Towards a Point-of-Care (POC) Diagnostic Platform for the Multiplex Electrochemiluminescent (ECL) Sensing of Mild Traumatic Brain Injury (mTBI) Biomarkers
Abstract
: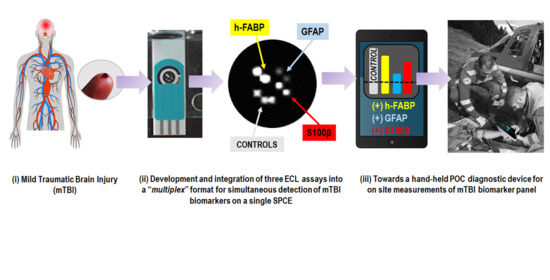
Graphical Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Apparatus
2.3. Methods
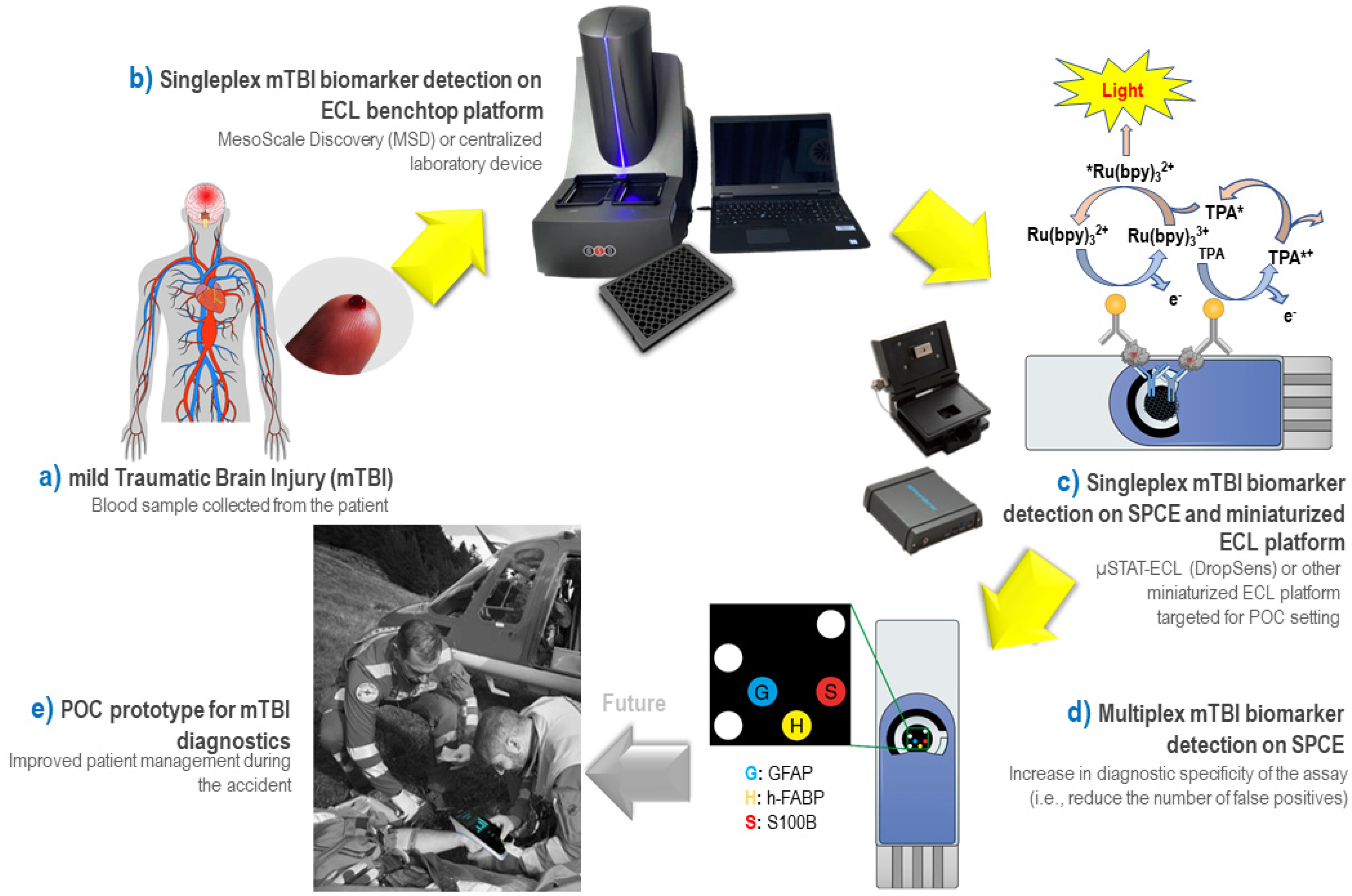
3. Results
3.1. Development of ECL “Singleplex” Assays for mTBI Biomarkers on MSD Platform
- Coating diluent (PBS 1× pH 7.4 or TRIS 50 mM pH 8.6, with addition of 0.1–5 mM CaCl2 for S100β assay)
- Blocking agent (0.1–2% of BSA or 0.1–2% casein in PBS 1× pH 7.4 or TRIS 50 mM pH 8.6, with/without addition of 0.1% Tween-20);
- Wash buffer (PBS 1× pH 7.4 or TRIS 50 mM pH 8.6, with 0.05–0.4% Tween-20);
- Detection antibody diluent (PBS 1× pH 7.4 or TRIS 50 mM pH 8.6, with/without addition of 0.1–1% of blocking agent and/or 0.06% Tween-20);
- Capture antibody (cAb) and detection antibody concentration (dAb) (cAb concentration range: 1–25 μg mL−1; dAb concentration range: 0.5–10 μg mL−1).
