Hammerstein–Wiener Multimodel Approach for Fast and Efficient Muscle Force Estimation from EMG Signals
Abstract
:1. Introduction
2. Related Studies
3. Methods
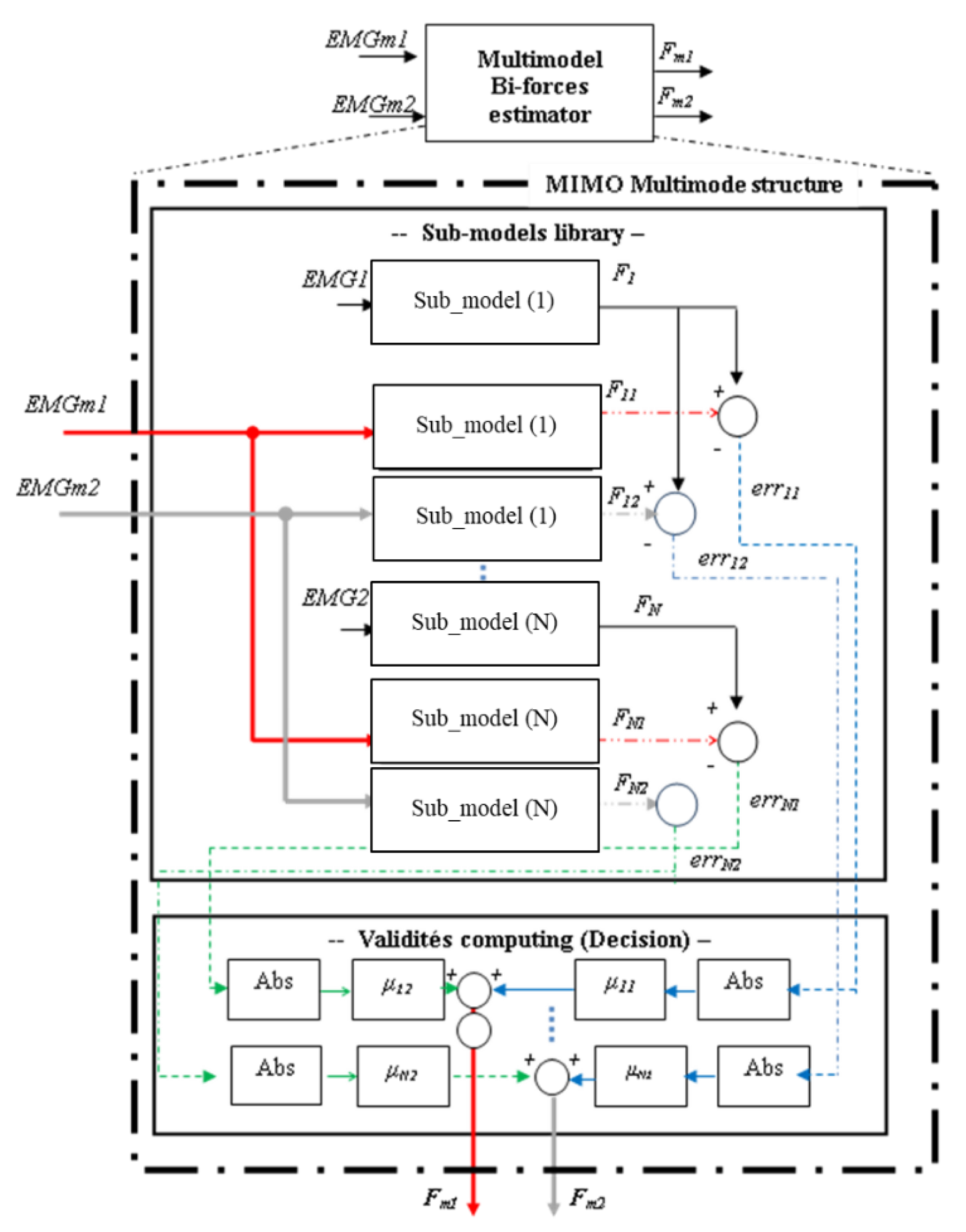
| EMGm1 and EMGm2: | Inputs of the multimodel bi-forces estimator. |
| Fm1 and Fm2: | Outputs of the multimodel bi-forces estimator. |
| EMGN: | Input of sub-model (i). |
| Fi: | Output of sub-model (i). |
| Fi1 and Fi2: | Output of sub-model (i) obtained by applying multimodel inputs, EMGm1 and EMGm2, respectively. |
| erri1 and erri2: | Errors of sub-model (i) computed between its real output, Fi, and outputs, Fi1 and Fi2 |
| μi1 and μi2: | Validities of sub-model (i) according to Fm1 and Fm2, respectively. |
- Step 1_Elaboration of sub-models: Definition of sub-models of the library: Sub-model (i): model defined for the measurements couple (EMGi, Fi).
