to the Coronavirus (COVID-19) Pandemic. Public Health 2020, 185, 221–222. [Google Scholar] [CrossRef] [PubMed]
- Seshadri, D.R.; Li, R.T.; Voos, J.E.; Rowbottom, J.R.; Alfes, C.M.; Zorman, C.A.; Drummond, C.K. Wearable Sensors for Monitoring the Internal and External Workload of the Athlete. NPJ Digit. Med. 2019, 2, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, J.; Torrente-Rodríguez, R.M.; Wang, M.; Gao, W. The Era of Digital Health: A Review of Portable and Wearable Affinity Biosensors. Adv. Funct. Mater. 2020, 30, 1906713. [Google Scholar] [CrossRef]
- Sharma, A.; Badea, M.; Tiwari, S.; Marty, J.L. Wearable Biosensors: An Alternative and Practical Approach in Healthcare and Disease Monitoring. Molecules 2021, 26, 748. [Google Scholar] [CrossRef] [PubMed]
- Windmiller, J.R.; Wang, J. Wearable Electrochemical Sensors and Biosensors: A Review. Electroanalysis 2013, 25, 29–46. [Google Scholar] [CrossRef]
- AJ Bandodkar, I.J.J.W. Wearable Chemical Sensors: Present Challenges and Future Prospects. ACS Sens. 2016, 1, 464–482. [Google Scholar] [CrossRef]
- Lee, H.; Hong, Y.J.; Baik, S.; Hyeon, T.; Kim, D.H. Enzyme-Based Glucose Sensor: From Invasive to Wearable Device. Adv. Healthc. Mater. 2018, 7, 1701150. [Google Scholar] [CrossRef] [Green Version]
- Sonawane, A.; Manickam, P.; Bhansali, S. Stability of Enzymatic Biosensors for Wearable Applications. IEEE Rev. Biomed. Eng. 2017, 10, 174–186. [Google Scholar] [CrossRef]
- Micura, R.; Höbartner, C. Fundamental Studies of Functional Nucleic Acids: Aptamers, Riboswitches, Ribozymes and DNAzymes. Chem. Soc. Rev. 2020, 49, 7331–7353. [Google Scholar] [CrossRef]
- Khorana, H.G. Nucleic Acid Synthesis. Pure Appl. Chem. 1968, 17, 349–382. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Cao, Z.; Lu, Y. Functional Nucleic Acid Sensors. Chem. Rev. 2009, 109, 1948–1998. [Google Scholar] [CrossRef] [PubMed]
- Famulok, M.; Hartig, J.S.; Mayer, G. Functional Aptamers and Aptazymes in Biotechnology, Diagnostics, and Therapy. Chem. Rev. 2007, 107, 3715–3743. [Google Scholar] [CrossRef] [PubMed]
- Paul, R.; Ostermann, E.; Wei, Q. Advances in Point-of-Care Nucleic Acid Extraction Technologies for Rapid Diagnosis of Human and Plant Diseases. Biosens. Bioelectron. 2020, 169, 112592. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Zhao, Z.; Lv, Y.-F.; Huan, S.-Y.; Fu, T.; Zhang, X.-B.; Shen, G.-L.; Yu, R.-Q. DNAzyme-Based Biosensors and Nanodevices. Chem. Commun. 2015, 51, 979–995. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.Q.; Soenksen, L.R.; Donghia, N.M.; Angenent-Mari, N.M.; de Puig, H.; Huang, A.; Lee, R.; Slomovic, S.; Galbersanini, T.; Lansberry, G.; et al. Wearable Materials with Embedded Synthetic Biology Sensors for Biomolecule Detection. Nat. Biotechnol. 2021, 39, 1366–1374. [Google Scholar] [CrossRef]
- Pandey, R.; Chang, D.; Smieja, M.; Hoare, T.; Li, Y.; Soleymani, L. Integrating Programmable DNAzymes with Electrical Readout for Rapid and Culture-Free Bacterial Detection Using a Handheld Platform. Nat. Chem. 2021, 13, 895–901. [Google Scholar] [CrossRef]