3.2. Development of ECL “Singleplex” Assays for mTBI Biomarkers on SPE
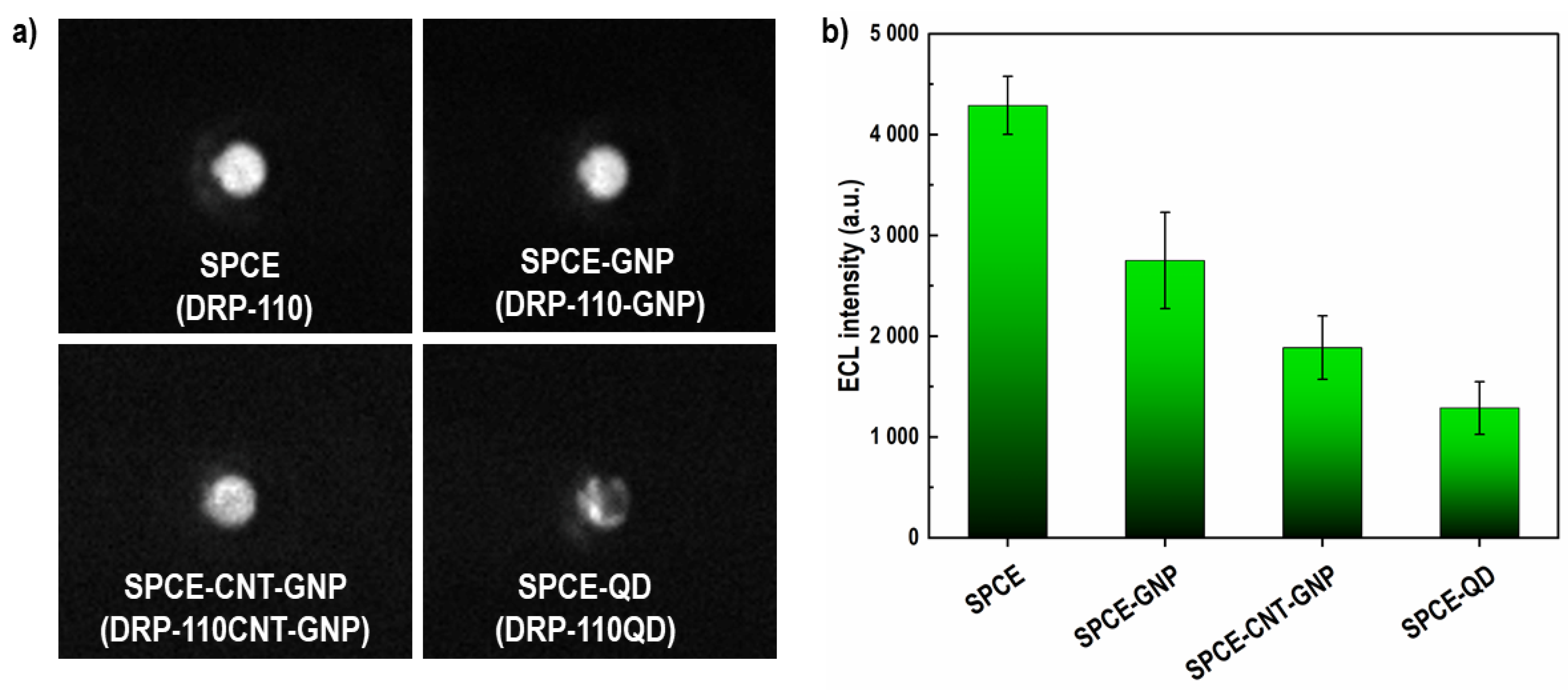
3.2.1. Choice of Electrode Material
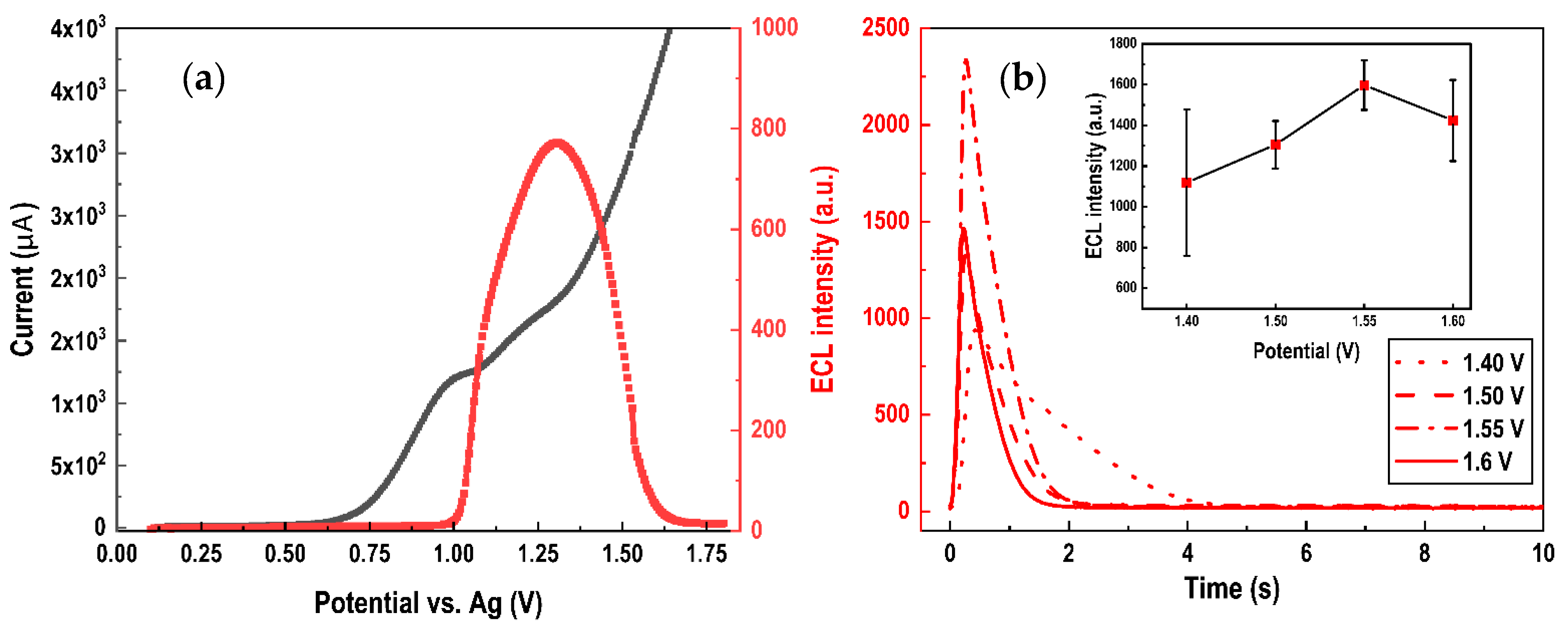
3.2.2. Optimization of SPCE-Based ECL Sensor
3.2.3. Optimization of the Assay Conditions
3.3. Standard Recovery Test
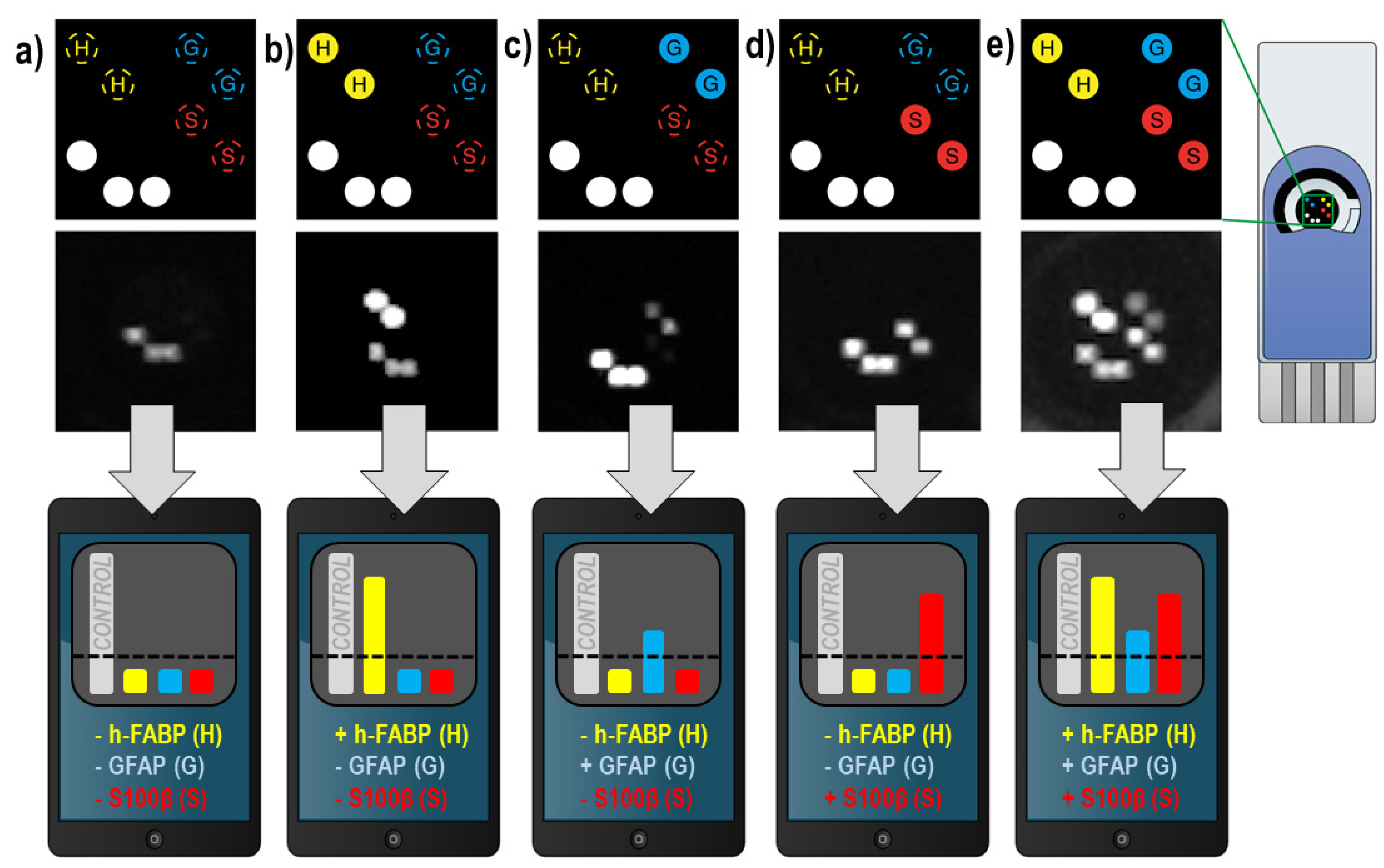
3.4. Multiplex ECL Assay for mTBI Biomarker Panel on SPCE
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Traumatic Brain Injury. Fact Sheets and Policy Brief. Available online: https://www.center-tbi.eu/ (accessed on 24 September 2020).