- Step 2_Normalised residues estimation: Computation of the normalised error of each sub-model (i): err’i1 and err’i2.
- Step 3_Validity Computing: Computation of the weight, also named validity, of each sub-model: μi1 and μi2.
- Step 4_Outputs Computing: Computation of outputs Fm1 and Fm2.
| ɸ(k) | : Observation vector, also named regression vector, contains inputs and outputs data of previous instants. ɸ(k)= ɸ(uk–1 , fk–1) |
| θ(k) | : Parameter vector θ = [θ1, θ2, …, θp] p is the number of parameters to be estimated. |
3.1. Experimental Approach
3.2. Data Analysis
- Scenario1: Predict force profiles from sub-models characterising the same kind of desired forces. For example, predicting a step from another instance of the step profile.
- Scenario2: Predict profiles of arbitrary forces from sub-models characterising 02 known forces. For example, using step and circle to estimate free profiles (vol).
- Scenario3: Predict profiles of arbitrary forces from sub-models characterising 03 known forces. This is the same as scenario 2, but with three standard profiles. For the different scenarios, we note that inputs/outputs of the multimodel approach are considered unknown, and any sub-model of the library does not represent them. Three performance measures: coefficient of determination (R2), root mean squared error (RMSE), and computational time (CT), were used to assess the proposed approach. For each performance metric, a two-way repeated measure analysis of variance (ANOVA, IBM SPSS Statistics 26) with factors methods (multimodel vs. ANN) and scenarios (1,2 and 3) was used to assess the performance of the proposed approach. Table 1 present the computational environment used in this paper.
| Number of feed-forward network layers: | 2 |
| Number of hidden layers: | 1 |
| Number of neurons in the hidden layer: | 7 |
| Type of activation function: | Tangent Sigmoid |
| Batch training method: | Levenberg–Marquardt |
| Number of output neurons: | 4 |

4. Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Allen, C.; Karam, K.Z.; Le Cam, P.; Hill, M.; Tindle, T. Application of virtual reality devices to the quantitative assessment of manual assembly forces in a factory environment. In Proceedings of the IECON ‘95—21st Annual Conference on IEEE Industrial Electronics, Orlando, FL, USA, 6–10 November 1995; Volume 2, pp. 1048–1053. [Google Scholar]
- Hill, A. The heat of shortening and dynamic constants of muscle. Proc. R. Soc. Lond. Ser. B Biol. Sci. 1938, 126, 136–195. [Google Scholar]
- Delp, S.L.; Anderson, F.C.; Arnold, A.S.; Loan, P.; Habib, A.; John, C.T.; Guendelman, E.; Thelen, D.G. OpenSim: Open-source software to create and analyse dynamic simulations of movement. IEEE Trans. Biomed. Eng. 2007, 54, 1940–1950. [Google Scholar] [CrossRef] [Green Version]
- Lai, A.; Schache, A.G.; Lin, Y.C.; Pandy, M.G. Tendon elastic strain energy in the human ankle plantar-flexors and its role with increased running speed. J. Exp. Biol. 2014, 217, 3159–3168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.S.M.; Arnold, A.S.; Miara, M.D.B.; Biewener, A.A.; Wakeling, J.M. Accuracy of gastrocnemius muscles forces in walking and running goats predicted by one-element and two-element Hill-type models. J. Biomech. 2013, 46, 2288–2295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben Messaoud, A.; Talmoudi Ben Aoun, S.; Lahmari Ksouri, M. A New Strategy of Validities’ Computation for Multimodel Approach: Experimental Validation. Int. J. Adv. Comput. Sci. Appl. 2017, 8, 233–241. [Google Scholar]