- Wang, B.; Zhao, C.; Wang, Z.; Yang, K.-A.; Cheng, X.; Liu, W.; Yu, W.; Lin, S.; Zhao, Y.; Cheung, K.M.; et al. Wearable aptamer-field-effect transistor sensing system for noninvasive cortisol monitoring. Sci. Adv. 2022, 8, eabk0967. [Google Scholar] [CrossRef]
- Takaloo, S.; Moghimi Zand, M. Wearable Electrochemical Flexible Biosensors: With the Focus on Affinity Biosensors. Sens. Biosens. Res. 2021, 32, 100403. [Google Scholar] [CrossRef]
- Gao, Y.; Nguyen, D.T.; Yeo, T.; bin Lim, S.; Xian Tan, W.; Edward Madden, L.; Jin, L.; Yong Kenan Long, J.; Abu Bakar Aloweni, F.; Jia Angela Liew, Y.; et al. A Flexible Multiplexed Immunosensor for Point-of-Care in Situ Wound Monitoring. Sci. Adv. 2021, 7, eabg9614. [Google Scholar] [CrossRef]
- Yang, B.; Kong, J.; Fang, X. Bandage-like Wearable Flexible Microfluidic Recombinase Polymerase Amplification Sensor for the Rapid Visual Detection of Nucleic Acids. Talanta 2019, 204, 685–692. [Google Scholar] [CrossRef]
- Li, X.; Zhao, X.; Yang, W.; Xu, F.; Chen, B.; Peng, J.; Huang, J.; Mi, S. Stretch-Driven Microfluidic Chip for Nucleic Acid Detection. Biotechnol. Bioeng. 2021, 118, 3559–3568. [Google Scholar] [CrossRef] [PubMed]
- Trinh, K.T.L.; Lee, N.Y. Fabrication of Wearable PDMS Device for Rapid Detection of Nucleic Acids via Recombinase Polymerase Amplification Operated by Human Body Heat. Biosensors 2022, 12, 72. [Google Scholar] [CrossRef]
- Bartholomeusz, D.A.; Boutte, R.W.; Andrade, J.D. Xurography: Rapid Prototyping of Microstructures Using a Cutting Plotter. J. Microelectromech. Syst. 2005, 14, 1364–1374. [Google Scholar] [CrossRef]
- Kong, M.; Li, Z.; Wu, J.; Hu, J.; Sheng, Y.; Wu, D.; Lin, Y.; Li, M.; Wang, X.; Wang, S. A Wearable Microfluidic Device for Rapid Detection of HIV-1 DNA Using Recombinase Polymerase Amplification. Talanta 2019, 205, 120155. [Google Scholar] [CrossRef]
- Xu, Y.; Zhao, G.; Zhu, L.; Fei, Q.; Zhang, Z.; Chen, Z.; An, F.; Chen, Y.; Ling, Y.; Guo, P.; et al. Pencil-Paper on-Skin Electronics. Proc. Natl. Acad. Sci. USA 2020, 117, 18292–18301. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Fei, Q.; Page, M.; Zhao, G.; Ling, Y.; Stoll, S.B.; Yan, Z. Paper-Based Wearable Electronics. iScience 2021, 24, 102736. [Google Scholar] [CrossRef] [PubMed]
- Kaur, N.; Toley, B.J. Paper-Based Nucleic Acid Amplification Tests for Point-of-Care Diagnostics. Analyst 2018, 143, 2213–2234. [Google Scholar] [CrossRef]
- Sadri, B.; Goswami, D.; Sala de Medeiros, M.; Pal, A.; Castro, B.; Kuang, S.; Martinez, R.V. Wearable and Implantable Epidermal Paper-Based Electronics. ACS Appl. Mater. Interfaces 2018, 10, 31061–31068. [Google Scholar] [CrossRef]
- Zhang, L.; Deng, H.; Yuan, R.; Yuan, Y. Electrochemical Lead(II) Biosensor by Using an Ion-Dependent Split DNAzyme and a Template-Free DNA Extension Reaction for Signal Amplification. Microchim. Acta 2019, 186, 709. [Google Scholar] [CrossRef]