- Mondello, S.; Muller, U.; Jeromin, A.; Streeter, J.; Hayes, R.L.; Wang, K.K.W. Blood-Based Diagnostics of Traumatic Brain Injuries. Expert Rev. Mol. Diagn. 2011, 11, 65–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Report to Congress: Traumatic Brain Injury in the United States|Concussion|Traumatic Brain Injury|CDC Injury Center. Available online: https://www.cdc.gov/traumaticbraininjury/pubs/tbi_report_to_congress.html (accessed on 15 April 2021).
- Mehta, R.; Trainee1, G.P.; Chinthapalli, K.; Neurologist2, C. Glasgow Coma Scale Explained. BMJ 2019, 365, l1296. [Google Scholar] [CrossRef] [PubMed]
- Dewan, M.C.; Rattani, A.; Gupta, S.; Baticulon, R.E.; Hung, Y.-C.; Punchak, M.; Agrawal, A.; Adeleye, A.O.; Shrime, M.G.; Rubiano, A.M.; et al. Estimating the Global Incidence of Traumatic Brain Injury. J. Neurosurg. 2018, 130, 1080–1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cnossen, M.C.; Winkler, E.A.; Yue, J.K.; Okonkwo, D.O.; Valadka, A.B.; Steyerberg, E.W.; Lingsma, H.F.; Manley, G.T. Development of a Prediction Model for Post-Concussive Symptoms Following Mild Traumatic Brain Injury: A TRACK-TBI Pilot Study. J. Neurotrauma 2017, 34, 2396–2409. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.M.; Ferraro, J.V.; Dikmen, S.S.; Temkin, N.R.; Bell, K.R. Accuracy of Mild Traumatic Brain Injury Diagnosis. Arch. Phys. Med. Rehabil. 2008, 89, 1550–1555. [Google Scholar] [CrossRef] [PubMed]
- Martinez, B.I.; Martinez, B.I.; Stabenfeldt, S.E. Current Trends in Biomarker Discovery and Analysis Tools for Traumatic Brain Injury. J. Biol. Eng. 2019, 16, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokobori, S.; Hosein, K.; Burks, S.; Sharma, I.; Gajavelli, S.; Bullock, R. Biomarkers for the Clinical Differential Diagnosis in Traumatic Brain Injury-a Systematic Review. CNS Neurosci. Ther. 2013, 19, 556–565. [Google Scholar] [CrossRef]
- Tomar, G.S.; Singh, G.P.; Lahkar, D.; Sengar, K.; Nigam, R.; Mohan, M.; Anindya, R. New Biomarkers in Brain Trauma. Clin. Chim. Acta 2018, 487, 325–329. [Google Scholar] [CrossRef]
- Sharma, R.; Rosenberg, A.; Bennett, E.R.; Laskowitz, D.T.; Acheson, S.K. A Blood-Based Biomarker Panel to Risk-Stratify Mild Traumatic Brain Injury. PLoS ONE 2017, 12, e0173798. [Google Scholar] [CrossRef] [Green Version]
- Blennow, K.; Brody, D.L.; Kochanek, P.M.; Levin, H.; McKee, A.; Ribbers, G.M.; Yaffe, K.; Zetterberg, H. Traumatic Brain Injuries. Nat. Rev. Dis. Primers 2016, 2, 1–19. [Google Scholar] [CrossRef]
- Krausz, A.D.; Korley, F.K.; Burns, M.A. The Current State of Traumatic Brain Injury Biomarker Measurement Methods. Biosensors 2021, 11, 319. [Google Scholar] [CrossRef] [PubMed]
- Undén, L.; Calcagnile, O.; Undén, J.; Reinstrup, P.; Bazarian, J. Validation of the Scandinavian Guidelines for Initial Management of Minimal, Mild and Moderate Traumatic Brain Injury in Adults. BMC Med. 2015, 13, 292–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohrt-Nissen, S.; Friis-Hansen, L.; Dahl, B.; Stensballe, J.; Romner, B.; Rasmussen, L.S. How Does Extracerebral Trauma Affect the Clinical Value of S100B Measurements? Emerg. Med. J. 2011, 28, 941–944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kahouadji, S.; Salamin, P.; Praz, L.; Coiffier, J.; Frochaux, V.; Durif, J.; Pereira, B.; Arlettaz, L.; Oris, C.; Sapin, V.; et al. S100B Blood Level Determination for Early Management of Ski-Related Mild Traumatic Brain Injury: A Pilot Study. Front. Neurol. 2020, 11, 856. [Google Scholar] [CrossRef]
- Hasselblatt, M.; Mooren, F.C.; von Ahsen, N.; Keyvani, K.; Fromme, A.; Schwarze-Eicker, K.; Senner, V.; Paulus, W. Serum S100beta Increases in Marathon Runners Reflect Extracranial Release Rather than Glial Damage. Neurology 2004, 62, 1634–1636. [Google Scholar] [CrossRef] [PubMed]
- Missler, U.; Wiesmann, M.; Wittmann, G.; Magerkurth, O.; Hagenström, H. Measurement of Glial Fibrillary Acidic Protein in Human Blood: Analytical Method and Preliminary Clinical Results. Clin. Chem. 1999, 45, 138–141. [Google Scholar] [CrossRef] [Green Version]
- Luoto, T.M.; Raj, R.; Posti, J.P.; Gardner, A.J.; Panenka, W.J.; Iverson, G.L. A Systematic Review of the Usefulness of Glial Fibrillary Acidic Protein for Predicting Acute Intracranial Lesions Following Head Trauma. Front. Neurol. 2017, 8, 652. [Google Scholar] [CrossRef] [Green Version]
- Mondello, S.; Sorinola, A.; Czeiter, E.; Vámos, Z.; Amrein, K.; Synnot, A.; Donoghue, E.; Sandor, J.; Wang, K.K.; Diaz-Arrastia, R. Blood-Based Protein Biomarkers for the Management of Traumatic Brain Injuries in Adults Presenting to Emergency Departments with Mild Brain Injury: A Living Systematic Review and Meta-Analysis. J. Neurotrauma 2021, 38, 1086–1106. [Google Scholar] [CrossRef] [Green Version]
- Di Pietro, V.; Yakoub, K.M.; Scarpa, U.; Di Pietro, C.; Belli, A. MicroRNA Signature of Traumatic Brain Injury: From the Biomarker Discovery to the Point-of-Care. Front. Neurol. 2018, 9, 429. [Google Scholar] [CrossRef] [Green Version]