- Duncombe, J.U. Infrared navigation—Part I: An assessment of feasibility. IEEE Trans. Electron Devices 1959, ED-11, 34–39. [Google Scholar]
- Lippold, O.C.J. The relation between integrated action potentials in a human muscle and its isometric tension. J. Physiol. 1952, 117, 492–499. [Google Scholar] [CrossRef]
- Lloyd, D.G.; Besier, T.F. An EMG-driven musculoskeletal model to estimate muscle forces and knee joint moments in vivo. J. Biomech. 2003, 36, 765–776. [Google Scholar] [CrossRef]
- Rodriguez Martinez, J.; Mannini, A.; Clemente, F.; Sabatini, A.M.; Cipriani, C. Grasp force estimation from the transient EMG using high-density surface recordings. J. Neural Eng. 2020, 17, 016052. [Google Scholar] [CrossRef]
- Wang, N.; Lao, K.; Zhang, X.; Lin, J.; Zhang, X. The recognition of grasping force using LDA. Biomed. Signal Process. Control 2018, 47, 393–400. [Google Scholar] [CrossRef]
- Baldacchino, T.; Jacobs, W.R.; Anderson, S.R.; Worden, K.; Rowson, J. Simultaneous force regression and movement classification of fingers via surface EMG within a unified Bayesian Framework. Front. Bioeng. Biotechnol. 2018, 6, 13. [Google Scholar] [CrossRef] [Green Version]
- Staudenmann, D.; Roeleveld, K.; Stegeman, D.F.; Van Dieën, J.H. Methodological aspects of SEMG recordings for force estimation—A tutorial and review. J. Electromyogr. Kinesiol. 2010, 20, 375–387. [Google Scholar] [CrossRef]
- Alkner, B.A.; Tesch, P.A.; Berg, H.E. Quadriceps EMG/force relationship in knee extension and leg press. Med. Sci. Sports Exerc. 2000, 2, 459–6332. [Google Scholar] [CrossRef]
- De Luca, C.J. The use of surface electromyography in biomechanics. J. Appl. Biomech. 1997, 2, 135–163. [Google Scholar] [CrossRef] [Green Version]
- Komi, P.V.; Buskirk, E.R. Reproducibility of electromyographic measurements with inserted wire electrodes and surface electrodes. Electromyography 1970, 4, 357–367. [Google Scholar]
- Potvin, J.R.; Brown, S.H. Less is more: High pass filtering, to remove up to 99% of the surface EMG signal power, improves EMG-based biceps brachii muscle force estimates. J. Electromyogr. Kinesiol. 2004, 3, 389–399. [Google Scholar] [CrossRef]
- Solomonow, M.; Baratta, R.; Shoji, H.; Ambrosia, R.D. The myoelectric signal of electrically stimulated muscle during recruitment: An inherent feedback parameter for a closed-loop control scheme. IEEE Trans. Biomed. Eng. 1986, 8, 35–45. [Google Scholar] [CrossRef]
- Vink, P.; Van Der Velde, E.A.; Verbout, A.J. A functional subdivision of the lumbar extensor musculature. Recruitment patterns and force-RA-EMG relationships under isometric conditions. Electromyogr. Clin. Neurophysiol. 1987, 8, 517–525. [Google Scholar]
- Milner-Brown, H.S.; Stein, R.B.; Yemm, R. The orderly recruitment of human motor units during voluntary isometric contractions. J. Physiol. 1973, 70, 230–359. [Google Scholar] [CrossRef]