- Wu, P.; Hwang, K.; Lan, T.; Lu, Y. A DNAzyme-Gold Nanoparticle Probe for Uranyl Ion in Living Cells. J. Am. Chem. Soc. 2013, 135, 5254–5257. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Hao, Z.; Yu, S.; Huang, C.; Pan, Y.; Zhao, X. A Wearable and Deformable Graphene-Based Affinity Nanosensor for Monitoring of Cytokines in Biofluids. Nanomaterials 2020, 10, 1503. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.; Wang, Z.; Li, Y.; Zhu, Y.; Wang, X.; de Moraes, C.G.; Pan, Y.; Zhao, X.; Lin, Q. Measurement of Cytokine Biomarkers Using an Aptamer-Based Affinity Graphene Nanosensor on a Flexible Substrate toward Wearable Applications. Nanoscale 2018, 10, 21681–21688. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, S.; Ye, Z.; Peng, D.; He, L.; Yan, F.; Yang, Y.; Zhang, H.; Zhang, Z. A Gold Electrode Modified with Amino-Modified Reduced Graphene Oxide, Ion Specific DNA and DNAzyme for Dual Electrochemical Determination of Pb(II) and Hg(II). Microchim. Acta 2015, 182, 2251–2258. [Google Scholar] [CrossRef]
- Wang, M.; Zhai, S.; Ye, Z.; He, L.; Peng, D.; Feng, X.; Yang, Y.; Fang, S.; Zhang, H.; Zhang, Z. An Electrochemical Aptasensor Based on a TiO2 /Three-Dimensional Reduced Graphene Oxide/PPy Nanocomposite for the Sensitive Detection of Lysozyme. Dalton Trans. 2015, 44, 6473–6479. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Xie, S.; Zhang, J.; Tang, D.; Tang, Y. An Electrochemical Impedance Biosensor for Hg2+ Detection Based on DNA Hydrogel by Coupling with DNAzyme-Assisted Target Recycling and Hybridization Chain Reaction. Biosens. Bioelectron. 2017, 98, 466–472. [Google Scholar] [CrossRef]
- Gao, J.; Gao, Y.; Han, Y.; Pang, J.; Wang, C.; Wang, Y.; Liu, H.; Zhang, Y.; Han, L. Ultrasensitive Label-Free MiRNA Sensing Based on a Flexible Graphene Field-Effect Transistor without Functionalization. ACS Appl. Electron. Mater. 2020, 2, 1090–1098. [Google Scholar] [CrossRef]
- Sheibani, S.; Capua, L.; Kamaei, S.; Akbari, S.S.A.; Zhang, J.; Guerin, H.; Ionescu, A.M. Extended Gate Field-Effect-Transistor for Sensing Cortisol Stress Hormone. Commun. Mater. 2021, 2, 10. [Google Scholar] [CrossRef]
- Liu, Q.; Liu, Y.; Wu, F.; Cao, X.; Li, Z.; Alharbi, M.; Abbas, A.N.; Amer, M.R.; Zhou, C. Highly Sensitive and Wearable In2O3 Nanoribbon Transistor Biosensors with Integrated On-Chip Gate for Glucose Monitoring in Body Fluids. ACS Nano 2018, 12, 1170–1178. [Google Scholar] [CrossRef]
- Seshadri, D.R.; Li, R.T.; Voos, J.E.; Rowbottom, J.R.; Alfes, C.M.; Zorman, C.A.; Drummond, C.K. Wearable Sensors for Monitoring the Physiological and Biochemical Profile of the Athlete. NPJ Digit. Med. 2019, 2, 72. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Fang, X.; Kong, J. In Situ Sampling and Monitoring Cell-Free DNA of the Epstein-Barr Virus from Dermal Interstitial Fluid Using Wearable Microneedle Patches. ACS Appl. Mater. Interfaces 2019, 11, 38448–38458. [Google Scholar] [CrossRef]
- Chen, B.; Li, Y.; Xu, F.; Yang, X. Powerful CRISPR-Based Biosensing Techniques and Their Integration With Microfluidic Platforms. Front. Bioeng. Biotechnol. 2022, 10, 851712. [Google Scholar] [CrossRef] [PubMed]
- Bamshad, A.; Cho, H.J. Laserjet Printed Micro/Nano Sensors and Microfluidic Systems: A Simple and Facile Digital Platform for Inexpensive, Flexible, and Low-Volume Devices. Adv. Mater. Technol. 2021, 6, 2100401. [Google Scholar] [CrossRef]