- Furuhashi, M.; Hotamisligil, G.S. Fatty Acid-Binding Proteins: Role in Metabolic Diseases and Potential as Drug Targets. Nat. Rev. Drug Discov. 2008, 7, 489–503. [Google Scholar] [CrossRef] [Green Version]
- Lagerstedt, L.; Egea-Guerrero, J.J.; Bustamante, A.; Montaner, J.; Rodriguez-Rodriguez, A.; El Rahal, A.; Turck, N.; Quintana, M.; Garcia-Armengol, R.; Prica, C.M.; et al. H-FABP: A New Biomarker to Differentiate between CT-Positive and CT-Negative Patients with Mild Traumatic Brain Injury. PLoS ONE 2017, 12, e0175572. [Google Scholar] [CrossRef] [PubMed]
- Bogoslovsky, T.; Gill, J.; Jeromin, A.; Davis, C.; Diaz-Arrastia, R. Fluid Biomarkers of Traumatic Brain Injury and Intended Context of Use. Diagnostics 2016, 6, 37. [Google Scholar] [CrossRef] [PubMed]
- Posti, J.P.; Takala, R.S.K.; Lagerstedt, L.; Dickens, A.M.; Hossain, I.; Mohammadian, M.; Ala-Seppälä, H.; Frantzén, J.; van Gils, M.; Hutchinson, P.J.; et al. Correlation of Blood Biomarkers and Biomarker Panels with Traumatic Findings on Computed Tomography after Traumatic Brain Injury. J. Neurotrauma 2019, 36, 2178–2189. [Google Scholar] [CrossRef] [PubMed]
- Lagerstedt, L.; Egea-Guerrero, J.J.; Bustamante, A.; Rodríguez-Rodríguez, A.; Rahal, A.E.; Quintana-Diaz, M.; García-Armengol, R.; Prica, C.M.; Andereggen, E.; Rinaldi, L.; et al. Combining H-FABP and GFAP Increases the Capacity to Differentiate between CT-Positive and CT-Negative Patients with Mild Traumatic Brain Injury. PLoS ONE 2018, 13, e0200394. [Google Scholar] [CrossRef] [PubMed]
- Bazarian, J.J.; Biberthaler, P.; Welch, R.D.; Lewis, L.M.; Barzo, P.; Bogner-Flatz, V.; Brolinson, P.G.; Büki, A.; Chen, J.Y.; Christenson, R.H.; et al. Serum GFAP and UCH-L1 for Prediction of Absence of Intracranial Injuries on Head CT (ALERT-TBI): A Multicentre Observational Study. Lancet Neurol. 2018, 17, 782–789. [Google Scholar] [CrossRef]
- Abbott Receives FDA 510(k) Clearance for the First Rapid Handheld Blood Test for Concussions. Available online: https://abbott.mediaroom.com/2021-01-11-Abbott-Receives-FDA-510-k-Clearance-for-the-First-Rapid-Handheld-Blood-Test-for-Concussions (accessed on 24 March 2021).
- Nanodiagnostics Nanosensor Technology|Our Solution. Available online: https://nanodiagnostics.com/our-solution/nanosensor-technology/ (accessed on 29 March 2021).
- Pankratova, N.; Jović, M.; Pfeifer, M.E. Electrochemical Sensing of Blood Proteins for Mild Traumatic Brain Injury (MTBI) Diagnostics and Prognostics: Towards a Point-of-Care Application. RSC Adv. 2021, 11, 17301–17319. [Google Scholar] [CrossRef] [PubMed]
- Gan, X.; Han, D.; Wang, J.; Liu, P.; Li, X.; Zheng, Q.; Yan, Y. A Highly Sensitive Electrochemiluminescence Immunosensor for H-FABP Determination Based on Self-Enhanced Luminophore Coupled with Ultrathin 2D Nickel Metal-Organic Framework Nanosheets. Biosens. Bioelectron. 2021, 171, 112735. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Zhang, X.; Wang, M.; Dong, Y.; Liu, J.; Zhu, Z.; Li, M.; Yang, D.; Shao, Y. Fabrication of Tris(Bipyridine)Ruthenium(II)-Functionalized Metal–Organic Framework Thin Films by Electrochemically Assisted Self-Assembly Technique for Electrochemiluminescent Immunoassay. Anal. Chem. 2018, 90, 11622–11628. [Google Scholar] [CrossRef]
- Fu, Z.; Wei, W.; Li, C.; Wang, Z. Electrochemiluminescent Immunosensing and Its Application in Biological and Pharmaceutical Analysis. Sci. Sin.-Chim 2011, 41, 773–784. [Google Scholar] [CrossRef]
- Hoffmann, U.; Espeter, F.; Weiss, C.; Ahmad-Nejad, P.; Lang, S.; Brueckmann, M.; Akin, I.; Neumaier, M.; Borggrefe, M.; Behnes, M. Ischemic Biomarker Heart-Type Fatty Acid Binding Protein (HFABP) in Acute Heart Failure—Diagnostic and Prognostic Insights Compared to NT-ProBNP and Troponin I. BMC Cardiovasc. Disord. 2015, 15, 50. [Google Scholar] [CrossRef] [Green Version]
- Abir, J.; Sondes, S.; Rania, E.; Latifa, K.; Mokhles, B.D.; Nedia, B.; Hadj, B.; Manel, M.; Souhir, K.; Hejer, G.; et al. Serum Heart Type Fatty Acid Binding Protein (H-FABP) Levels in Metabolic Syndrome. Int. J. Pharm. Sci. Res. 2017, 8, 1441–1448. [Google Scholar]
- Wiesmann, M.; Missler, U.; Gottmann, D.; Gehring, S. Plasma S-100b Protein Concentration in Healthy Adults Is Age-and Sex-Independent. Clin. Chem. 1998, 44, 1056–1058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adrian, H.; Marten, K.; Salla, N.; Lasse, V. Biomarkers of Traumatic Brain Injury: Temporal Changes in Body Fluids. eNeuro 2016, 3, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordillo-Escobar, E.; Egea-Guerrero, J.J.; Rodríguez-Rodríguez, A.; Murillo-Cabezas, F. Usefulness of Biomarkers in the Prognosis of Severe Head Injuries. Med. Intensiva 2016, 40, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Lewis, L.M.; Schloemann, D.T.; Lindburg, M.; Papa, L.; Fucetola, R.P.; Bazarian, J.; Welch, R.D. Utility of Serum Biomarkers in the Diagnosis and Stratification of Mild Traumatic Brain Injury. Acad. Emerg. Med. 2017, 24, 710–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MSD Technology Platform. Available online: https://www.mesoscale.com/~/media/files/brochures/techbrochure.pdf (accessed on 24 September 2020).