- Kukulka, C.G.; Clamann, H.P. Comparison of the recruitment and discharge properties of motor units in human brachial biceps and adductor pollicis during isometric contractions. Medicine 1981, 1, 45–55. [Google Scholar] [CrossRef]
- Woods, J.J.; Bigland-Ritchie, B. Linear and nonlinear surface EMG/force relationships in human muscles. Am. J. Phys. Med. Rehabil. 1983, 62, 287–299. [Google Scholar]
- Kamavuako, E.N.; Rosenvang, J.C. Hysteresis in the electromyography-force relationship: Toward an optimal model for the estimation of force. Muscle Nerve 2012, 46, 755–758. [Google Scholar] [CrossRef]
- Calvert, T.W.; Chapman, A.E. The relationship between the surface EMG and force transients in muscle: Simulation and experimental studies. Proc. IEEE 1977, 65, 682–689. [Google Scholar] [CrossRef]
- Bottomley, H. Myo-electric control of powered prostheses. J. Bone Joint Surg. Br. 1965, 47, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Kamavuako, E.N.; Scheme, E.J.; Englehart, K.B. Wrist torque estimation during simultaneous and continuously changing movements: Surface versus untargeted intramuscular EMG. J. Neurophysiol. 2013, 11, 2658–2665. [Google Scholar] [CrossRef]
- Hahne, J.M.; Bießmann, F.; Jiang, N.; Rehbaum, H.; Farina, D.; Meinecke, F.C.; Müller, K.-R.; Parra, L.C. Linear and Nonlinear Regression Techniques for Simultaneous and Proportional Myoelectric Control. IEEE Trans. Neural Syst. Rehabil. Eng. 2014, 22, 269–279. [Google Scholar] [CrossRef]
- Farina, D.; Jiang, N.; Rehbaum, H.; Holobar, A.; Graimann, B.; Dietl, H.; Aszmann, O.C. The extraction of neural information from the surface EMG for the control of upper-limb prostheses: Emerging avenues and challenges. IEEE Trans. Neural Syst. Rehabil. Eng. 2014, 22, 797–809. [Google Scholar] [CrossRef]
- Scott, N.R. Myoelectric control of prostheses and orthoses. Bull. Prosthet. Res. 1967, 93–114. [Google Scholar]
- Luo, J.; Liu, C.; Yang, C. Estimation of EMG-Based Force Using a Neural-Network-Based Approach. IEEE Access 2019, 7, 64856–64865. [Google Scholar] [CrossRef]
- Wimalasena, L.N.; Braun, J.F.; Keshtkaran, M.R.; Hofmann, D.; Gallego, J.L.; Alessandro, C.; Tresch, M.C.; Miller, L.E.; Pandarinath, C. Estimating muscle activation from EMG using deep learning-based dynamical systems models. Cold Spring Harb. Lab. 2021. [Google Scholar] [CrossRef]
- Geethanjali, P. Myoelectric control of prosthetic hands: A state-of-the-art review. Med. Dev. 2016, 9, 247–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamavuako, E.N.; Rosenvang, J.C.; Bøg, M.F.; Smidstrup, A.; Erkocevic, E.; Niemeier, M.J.; Jensen, W.; Farina, D. Influence of the feature space on the estimation of hand grasping force from intramuscular EMG. Biomed. Signal Process. Control 2013, 8, 1–5. [Google Scholar] [CrossRef]
- Kuriki, H.U.; De Azevedo, F.M.; Takahashi, L.S.O.; Mello, E.M.; Filho, R.D.F.N.; Alves, N. The Relationship between Electromyography and Muscle Force. In EMG Methods for Evaluating Muscle and Nerve Function; IntechOpen: London, UK, 2012; pp. 31–54. [Google Scholar]