- Ponce Wong, R.D.; Posner, J.D.; Santos, V.J. Flexible Microfluidic Normal Force Sensor Skin for Tactile Feedback. Sens. Actuators A Phys. 2012, 179, 62–69. [Google Scholar] [CrossRef]
- Park, S.-J.; Kim, J.; Chu, M.; Khine, M. Highly Flexible Wrinkled Carbon Nanotube Thin Film Strain Sensor to Monitor Human Movement. Adv. Mater. Technol. 2016, 1, 1600053. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Mukasa, D.; Zhang, H.; Gao, W. Self-Powered Wearable Biosensors. Acc. Mater. Res. 2021, 2, 184–197. [Google Scholar] [CrossRef]
- Zou, Y.; Raveendran, V.; Chen, J. Wearable Triboelectric Nanogenerators for Biomechanical Energy Harvesting. Nano Energy 2020, 77, 105303. [Google Scholar] [CrossRef]
- Fan, W.; Shen, Z.; Zhang, Q.; Liu, F.; Fu, C.; Zhu, T.; Zhao, X. High-Power-Density Wearable Thermoelectric Generators for Human Body Heat Harvesting. ACS Appl. Mater. Interfaces 2022, 14, 21224–21231. [Google Scholar] [CrossRef] [PubMed]
- Komkova, M.A.; Karyakina, E.E.; Karyakin, A.A. Noiseless Performance of Prussian Blue Based (Bio)Sensors through Power Generation. Anal. Chem. 2017, 89, 6290–6294. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.; GhavamiNejad, A.; GhavamiNejad, P.; Samarikhalaj, M.; Giacca, A.; Poudineh, M. Hydrogel Microneedle-Assisted Assay Integrating Aptamer Probes and Fluorescence Detection for Reagentless Biomarker Quantification. ACS Sens. 2022, 7, 2387–2399. [Google Scholar] [CrossRef]
- Labuda, J.; Oliveira Brett, A.M.; Evtugyn, G.; Fojta, M.; Mascini, M.; Ozsoz, M.; Palchetti, I.; Paleček, E.; Wang, J. Electrochemical Nucleic Acid-Based Biosensors: Concepts, Terms, and Methodology (IUPAC Technical Report). Pure Appl. Chem. 2010, 82, 1161–1187. [Google Scholar] [CrossRef]
- Chambers, J.P.; Arulanandam, B.P.; Matta, L.L.; Weis, A.; Valdes, J.J. Biosensor Recognition Elements. Curr. Issues Mol. Biol. 2008, 10, 1–12. [Google Scholar] [PubMed]
- Morales, M.A.; Halpern, J.M. Guide to Selecting a Biorecognition Element for Biosensors. Bioconjug. Chem. 2018, 29, 3231–3239. [Google Scholar] [CrossRef] [PubMed]
- Howard, G.C.; Bethell, D.R. Basic Methods in Antibody Production and Characterization; CRC Press: Boca Raton, FL, USA, 2001; pp. 51–68. [Google Scholar]
- Bradbury, A.R.M.; Trinklein, N.D.; Thie, H.; Wilkinson, I.C.; Tandon, A.K.; Anderson, S.; Bladen, C.L.; Jones, B.; Aldred, S.F.; Bestagno, M.; et al. When Monoclonal Antibodies Are Not Monospecific: Hybridomas Frequently Express Additional Functional Variable Regions. MAbs 2018, 10, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Jeerapan, I.; Sempionatto, J.R.; Barfidokht, A.; Mishra, R.K.; Campbell, A.S.; Hubble, L.J.; Wang, J. Wearable Bioelectronics: Enzyme-Based Body-Worn Electronic Devices. Acc. Chem. Res. 2018, 51, 2820–2828. [Google Scholar] [CrossRef]
- Singh, R.S.; Singh, T.; Singh, A.K. Enzymes as Diagnostic Tools. In Advances in Enzyme Technology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 225–271. [Google Scholar]