- Neves, M.M.P.S.; Bobes-Limenes, P.; Pérez-Junquera, A.; González-García, M.B.; Hernández-Santos, D.; Fanjul-Bolado, P. Miniaturized Analytical Instrumentation for Electrochemiluminescence Assays: A Spectrometer and a Photodiode-Based Device. Anal. Bioanal. Chem. 2016, 408, 7121–7127. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.P.; Shaw, G.S. A Novel Calcium-Sensitive Switch Revealed by the Structure of Human S100B in the Calcium-Bound Form. Structure 1998, 6, 211–222. [Google Scholar] [CrossRef] [Green Version]
- Gonçalves, D.S.; Lenz, G.; Karl, J.; Gonçalves, C.A.; Rodnight, R. Extracellular S100B Protein Modulates ERK in Astrocyte Cultures. Neuroreport 2000, 11, 807–809. [Google Scholar] [CrossRef] [Green Version]
- Green, A.J.; Keir, G.; Thompson, E.J. A Specific and Sensitive ELISA for Measuring S-100b in Cerebrospinal Fluid. J. Immunol. Methods 1997, 205, 35–41. [Google Scholar] [CrossRef]
- Tramontina, F.; Karl, J.; Gottfried, C.; Mendez, A.; Gonçalves, D.; Portela, L.V.; Gonçalves, C.-A. Digitonin-Permeabilization of Astrocytes in Culture Monitored by Trypan Blue Exclusion and Loss of S100B by ELISA. Brain Res. Protoc. 2000, 6, 86–90. [Google Scholar] [CrossRef]
- Tort, A.B.L.; Dietrich, M.O.; Gonçalves, C.A.; Souza, D.O.; Portela, L.V.C. Influence of Anticoagulants on the Measurement of S100B Protein in Blood. Clin. Biochem. 2003, 36, 629–632. [Google Scholar] [CrossRef]
- Valenti, G.; Fiorani, A.; Li, H.; Sojic, N.; Paolucci, F. Essential Role of Electrode Materials in Electrochemiluminescence Applications. ChemElectroChem 2016, 3, 1990–1997. [Google Scholar] [CrossRef]
- Workman, S.; Richter, M.M. The Effects of Nonionic Surfactants on the Tris(2,2‘-Bipyridyl)Ruthenium(II)–Tripropylamine Electrochemiluminescence System. Anal. Chem. 2000, 72, 5556–5561. [Google Scholar] [CrossRef] [PubMed]
- Kerr, E.; Alexander, R.; Francis, P.S.; Guijt, R.M.; Barbante, G.J.; Doeven, E.H. A Comparison of Commercially Available Screen-Printed Electrodes for Electrogenerated Chemiluminescence Applications. Front. Chem. 2021, 8, 628483. [Google Scholar] [CrossRef] [PubMed]
- Banks, C.E.; Davies, T.J.; Wildgoose, G.G.; Compton, R.G. Electrocatalysis at Graphite and Carbon Nanotube Modified Electrodes: Edge-Plane Sites and Tube Ends Are the Reactive Sites. Chem. Commun. 2005, 829–841. [Google Scholar] [CrossRef] [PubMed]
- Fanjul-Bolado, P.; Hernández-Santos, D.; Lamas-Ardisana, P.J.; Martín-Pernía, A.; Costa-García, A. Electrochemical Characterization of Screen-Printed and Conventional Carbon Paste Electrodes. Electrochim. Acta 2008, 53, 3635–3642. [Google Scholar] [CrossRef]
- Bard, A.J.; Faulkner, L.R. Electrochemical Methods: Fundamentals and Applications, 2nd ed.; John Wiley & Sons: New York, NY, USA, 2001. [Google Scholar]
- Zu, Y.; Bard, A.J. Electrogenerated Chemiluminescence. 67. Dependence of Light Emission of the Tris(2,2‘)Bipyridylruthenium(II)/Tripropylamine System on Electrode Surface Hydrophobicity. Anal. Chem. 2001, 73, 3960–3964. [Google Scholar] [CrossRef] [PubMed]
- Button, E.B.; Cheng, W.H.; Barron, C.; Cheung, H.; Bashir, A.; Cooper, J.; Gill, J.; Stukas, S.; Baron, D.C.; Robert, J.; et al. Development of a Novel, Sensitive Translational Immunoassay to Detect Plasma Glial Fibrillary Acidic Protein (GFAP) after Murine Traumatic Brain Injury. Alz. Res. Ther. 2021, 13, 58. [Google Scholar] [CrossRef]
- Elecsys® S100. Available online: https://diagnostics.roche.com/global/en/products/params/elecsys-s100.html (accessed on 27 August 2021).
- Natarajan, S.; Joseph, J. A Novel Time-Resolved Fluorescent Lateral Flow Immunoassay for Quantitative Detection of the Trauma Brain Injury Biomarker-Glial Fibrillary Acidic Protein. Sens. Diagn. 2022, 1, 193–197. [Google Scholar] [CrossRef]
- Savin, M.; Mihailescu, C.-M.; Matei, I.; Stan, D.; Moldovan, C.A.; Ion, M.; Baciu, I. A Quantum Dot-Based Lateral Flow Immunoassay for the Sensitive Detection of Human Heart Fatty Acid Binding Protein (HFABP) in Human Serum. Talanta 2018, 178, 910–915. [Google Scholar] [CrossRef]
- Gao, X.; Boryczka, J.; Zheng, P.; Kasani, S.; Yang, F.; Engler-Chiurazzi, E.B.; Simpkins, J.W.; Wigginton, J.G.; Wu, N. A “Hot Spot”-Enhanced Paper Lateral Flow Assay for Ultrasensitive Detection of Traumatic Brain Injury Biomarker S-100β in Blood Plasma. Biosens. Bioelectron. 2021, 177, 112967. [Google Scholar] [CrossRef] [PubMed]
- Mincu, N.-B.; Lazar, V.; Stan, D.; Mihailescu, C.M.; Iosub, R.; Mateescu, A.L. Screen-Printed Electrodes (SPE) for In Vitro Diagnostic Purpose. Diagnostics 2020, 10, 517. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Periñán, E.; Gutiérrez-Sánchez, C.; García-Mendiola, T.; Lorenzo, E. Electrochemiluminescence Biosensors Using Screen-Printed Electrodes. Biosensors 2020, 10, 118. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Li, H.; Lau, C.; Zhang, Y. Cross-Talk-Free Simultaneous Fluoroimmunoassay of Two Biomarkers Based on Dual-Color Quantum Dots. Anal. Chim. Acta 2011, 698, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zheng, L.; Cai, G.; Liu, N.; Liao, M.; Li, Y.; Zhang, X.; Lin, J. A Microfluidic Biosensor for Online and Sensitive Detection of Salmonella Typhimurium Using Fluorescence Labeling and Smartphone Video Processing. Biosens. Bioelectron. 2019, 140, 111333. [Google Scholar] [CrossRef]
- Wu, Y.; Xue, P.; Kang, Y.; Hui, K.M. Paper-Based Microfluidic Electrochemical Immunodevice Integrated with Nanobioprobes onto Graphene Film for Ultrasensitive Multiplexed Detection of Cancer Biomarkers. Anal. Chem. 2013, 85, 8661–8668. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Rong, Q.; Ma, Z.; Han, H. One-Step Synthesis of Redox-Active Polymer/AU Nanocomposites for Electrochemical Immunoassay of Multiplexed Tumor Markers. Biosens. Bioelectron. 2015, 65, 307–313. [Google Scholar] [CrossRef]
- Otsuki, S.; Ishikawa, M. Wavelength-Scanning Surface Plasmon Resonance Imaging for Label-Free Multiplexed Protein Microarray Assay. Biosens. Bioelectron. 2010, 26, 202–206. [Google Scholar] [CrossRef]
- Yang, Z.; Zong, C.; Yan, F.; Ju, H. Automated Chemiluminescent Dual-Analyte Immunoassay Based on Resolved Immunosensing Channels. Talanta 2010, 82, 1462–1467. [Google Scholar] [CrossRef]
- Lv, W.; Ye, H.; Yuan, Z.; Liu, X.; Chen, X.; Yang, W. Recent Advances in Electrochemiluminescence-Based Simultaneous Detection of Multiple Targets. TrAC Trends Anal. Chem. 2020, 123, 115767. [Google Scholar] [CrossRef]