- Lee, S.W.; Kim, J.H.; Jun, J.; Ha, J.W.; Zhang, B.T. Overcoming Catastrophic Forgetting by Incremental Moment Matching. In Proceedings of the 31st Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Broderick, T.; Boyd, N.; Wibisono, A.; Wilson, A.C.; Jordan, M.I. Streaming Variational Bayes. In Proceedings of the Advances in Neural Information Processing Systems, Lake Tahoe, CA, USA, 5–10 December 2013; pp. 1727–1735. [Google Scholar]
- Li, Z.; Hoiem, D. Learning without forgetting. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 8–16 October 2016; pp. 614–629. [Google Scholar]
- Kirkpatrick, J.; Pascanu, R.; Rabinowitz, N.; Veness, J.; Desjardins, G.; Rusu, A.A.; Milan, K.; Quan, J.; Ramalho, T.; Barwinska, A.G. Overcoming catastrophic forgetting in neural networks. Proc. Natl. Acad. Sci. USA 2017, 13, 3521–3526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, Y.; Chai, T. Nonlinear multivariable adaptive control using multiple models and neural networks. Automatica 2007, 43, 1101–1110. [Google Scholar] [CrossRef]
- Elfelly, N.; Dieulot, J.-Y.; Benrejeb, M.; Borne, P. A new approach for multimodel identification of complex systems based on both neural and fuzzy clustering algorithms. Eng. Appl. Artif. Intell. 2010, 23, 1064–1071. [Google Scholar] [CrossRef]
- Elfelly, N.; Dieulot, J.-Y.; Borne, P. Neural approach for the multimodel representation of complex processes. Int. J. Comput. Commun. Control 2008, 3, 39–50. [Google Scholar] [CrossRef] [Green Version]
- Xue, Z.K.; Li, S.Y. Multimodel modelling and predictive control based on local model networks. Control Intell. Syst. 2006, 34, 105–112. [Google Scholar]
- Chihi, I.; Abdelkrim, A.; Benrejeb, M. Multimodel approach to characterise human handwriting motion. Biol. Cybern. 2016, 110, 17–30. [Google Scholar] [CrossRef]
- Adeniran, A.A.; El Ferik, S. Modeling and Identification of Nonlinear Systems: A Review of the Multimodel Approach—Part 1. IEEE Trans. Syst. Man Cybern. Syst. 2017, 47, 1149–1159. [Google Scholar] [CrossRef]
- Greblicki, W. Nonparametric identification of Wiener systems by orthogonal series. IEEE Trans. Autom. Control 1994, 39, 2077–2086. [Google Scholar] [CrossRef]
- Voros, J. Identification of Nonlinear Dynamic Systems Using Extended Hammerstein and Wiener Models. Control-Theory Adv. Technol. 1995, 10, 1203–1212. [Google Scholar]
- Kumar, P.; Potluri, C.; Sebastian, A.; Chiu, S.; Urfer, A.; Naidu, D.S.; Schoen, M.P. An adaptive multi sensor data fusion with hybrid nonlinear ARX and Wiener–Hammerstein models for skeletal muscle force estimation. WSEAS Trans. Syst. 2010, 9, 1050–1062. [Google Scholar]
- Abbasi-Asl, R.; Khorsandi, R.; Farzampour, S.; Zahedi, E. Estimation of Muscle Force with EMG Signals Using Hammerstein-Wiener Model. Biomed. IFMBE Proc. 2011, 35, 157–160. [Google Scholar]
- Eskinat, E.; Johnson, S.H.; Luyben, W.L. Use of Hammerstein models in identification of nonlinear systems. AIChE J. 1991, 37, 255–268. [Google Scholar] [CrossRef]
- Sebastian, A.; Kumar, P.; Schoen, M.P. Modelling surface electromyogram dynamics using Hammerstein-Wiener models with comparison of IIR and spatial filtering techniques. Int. J. Circuits Syst. Signal Process. 2011, 5, 545–556. [Google Scholar]