- Campbell, A.S.; Kim, J.; Wang, J. Wearable Electrochemical Alcohol Biosensors. Curr. Opin. Electrochem. 2018, 10, 126–135. [Google Scholar] [CrossRef]
- Kvassman, J.; Pettersson, G. Effect of PH on Coenzyme Binding to Liver Alcohol Dehydrogenase. Eur. J. Biochem. 1979, 100, 115–123. [Google Scholar] [CrossRef]
- Sato, K.; Kang, W.H.; Saga, K.; Sato, K.T. Biology of Sweat Glands and Their Disorders. I. Normal Sweat Gland Function. J. Am. Acad. Dermatol. 1989, 20, 537–563. [Google Scholar] [CrossRef]
- Wang, L.; Sun, Y. Engineering Organophosphate Hydrolase for Enhanced Biocatalytic Performance: A Review. Biochem. Eng. J. 2021, 168, 107945. [Google Scholar] [CrossRef]
- Yoo, E.-H.; Lee, S.-Y. Glucose Biosensors: An Overview of Use in Clinical Practice. Sensors 2010, 10, 4558–4576. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Li, P.; Chu, H.C.; Lo, P.K. Nucleic Acids and Their Analogues for Biomedical Applications. Biosensors 2022, 12, 93. [Google Scholar] [CrossRef]
- Du, Y.; Dong, S. Nucleic Acid Biosensors: Recent Advances and Perspectives. Anal. Chem. 2017, 89, 189–215. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yang, D.; Schluesener, H.J.; Zhang, Z. Advances in SELEX and Application of Aptamers in the Central Nervous System. Biomol. Eng. 2007, 24, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Bora, U. Nucleic Acid Based Biosensors for Clinical Applications. Biosens. J. 2013, 2, 1–8. [Google Scholar] [CrossRef]
- Zeng, R.; Wang, W.; Chen, M.; Wan, Q.; Wang, C.; Knopp, D.; Tang, D. CRISPR-Cas12a-Driven MXene-PEDOT:PSS Piezoresistive Wireless Biosensor. Nano Energy 2021, 82, 105711. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, S.; Guo, W.; Li, B.; Yang, Y.; Xie, B.; Li, K.; Zhang, L. Recent Advances on Functional Nucleic-Acid Biosensors. Sensors 2021, 21, 7109. [Google Scholar] [CrossRef]
- Chen, T.-Y.; Loan, P.T.K.; Hsu, C.-L.; Lee, Y.-H.; Tse-Wei Wang, J.; Wei, K.-H.; Lin, C.-T.; Li, L.-J. Label-Free Detection of DNA Hybridization Using Transistors Based on CVD Grown Graphene. Biosens. Bioelectron. 2013, 41, 103–109. [Google Scholar] [CrossRef]
- Wang, Z.; Hao, Z.; Wang, X.; Huang, C.; Lin, Q.; Zhao, X.; Pan, Y. A Flexible and Regenerative Aptameric Graphene–Nafion Biosensor for Cytokine Storm Biomarker Monitoring in Undiluted Biofluids toward Wearable Applications. Adv. Funct. Mater. 2021, 31, 2005958. [Google Scholar] [CrossRef]
- Ganguly, A.; Lin, K.C.; Muthukumar, S.; Prasad, S. Autonomous, Real-Time Monitoring Electrochemical Aptasensor for Circadian Tracking of Cortisol Hormone in Sub-Microliter Volumes of Passively Eluted Human Sweat. ACS Sens. 2021, 6, 63–72. [Google Scholar] [CrossRef]
- Pali, M.; Jagannath, B.; Lin, K.C.; Upasham, S.; Sankhalab, D.; Upashama, S.; Muthukumar, S.; Prasad, S. CATCH (Cortisol Apta WATCH): ‘Bio-Mimic Alarm’ to Track Anxiety, Stress, Immunity in Human Sweat. Electrochim. Acta 2021, 390, 138834. [Google Scholar] [CrossRef]
- Mugo, S.M.; Alberkant, J.; Bernstein, N.; Zenkina, O.V. Flexible Electrochemical Aptasensor for Cortisol Detection in Human Sweat. Anal. Methods 2021, 13, 4169–4173. [Google Scholar] [CrossRef]