- Papa, L.; Brophy, G.M.; Welch, R.D.; Lewis, L.M.; Braga, C.F.; Tan, C.N.; Ameli, N.J.; Lopez, M.A.; Haeussler, C.A.; Giordano, D.I.M. Time Course and Diagnostic Accuracy of Glial and Neuronal Blood Biomarkers GFAP and UCH-L1 in a Large Cohort of Trauma Patients with and without Mild Traumatic Brain Injury. JAMA Neurol. 2016, 73, 551–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arya, S.K.; Pui, T.S.; Wong, C.C.; Kumar, S.; Rahman, A.R.A. Effects of the Electrode Size and Modification Protocol on a Label-Free Electrochemical Biosensor. Langmuir 2013, 29, 6770–6777. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Besar, K.; LeCover, R.; Dulloor, P.; Sinha, J.; Martínez Hardigree, J.F.; Pick, C.; Swavola, J.; Everett, A.D.; Frechette, J.; et al. Label-Free Brain Injury Biomarker Detection Based on Highly Sensitive Large Area Organic Thin Film Transistor with Hybrid Coupling Layer. Chem. Sci. 2014, 5, 416–426. [Google Scholar] [CrossRef]
- Wang, T.; Fang, Y.; He, Z. Electrochemical Quantitative Detection of Glial Fibrillary Acidic Protein Based on Molecularly Imprinted Polymer Sensor. Int. J. Electrochem. Sci. 2017, 12, 7341–7350. [Google Scholar] [CrossRef]
- Song, J.; Dailey, J.; Li, H.; Jang, H.-J.; Zhang, P.; Wang, J.T.-H.; Everett, A.D.; Katz, H.E. Extended Solution Gate OFET-Based Biosensor for Label-Free Glial Fibrillary Acidic Protein Detection with Polyethylene Glycol-Containing Bioreceptor Layer. Adv. Funct. Mater. 2017, 27, 1606506. [Google Scholar] [CrossRef] [PubMed]
- Khetani, S.; Ozhukil Kollath, V.; Kundra, V.; Nguyen, M.D.; Debert, C.; Sen, A.; Karan, K.; Sanati-Nezhad, A. Polyethylenimine Modified Graphene-Oxide Electrochemical Immunosensor for the Detection of Glial Fibrillary Acidic Protein in Central Nervous System Injury. ACS Sens. 2018, 3, 844–851. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Xu, G.; Wei, F.; Cen, Y.; Song, Y.; Ma, Y.; Xu, X.; Shi, M.; Sohail, M.; Hu, Q. Carbon Dots Based Immunosorbent Assay for the Determination of GFAP in Human Serum. Nanotechnology 2018, 29, 145501. [Google Scholar] [CrossRef]
- Cardinell, B.A.; Addington, C.P.; Stabenfeldt, S.E.; La Belle, J.T. Multi-Biomarker Detection Following Traumatic Brain Injury. Crit. Rev. Biomed. Eng. 2019, 47, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Agostini, M.; Amato, F.; Vieri, M.L.; Greco, G.; Tonazzini, I.; Baroncelli, L.; Caleo, M.; Vannini, E.; Santi, M.; Signore, G.; et al. Glial-Fibrillary-Acidic-Protein (GFAP) Biomarker Detection in Serum-Matrix: Functionalization Strategies and Detection by an Ultra-High-Frequency Surface-Acoustic-Wave (UHF-SAW) Lab-on-Chip. Biosens. Bioelectron. 2021, 172, 112774. [Google Scholar] [CrossRef]
- Krausz, A.D.; Korley, F.K.; Burns, M.A. A Variable Height Microfluidic Device for Multiplexed Immunoassay Analysis of Traumatic Brain Injury Biomarkers. Biosensors 2021, 11, 320. [Google Scholar] [CrossRef] [PubMed]
- GFAP ELISA Kit|Sigma-Aldrich. Available online: https://www.sigmaaldrich.com/HK/zh/life-science/supelco (accessed on 30 August 2021).
- R-PLEX Human GFAP Antibody Set|Meso Scale Discovery. Available online: https://www.mesoscale.com/en/products/r-plex-human-gfap-antibody-set-f211m/ (accessed on 31 August 2021).
- Available online: https://www.Banyanbio.Com/Assets/Files/000175_vA-IFU-Banyan-BTI.Pdf (accessed on 27 April 2020).
- Simoa Technology. Available online: https://www.Quanterix.Com/Technology (accessed on 27 April 2020).
- TBI Plasma Cartridge|Abbott Point of Care. Available online: https://www.pointofcare.abbott/us/en/offerings/istat/istat-test-cartridges/TBI-Plasma (accessed on 30 August 2021).
- Okonkwo, D.O.; Puffer, R.C.; Puccio, A.M.; Yuh, E.L.; Yue, J.K.; Diaz-Arrastia, R.; Korley, F.K.; Wang, K.K.W.; Sun, X.; Taylor, S.R.; et al. Point-of-Care Platform Blood Biomarker Testing of Glial Fibrillary Acidic Protein versus S100 Calcium-Binding Protein B for Prediction of Traumatic Brain Injuries: A Transforming Research and Clinical Knowledge in Traumatic Brain Injury Study. J. Neurotrauma 2020, 37, 2460–2467. [Google Scholar] [CrossRef] [PubMed]
- Korley, F.K.; Datwyler, S.A.; Jain, S.; Sun, X.; Beligere, G.; Chandran, R.; Marino, J.A.; McQuiston, B.; Zhang, H.; Caudle, K.L.; et al. Comparison of GFAP and UCH-L1 Measurements from Two Prototype Assays: The Abbott i-STAT and ARCHITECT Assays. Neurotrauma Rep. 2021, 2, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.J.; Hu, J. Magnetic Immunosensor and Method of Use. U.S. Patent 9,958,440, 2018. [Google Scholar]
- The NanoDxTM System for TBI|Our Solution. Available online: https://nanodiagnostics.com/our-solution/the-nanodx-system-for-tbi/ (accessed on 30 August 2021).
- Feng, L.-N.; Bian, Z.-P.; Peng, J.; Jiang, F.; Yang, G.-H.; Zhu, Y.-D.; Yang, D.; Jiang, L.-P.; Zhu, J.-J. Ultrasensitive Multianalyte Electrochemical Immunoassay Based on Metal Ion Functionalized Titanium Phosphate Nanospheres. Anal. Chem. 2012, 84, 7810–7815. [Google Scholar] [CrossRef] [PubMed]
- Stan, D.; Mihailescu, C.-M.; Iosub, R.; Moldovan, C.; Savin, M.; Baciu, I. Electrochemical Studies of Homogeneous Self-Assembled Monolayers versus Mixed Self-Assembled Monolayers on Gold Electrode for “Label Free” Detection of Heart Fatty Acid Binding Protein. Thin Solid Film. 2012, 526, 143–149. [Google Scholar] [CrossRef]
- Carless, D.R.; Wnęk, M.; Knox, C.; Harrison, K.R.; Calder, N.; Hall, A.S.; Barth, J.H. Clinical and Analytical Evaluation of an Immunoturbidimetric Heart-Type Fatty Acid-Binding Protein Assay. Scand. J. Clin. Lab. Investig. 2013, 73, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Mihailescu, C.-M.; Stan, D.; Iosub, R.; Moldovan, C.; Savin, M. A Sensitive Capacitive Immunosensor for Direct Detection of Human Heart Fatty Acid-Binding Protein (h-FABP). Talanta 2015, 132, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Xu, A.; Liu, L.; Sui, Y.; Li, Y.; Tan, Y.; Chen, C.; Xie, Q. Selective Staining of CdS on ZnO Biolabel for Ultrasensitive Sandwich-Type Amperometric Immunoassay of Human Heart-Type Fatty-Acid-Binding Protein and Immunoglobulin G. Biosens. Bioelectron. 2017, 91, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Crapnell, R.D.; Canfarotta, F.; Czulak, J.; Johnson, R.; Betlem, K.; Mecozzi, F.; Down, M.P.; Eersels, K.; van Grinsven, B.; Cleij, T.J.; et al. Thermal Detection of Cardiac Biomarkers Heart-Fatty Acid Binding Protein and ST2 Using a Molecularly Imprinted Nanoparticle-Based Multiplex Sensor Platform. ACS Sens. 2019, 4, 2838–2845. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Guo, L.; Hu, Y.; Li, Z.; Liu, J.; He, J.; Cui, H. Multiplexed Chemiluminescence Determination of Three Acute Myocardial Infarction Biomarkers Based on Microfluidic Paper-Based Immunodevice Dual Amplified by Multifunctionalized Gold Nanoparticles. Talanta 2020, 207, 120346. [Google Scholar] [CrossRef] [PubMed]
- TBICheck—The Fastest Diagnostic Test for Traumatic Brain Injury. Available online: https://tbicheck.com/home (accessed on 30 August 2021).