- Zhu, Y. Estimation of an N-L-N Hammerstein-Wiener Model. Automatica 2002, 38, 1607–1614. [Google Scholar] [CrossRef]
- Mete, S.; Ozer, S.; Zorlu, H. System identification using Hammerstein model. In Proceedings of the Signal Processing and Communications Applications Conference, Trabzon, Turkey, 23–25 April 2014; pp. 1303–1306. [Google Scholar]
- Ozer, S.; Zorlu, H.; Mete, S. System identification application using Hammerstein model. Indian Acad. Sci. 2016, 41, 597–6056. [Google Scholar] [CrossRef] [Green Version]
- Chihi, I.; Sidhom, L.; Trabelsi, M. Nonlinear Hammerstein-Wiener model-based Fault Detection Approach for Cascaded H-Bridge Multilevel Inverters. In Proceedings of the IEEE-GCC Conference & Exhibition (IEEE-GCC 2019), IECON, Kuwait, Kuwait, 19–23 April 2019. [Google Scholar]
- Wang, Z.; Georgakis, C. Identification of Hammerstein-Weiner models for nonlinear MPC from infrequent measurements in batch processes. J. Process Control 2019, 82, 58–69. [Google Scholar] [CrossRef]
- Kamavuako, E.N.; Englehart, K.B.; Jensen, W.; Farina, D. Simultaneous and Proportional Force Estimation in Multiple Degrees of Freedom from Intramuscular EMG. IEEE Trans. Biomed. Eng. 2012, 59, 1804–1807. [Google Scholar] [CrossRef]
- Djigan, V.I. Multichannel parallelisable sliding window RLS and fast RLS algorithms with linear constraints. Int. J. Adapt. Control Signal Process. 2006, 86, 776–791. [Google Scholar]
- Ding, F.; Ding, J. Least-squares parameter estimation for systems with irregularly missing data. Int. J. Adapt. Control Signal Process. 2010, 24, 540–553. [Google Scholar] [CrossRef]
- Narenda, K.-S.; Balakrishman, J. Adaptive control using multiple models. IEEE Trans. Autom. Control 1997, 42, 171–187. [Google Scholar] [CrossRef]
- Pappas, S.P.; Leros, A.K.; Katsikas, S.K. Joint order and parameter estimation of multivariate autoregressive models using multimodel partitioning theory. Digit. Signal Process. 2006, 16, 782–795. [Google Scholar] [CrossRef]







| Experimental Environment | Proprieties |
|---|---|
| Operating system | Windows 10 Professionnel |
| Processor | Intel(R) Core(TM) i7-8565U CPU @ 1.80 GHz 1.99 GHz |
| Processor generation | 8th Gen |
| Installed RAM | 8.00 Go, (7.88 Go usable) |
| System Type | 64 bits operating system, x64-based process |
| Graphics card | NVIDIA GeForce MX110 |
| Software | Matlab 2017 |
| Scenario-1 | Scenario-2 | Scenario-3 | ||||
|---|---|---|---|---|---|---|
| mm | ANN | mm | ANN | mm | ANN | |
| Sub-1 | 0.9592 | 0.6374 | 0.9742 | 0.7479 | 0.7922 | 0.5060 |
| Sub-2 | 0.8923 | 0.8443 | 0.8979 | 0.8987 | 0.6568 | 0.5625 |
| Sub-3 | 0.8878 | 0.8620 | 0.9466 | 0.9329 | 0.7997 | 0.5780 |
| Sub-4 | 0.9754 | 0.8258 | 0.8299 | 0.8855 | 0.6015 | 0.6054 |
| Sub-5 | 0.9330 | 0.7038 | 0.8867 | 0.8452 | 0.8517 | 0.6014 |
| Sub-6 | 0.8963 | 0.8919 | 0.9038 | 0.8619 | 0.9090 | 0.6830 |
| Sub-7 | 0.8285 | 0.5417 | 0.8087 | 0.8317 | 0.8972 | 0.5419 |
| Sub-8 | 0.9584 | 0.8092 | 0.9684 | 0.8858 | 0.9062 | 0.8354 |
| Sub-9 | 0.9539 | 0.7776 | 0.9033 | 0.5931 | 0.8454 | 0.7360 |
| Sub-10 | 0.9326 | 0.6403 | 0.8026 | 0.8326 | 0.8912 | 0.6434 |
| mean | 0.8927 | 0.7127 | 0.8427 | 0.6485 | 0.7559 | 0.3810 |