- Liu, Q.; Zhao, C.; Chen, M.; Liu, Y.; Zhao, Z.; Wu, F.; Li, Z.; Weiss, P.S.; Andrews, A.M.; Zhou, C. Flexible Multiplexed In2O3 Nanoribbon Aptamer-Field-Effect Transistors for Biosensing. iScience 2020, 23, 101469. [Google Scholar] [CrossRef] [PubMed]
- An, J.E.; Kim, K.H.; Park, S.J.; Seo, S.E.; Kim, J.; Ha, S.; Bae, J.; Kwon, O.S. Wearable Cortisol Aptasensor for Simple and Rapid Real-Time Monitoring. ACS Sens. 2022, 7, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Suo, C.; Brown, T.; Wang, T.; Teichmann, S.A.; Bassett, A.R. INSIGHT: A Population-Scale COVID-19 Testing Strategy Combining Point-of-Care Diagnosis with Centralized High-Throughput Sequencing. Sci. Adv. 2021, 7, eabe5054. [Google Scholar] [CrossRef]
- Bokelmann, L.; Nickel, O.; Maricic, T.; Pääbo, S.; Meyer, M.; Borte, S.; Riesenberg, S. Point-of-Care Bulk Testing for SARS-CoV-2 by Combining Hybridization Capture with Improved Colorimetric LAMP. Nat. Commun. 2021, 12, 1467. [Google Scholar] [CrossRef] [PubMed]
- Toley, B.J.; Covelli, I.; Belousov, Y.; Ramachandran, S.; Kline, E.; Scarr, N.; Vermeulen, N.; Mahoney, W.; Lutz, B.R.; Yager, P. Isothermal Strand Displacement Amplification (ISDA): A Rapid and Sensitive Method of Nucleic Acid Amplification for Point-of-Care Diagnosis. Analyst 2015, 140, 7540–7549. [Google Scholar] [CrossRef]
- Ciftci, S.; Neumann, F.; Abdurahman, S.; Appelberg, K.S.; Mirazimi, A.; Nilsson, M.; Madaboosi, N. Digital Rolling Circle Amplification–Based Detection of Ebola and Other Tropical Viruses. J. Mol. Diagn. 2020, 22, 272–283. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, B.B.; Veigas, B.; Baptista, P.V. Isothermal Amplification of Nucleic Acids: The Race for the Next “Gold Standard”. Front. Sens. 2021, 2, 752600. [Google Scholar] [CrossRef]
- Müller, S.; Strohbach, D.; Wolf, J. Sensors Made of RNA: Tailored Ribozymes for Detection of Small Organic Molecules, Metals, Nucleic Acids and Proteins. IEE Proc. Nanobiotechnol. 2006, 153, 31. [Google Scholar] [CrossRef]
- Yao, C.; Zhu, T.; Qi, Y.; Zhao, Y.; Xia, H.; Fu, W. Development of a Quartz Crystal Microbalance Biosensor with Aptamers as Bio-Recognition Element. Sensors 2010, 10, 5859–5871. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Zhu, Q.; Zhou, X.; Wang, R.; Yang, Z. Reusable, Facile, and Rapid Aptasensor Capable of Online Determination of Trace Mercury. Environ. Int. 2021, 146, 106181. [Google Scholar] [CrossRef]
- Ly, T.T.; Ruan, Y.; Du, B.; Jia, P.; Zhang, H. Fibre-Optic Surface Plasmon Resonance Biosensor for Monoclonal Antibody Titer Quantification. Biosensors 2021, 11, 383. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-O.; So, H.-M.; Jeon, E.-K.; Chang, H.; Won, K.; Kim, Y.H. Aptamers as Molecular Recognition Elements for Electrical Nanobiosensors. Anal. Bioanal. Chem. 2008, 390, 1023–1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, M.; Chang, Q.; Zhang, L.; Huang, Z.; Song, C.; Chen, Y.; Wu, X.; Lu, Y. Ultra-sensitive Detecting OPs-Isocarbophos Using Photoinduced Regeneration of Aptamer-based Electrochemical Sensors. Electroanalysis 2022, 34, 995–1000. [Google Scholar] [CrossRef]