- Human H-FABP ELISA Kit (Ab243682)|Abcam. Available online: https://www.abcam.com/Human-H-FABP-ELISA-Kit-ab243682.html?gclsrc=aw.ds|aw.ds&gclid=Cj0KCQjwg7KJBhDyARIsAHrAXaH7SK6R4I_1DbOvLN1pxlnqW42hdrT0Nk4Yjew7t-ynxwT-aoeUn4EaAuTJEALw_wcB (accessed on 30 August 2021).
- R-PLEX Human FABP3/H-FABP Antibody Set|Meso Scale Discovery. Available online: https://www.mesoscale.com/en/products/r-plex-human-fabp3-h-fabp-antibody-set-f214t/ (accessed on 31 August 2021).
- Missler, U.; Wiesmann, M.; Friedrich, C.; Kaps, M. S-100 Protein and Neuron-Specific Enolase Concentrations in Blood as Indicators of Infarction Volume and Prognosis in Acute Ischemic Stroke. Stroke 1997, 28, 1956–1960. [Google Scholar] [CrossRef] [PubMed]
- PII: S0022-1759(97)00050-1|Elsevier Enhanced Reader. Available online: https://reader.elsevier.com/reader/sd/pii/S0022175997000501?token=A464853B1ED8E1A9E89C9B8F041BB4E0DF5188385648EF4C35723E513BFC3977CFC76D86BA77D9A85AFD7B71FF48711E&originRegion=eu-west-1&originCreation=20210827152016 (accessed on 27 August 2021).
- Leite, M.C.; Galland, F.; Brolese, G.; Guerra, M.C.; Bortolotto, J.W.; Freitas, R.; de Almeida, L.M.V.; Gottfried, C.; Gonçalves, C.-A. A Simple, Sensitive and Widely Applicable ELISA for S100B: Methodological Features of the Measurement of This Glial Protein. J. Neurosci. Methods 2008, 169, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, H.; Chen, J.; Liu, C.; Li, W.; Kong, J.; Yang, P.; Liu, B. A Sensitive Microchip-Based Immunosensor for Electrochemical Detection of Low-Level Biomarker S100B. Electroanalysis 2013, 25, 1050–1055. [Google Scholar] [CrossRef]
- Wang, X.; Li, H.; Li, X.; Chen, Y.; Yin, Y.; Li, G. Electrochemical Assay of Melanoma Biomarker in Human Blood. Electrochem. Commun. 2014, 39, 12–14. [Google Scholar] [CrossRef]
- Mikuła, E.; Wysłouch-Cieszyńska, A.; Zhukova, L.; Puchalska, M.; Verwilst, P.; Dehaen, W.; Radecki, J.; Radecka, H. Voltammetric Detection of S100B Protein Using His-Tagged Receptor Domains for Advanced Glycation End Products (RAGE) Immobilized onto a Gold Electrode Surface. Sensors 2014, 14, 10650–10663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.; Searson, P.C. Magnetic Bead-Quantum Dot Assay for Detection of a Biomarker for Traumatic Brain Injury. Nanoscale 2015, 7, 17820–17826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurzątkowska, K.; Jankowska, A.; Wysłouch-Cieszyńska, A.; Zhukova, L.; Puchalska, M.; Dehaen, W.; Radecka, H.; Radecki, J. Voltammetric Detection of the S100B Protein Using His-Tagged RAGE Domain Immobilized onto a Gold Electrode Modified with a Dipyrromethene—Cu(II) Complex and Different Diluents. J. Electroanal. Chem. 2016, 767, 76–83. [Google Scholar] [CrossRef]
- Khetani, S.; Aburashed, R.; Singh, A.; Sen, A.; Sanati-Nezhad, A. Immunosensing of S100β Biomarker for Diagnosis of Spinal Cord Injuries (SCI). Sens. Actuators B Chem. 2017, 247, 163–169. [Google Scholar] [CrossRef]
- Kuo, Y.-C.; Lee, C.-K.; Lin, C.-T. Improving Sensitivity of a Miniaturized Label-Free Electrochemical Biosensor Using Zigzag Electrodes. Biosens. Bioelectron. 2018, 103, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, P.; Mao, L.; Hou, Y.; Li, D. Determination of Brain Injury Biomarkers by Surface-Enhanced Raman Scattering Using Hollow Gold Nanospheres. RSC Adv. 2018, 8, 3143–3150. [Google Scholar] [CrossRef] [Green Version]
- Mathew, A.S.; Shi, X.; Yau, S.-T. Detection of a Traumatic Brain Injury Biomarker at the 10 Fg/ML Level. Mol. Diagn. Ther. 2018, 22, 729–735. [Google Scholar] [CrossRef]
- Sun, J.; Zhao, Y.; Hou, Y.; Li, H.; Yang, M.; Wang, Y.; Sun, B. Multiplexed Electrochemical and SERS Dual-Mode Detection of Stroke Biomarkers: Rapid Screening with High Sensitivity. New J. Chem. 2019, 43, 13381–13387. [Google Scholar] [CrossRef]
- Tabrizi, M.A.; Ferré-Borrull, J.; Kapruwan, P.; Marsal, L.F. A Photoelectrochemical Sandwich Immunoassay for Protein S100β, a Biomarker for Alzheimer’s Disease, Using an ITO Electrode Modified with a Reduced Graphene Oxide-Gold Conjugate and CdS-Labeled Secondary Antibody. Microchim. Acta 2019, 186, 117. [Google Scholar] [CrossRef] [PubMed]
- Harpaz, D.; Koh, B.; Marks, R.S.; Seet, R.C.S.; Abdulhalim, I.; Tok, A.I.Y. Point-of-Care Surface Plasmon Resonance Biosensor for Stroke Biomarkers NT-ProBNP and S100β Using a Functionalized Gold Chip with Specific Antibody. Sensors 2019, 19, 2533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, L.; Ding, C.; Guo, Y.; Wang, Y.; Ding, Y. Sensitively Detecting MTBI Biomarker S100B by Using Peptide-Modified Ratiometric Fluorescent C/AuNCs Nanoprobe. Anal Bioanal Chem 2020, 412, 3695–3702. [Google Scholar] [CrossRef] [PubMed]
- Hassanain, W.A.; Sivanesan, A.; Ayoko, G.A.; Izake, E.L. Rapid Electrochemical Nanosensing of S100ß in Blood. J. Electrochem. Soc. 2020, 167, 067518. [Google Scholar] [CrossRef]
- Delefortrie, Q.; Lejeune, F.; Kerzmann, B.; Levy, R.; Adam, J.-F.; Sottiaux, T.; Grimmelprez, A.; Vankerkhoven, P.; Hachimi-Idrissi, S. Evaluation of the Roche® Elecsys and the Diasorin® Liaison S100 Kits in the Management of Mild Head Injury in the Emergency Room. Clin. Biochem. 2018, 52, 123–130. [Google Scholar] [CrossRef] [PubMed]
- CanAg® S100 EIA|Fujirebio. Available online: https://www.fujirebio.com/en/products-solutions/canag-s100-eia (accessed on 27 August 2021).