| Max | 0.9754 | 0.8970 | 0.9742 | 0.9329 | 0.9090 | 0.8354 |
| STD | 0.0043 | 0.0180 | 0.0099 | 0.0884 | 0.0210 | 0.1480 |
| Scenario-1 | Scenario-2 | Scenario-3 | ||||
|---|---|---|---|---|---|---|
| mm | ANN | mm | ANN | mm | ANN | |
| Sub-1 | 0.0186 | 0.0554 | 0.0201 | 0.0190 | 0.0353 | 0.0671 |
| Sub-2 | 0.0348 | 0.0348 | 0.0123 | 0.0215 | 0.0471 | 0.0434 |
| Sub-3 | 0.0211 | 0.0227 | 0.0230 | 0.0255 | 0.0232 | 0.0360 |
| Sub-4 | 0.0173 | 0.0460 | 0.0352 | 0.0288 | 0.0477 | 0.0658 |
| Sub-5 | 0.0249 | 0.0524 | 0.0217 | 0.0389 | 0.0261 | 0.3050 |
| Sub-6 | 0.0300 | 0.0249 | 0.0146 | 0.0194 | 0.0193 | 0.0323 |
| Sub-7 | 0.0242 | 0.0395 | 0.0227 | 0.0213 | 0.0207 | 0.0437 |
| Sub-8 | 0.0167 | 0.0358 | 0.0244 | 0.0465 | 0.0275 | 0.0383 |
| Sub-9 | 0.0210 | 0.0481 | 0.0107 | 0.0219 | 0.0356 | 0.0440 |
| Sub-10 | 0.0261 | 0.0602 | 0.0549 | 0.0508 | 0.0308 | 0.0557 |
| mean | 0.0304 | 0.0498 | 0.0318 | 0.0387 | 0.0396 | 0.0868 |
| min | 0.0167 | 0.0227 | 0.0107 | 0.0190 | 0.0193 | 0.0323 |
| STD | 1.7450 e-04 | 2.2470 e-04 | 0.0003 | 0.00035 | 0.00025 | 0.0091 |
| Scenario-1 | Scenario-2 | Scenario-3 | ||||
|---|---|---|---|---|---|---|
| mm | ANN | mm | ANN | mm | ANN | |
| Sub-1 | 0.1114 | 0.3606 | 0.0859 | 0.2210 | 0.0859 | 0.2210 |
| Sub-2 | 0.1014 | 0.6231 | 0.1191 | 0.8610 | 0.0977 | 1.1734 |
| Sub-3 | 0.0989 | 0.8061 | 0.0986 | 0.6714 | 0.0884 | 0.8842 |
| Sub-4 | 0.0911 | 0.5401 | 0.1067 | 0.7296 | 0.1245 | 0.8619 |
| Sub-5 | 0.0872 | 0.4602 | 0.1155 | 0.4712 | 0.0839 | 0.2351 |
| Sub-6 | 0.0952 | 0.4128 | 0.1061 | 0.5638 | 0.1028 | 0.5850 |
| Sub-7 | 0.0936 | 1.7428 | 0.0930 | 0.8783 | 0.1219 | 1.9918 |
| Sub-8 | 0.1006 | 0.9940 | 0.1589 | 1.1822 | 0.1477 | 2.1794 |
| Sub-9 | 0.1858 | 0.5114 | 0.1545 | 1.4397 | 0.1822 | 0.9611 |
| Sub-10 | 0.1089 | 0.7438 | 0.1041 | 0.7531 | 0.0833 | 0.7904 |
| Mean | 0.1074 | 0.7195 | 0.1142 | 0.7771 | 0.1118 | 0.9783 |
| Max | 0.1858 | 1.7428 | 0.1589 | 1.1822 | 0.1822 | 2.1794 |
| Min | 0.0872 | 0.3606 | 0.0859 | 0.2210 | 0.0833 | 0.2210 |
| mm | ANN | |
|---|---|---|
| Maximum Possible Array Bytes | 2.8949 × 109 | 3.3673 × 109 |
| Memory Available All Arrays | 2.8949 × 109 | 3.3673 × 109 |
| Memory Used MATLAB | 3.9115 × 109 | 4.9214 × 109 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chihi, I.; Sidhom, L.; Kamavuako, E.N. Hammerstein–Wiener Multimodel Approach for Fast and Efficient Muscle Force Estimation from EMG Signals. Biosensors 2022, 12, 117. https://doi.org/10.3390/bios12020117
Chihi I, Sidhom L, Kamavuako EN. Hammerstein–Wiener Multimodel Approach for Fast and Efficient Muscle Force Estimation from EMG Signals. Biosensors. 2022; 12(2):117. https://doi.org/10.3390/bios12020117
Chicago/Turabian StyleChihi, Ines, Lilia Sidhom, and Ernest Nlandu Kamavuako. 2022. "Hammerstein–Wiener Multimodel Approach for Fast and Efficient Muscle Force Estimation from EMG Signals" Biosensors 12, no. 2: 117. https://doi.org/10.3390/bios12020117
APA StyleChihi, I., Sidhom, L., & Kamavuako, E. N. (2022). Hammerstein–Wiener Multimodel Approach for Fast and Efficient Muscle Force Estimation from EMG Signals. Biosensors, 12(2), 117. https://doi.org/10.3390/bios12020117