- Knott, G.J.; Doudna, J.A. CRISPR-Cas Guides the Future of Genetic Engineering. Science 2018, 361, 866–869. [Google Scholar] [CrossRef] [Green Version]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic Acid Detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef] [Green Version]
- Kaminski, M.M.; Abudayyeh, O.O.; Gootenberg, J.S.; Zhang, F.; Collins, J.J. CRISPR-Based Diagnostics. Nat. Biomed. Eng. 2021, 5, 643–656. [Google Scholar] [CrossRef]
- Al Mamun, M.; Wahab, Y.A.; Hossain, M.A.M.; Hashem, A.; Johan, M.R. Electrochemical Biosensors with Aptamer Recognition Layer for the Diagnosis of Pathogenic Bacteria: Barriers to Commercialization and Remediation. TrAC Trends Anal. Chem. 2021, 145, 116458. [Google Scholar] [CrossRef]
- Chen, C.; Wang, J. Optical Biosensors: An Exhaustive and Comprehensive Review. Analyst 2020, 145, 1605–1628. [Google Scholar] [CrossRef]
- Damborský, P.; Švitel, J.; Katrlík, J. Optical Biosensors. Essays Biochem. 2016, 60, 91–100. [Google Scholar] [CrossRef] [Green Version]
- Sedki, M.; Shen, Y.; Mulchandani, A. Nano-FET-Enabled Biosensors: Materials Perspective and Recent Advances in North America. Biosens. Bioelectron. 2020, 176, 112941. [Google Scholar] [CrossRef]
- Wu, Y.; Ghoraani, B. Biological Signal Processing and Analysis for Healthcare Monitoring. Sensors 2022, 22, 5341. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Lieber, C.M. Nano-Bioelectronics. Chem. Rev. 2016, 116, 215–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vu, C.A.; Chen, W.Y. Field-Effect Transistor Biosensors for Biomedical Applications: Recent Advances and Future Prospects. Sensors 2019, 19, 4214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadighbayan, D.; Hasanzadeh, M.; Ghafar-Zadeh, E. Biosensing Based on Field-Effect Transistors (FET): Recent Progress and Challenges. TrAC-Trends Anal. Chem. 2020, 133, 116067. [Google Scholar] [CrossRef]
- Nakatsuka, N.; Yang, K.-A.; Abendroth, J.M.; Cheung, K.M.; Xu, X.; Yang, H.; Zhao, C.; Zhu, B.; Rim, Y.S.; Yang, Y.; et al. Aptamer-Field-Effect Transistors Overcome Debye Length Limitations for Small-Molecule Sensing. Science 2018, 362, 319–324. [Google Scholar] [CrossRef]
- Stern, E.; Wagner, R.; Sigworth, F.J.; Breaker, R.; Fahmy, T.M.; Reed, M.A. Importance of the Debye Screening Length on Nanowire Field Effect Transistor Sensors. Nano Lett. 2007, 7, 3405–3409. [Google Scholar] [CrossRef] [Green Version]
- Zea, M.; Bellagambi, F.G.; ben Halima, H.; Zine, N.; Jaffrezic-Renault, N.; Villa, R.; Gabriel, G.; Errachid, A. Electrochemical Sensors for Cortisol Detections: Almost There. TrAC Trends Anal. Chem. 2020, 132, 116058. [Google Scholar] [CrossRef]
- Menon, S.; Mathew, M.R.; Sam, S.; Keerthi, K.; Kumar, K.G. Recent Advances and Challenges in Electrochemical Biosensors for Emerging and Re-Emerging Infectious Diseases. J. Electroanal. Chem. 2020, 878, 114596. [Google Scholar] [CrossRef]
- Rafat, N.; Satoh, P.; Worden, R.M. Electrochemical Biosensor for Markers of Neurological Esterase Inhibition. Biosensors 2021, 11, 459. [Google Scholar] [CrossRef]
- Yang, B.; Fang, X.; Kong, J. Engineered Microneedles for Interstitial Fluid Cell-Free DNA Capture and Sensing Using Iontophoretic Dual-Extraction Wearable Patch. Adv. Funct. Mater. 2020, 30, 2000591. [Google Scholar] [CrossRef]
- Kim, J.; Sempionatto, J.R.; Imani, S.; Hartel, M.C.; Barfidokht, A.; Tang, G.; Campbell, A.S.; Mercier, P.P.; Wang, J. Simultaneous Monitoring of Sweat and Interstitial Fluid Using a Single Wearable Biosensor Platform. Adv. Sci. 2018, 5, 1800880. [Google Scholar] [CrossRef] [PubMed]
- Bariya, M.; Nyein, H.Y.Y.; Javey, A. Wearable Sweat Sensors. Nat. Electron. 2018, 1, 160–171. [Google Scholar] [CrossRef]
- Manjakkal, L.; Yin, L.; Nathan, A.; Wang, J.; Dahiya, R. Energy Autonomous Sweat-Based Wearable Systems. Adv. Mater. 2021, 33, 2100899. [Google Scholar] [CrossRef] [PubMed]
- Anwer, A.H.; Khan, N.; Ansari, M.Z.; Baek, S.-S.; Yi, H.; Kim, S.; Noh, S.M.; Jeong, C. Recent Advances in Touch Sensors for Flexible Wearable Devices. Sensors 2022, 22, 4460. [Google Scholar] [CrossRef] [PubMed]
- Sekar, M.; Sriramprabha, R.; Sekhar, P.K.; Bhansali, S.; Ponpandian, N.; Pandiaraj, M.; Viswanathan, C. Review—Towards Wearable Sensor Platforms for the Electrochemical Detection of Cortisol. J. Electrochem. Soc. 2020, 167, 067508. [Google Scholar] [CrossRef]
- Shaver, A.; Arroyo-Currás, N. The Challenge of Long-Term Stability for Nucleic Acid-Based Electrochemical Sensors. Curr. Opin. Electrochem. 2022, 32, 100902. [Google Scholar] [CrossRef]
- Singh, N.K.; Chung, S.; Sveiven, M.; Hall, D.A. Cortisol Detection in Undiluted Human Serum Using a Sensitive Electrochemical Structure-Switching Aptamer over an Antifouling Nanocomposite Layer. ACS Omega 2021, 6, 27888–27897. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Janghorban, M.; Aradanas, I.; Kazemi, S.; Ngaju, P.; Pandey, R. Recent Advances, Opportunities, and Challenges in Developing Nucleic Acid Integrated Wearable Biosensors for Expanding the Capabilities of Wearable Technologies in Health Monitoring. Biosensors 2022, 12, 986. https://doi.org/10.3390/bios12110986
Janghorban M, Aradanas I, Kazemi S, Ngaju P, Pandey R. Recent Advances, Opportunities, and Challenges in Developing Nucleic Acid Integrated Wearable Biosensors for Expanding the Capabilities of Wearable Technologies in Health Monitoring. Biosensors. 2022; 12(11):986. https://doi.org/10.3390/bios12110986
Chicago/Turabian StyleJanghorban, Mohammad, Irvyne Aradanas, Sara Kazemi, Philippa Ngaju, and Richa Pandey. 2022. "Recent Advances, Opportunities, and Challenges in Developing Nucleic Acid Integrated Wearable Biosensors for Expanding the Capabilities of Wearable Technologies in Health Monitoring" Biosensors 12, no. 11: 986. https://doi.org/10.3390/bios12110986
APA StyleJanghorban, M., Aradanas, I., Kazemi, S., Ngaju, P., & Pandey, R. (2022). Recent Advances, Opportunities, and Challenges in Developing Nucleic Acid Integrated Wearable Biosensors for Expanding the Capabilities of Wearable Technologies in Health Monitoring. Biosensors, 12(11), 986. https://doi.org/10.3390/bios12110986