- Human S100B ELISA, EZHS100B-33K This Human S100B ELISA Kit Is to Be Used for the Quantification of Human S100B in Cerebrospinal Fluid, Serum & Plasma.|Sigma-Aldrich. Available online: http://www.sigmaaldrich.com/ (accessed on 30 August 2021).
- S100B ELISA Kit (Ab234573)|Abcam. Available online: https://www.abcam.com/s100b-elisa-kit-ab234573.html (accessed on 30 August 2021).
- Human S100B ELISA Kit by Bioss, Cat. No. BSKH1066|Lucerna-Chem AG. Available online: https://lucerna-chem.ch/shop/3438089/human-s100b-elisa-kit (accessed on 30 August 2021).
- S100b ELISA Kit (Rabbit) (GWB-KBBVZ1)|Quantitative Sandwich ELISA. Available online: https://www.genwaybio.com/s100b-elisa-kit-rabbit-gwb-kbbvz1 (accessed on 30 August 2021).


| Biomarker | Physiological Concentration 1 | ||
|---|---|---|---|
| Abbreviation | Full Name | Normal | Mild TBI |
| GFAP | Glial fibrillary acidic protein | RG: 2–49 pg mL−1 [18] MD: 4 pg mL−1 [18] | ≥33 pg mL−1 [18] CO: 22 pg mL−1 [27] |
| h-FABP | Heart-fatty acidic binding protein | <5.5 ng mL−1 [34] MN: 3.78 ng mL−1 [35] | CO: 2.62 ng mL−1 (HS/HP) [23] |
| S100β | S100β calcium-binding protein | MD: 50 pg mL−1 (HP) [36] <0.11 pg mL−1 [37] | ≥100 pg mL−1 (HP) [38] >75 pg mL−1 [39] CO: 42 pg mL−1 (HS/HP) [23] |
| mTBI Biomarker | cAb Diluent | Blocking Agent | Assay Diluent | Wash Buffer | dAb Diluent | cAb Conc. (μg mL−1) | dAb Conc. (μg mL−1) | LOD (pg mL−1) | Range (pg mL−1) |
|---|---|---|---|---|---|---|---|---|---|
| GFAP | PBS 1× | 1% BSA; 0.06% Tween-20; PBS 1× | MQ water | 0.06% Tween-20; PBS 1× | 1% BSA; PBS 1× | 25.00 | 2.50 | 6.94 [6.9–10.0; CI:95%] | 0–10,000 |
| h-FABP | PBS 1× | 1% BSA; 0.06% Tween-20; PBS 1× | PBS 1× | 0.06% Tween-20; PBS 1× | 1% BSA; PBS 1× | 5.00 | 0.50 | 1.35 [0–4.4; CI:95%] | 0–10,000 |
| S100β | 5 mM CaCl2; 50 mM TRIS | 2% BSA; 1 mM CaCl2; PBS 1× | 0.1% BSA; 1 mM CaCl2; 50 mM TRIS | 0.06% Tween-20; PBS 1× | 0.1% BSA; 1 mM CaCl2; 50 mM TRIS | 10.00 | 5.00 | 15.73 [11.0–20.5; CI:95%] | 0–10,000 |
| SPE | Product Number | A (cm2) * | ΔE (mV) | Ia/Ic |
|---|---|---|---|---|
| SPCE | DRP-110 | 0.1540 | 150 | 1.04 |
| SPCE-GNP | DRP-110-GNP | 0.1580 | 180 | 1.00 |
| SPCE-CNT-GNP | DRP-110CNT-GNP | 0.1470 | 285 | 1.03 |
| SPCE-QD | DRP-110QD | 0.0270 | 270 | 1.00 |
| mTBI Biomarker | cAb Diluent | Blocking Agent | Assay Diluent | Wash Buffer | dAb Diluent | cAb Concentration (μg mL−1) | dAb Concentration (μg mL−1) | LOD | Dynamic Range |
|---|---|---|---|---|---|---|---|---|---|
| GFAP | PBS 1× | 1% BSA; 0.06% Tween-20; PBS 1× | MQ water | 0.06% Tween-20; PBS 1× | 1% BSA; PBS 1× | 25.00 | 2.50 | 0.59 ng mL−1 | 0–50 ng mL−1 |
| h-FABP | PBS 1× | 1% BSA; 0.06% Tween-20; PBS 1× | PBS 1× | 0.06% Tween-20; PBS 1× | 1% BSA; PBS 1× | 5.00 | 0.50 | 0.44 ng mL−1 | 0–50 ng mL−1 |
| S100β | 5 mM CaCl2; 50 mM TRIS | 2% BSA; 1 mM CaCl2; PBS 1× | 1 mM CaCl2; 50 mM TRIS | 0.06% Tween-20; PBS 1× | 0.1% BSA; 1 mM CaCl2; 50 mM TRIS | 25.00 | 12.50 | 1.34 ng mL−1 | 0–50 ng mL−1 |
| Sample | Added Biomarker | Spiked Concentration (ng mL−1) | Detected Concentration by MSD (ng mL−1) | Recovery (%) | Detected Concentration by µSTAT-ECL (ng mL−1) | Recovery (%) |
|---|---|---|---|---|---|---|
| Human serum | h-FABP | 0 | 0 | / | 0 | / |
| 2.50 | 2.30 | 92% | 1.99 | 79% | ||
| 5.00 | 4.37 | 87% | 5.33 | 106% | ||
| GFAP | 0 | 0 | / | 0 | / | |
| 2.50 | 2.23 | 89% | 2.62 | 105% | ||
| 5.00 | 5.30 | 106% | 4.34 | 87% | ||
| S100β | 0 | 0 | / | 0 | / | |
| 2.50 | 2.43 | 96% | 2.11 | 84% | ||
| 5.00 | 6.41 | 128% | 5.92 | 118% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jović, M.; Prim, D.; Saini, E.; Pfeifer, M.E. Towards a Point-of-Care (POC) Diagnostic Platform for the Multiplex Electrochemiluminescent (ECL) Sensing of Mild Traumatic Brain Injury (mTBI) Biomarkers. Biosensors 2022, 12, 172. https://doi.org/10.3390/bios12030172
Jović M, Prim D, Saini E, Pfeifer ME. Towards a Point-of-Care (POC) Diagnostic Platform for the Multiplex Electrochemiluminescent (ECL) Sensing of Mild Traumatic Brain Injury (mTBI) Biomarkers. Biosensors. 2022; 12(3):172. https://doi.org/10.3390/bios12030172
Chicago/Turabian StyleJović, Milica, Denis Prim, Edis Saini, and Marc Emil Pfeifer. 2022. "Towards a Point-of-Care (POC) Diagnostic Platform for the Multiplex Electrochemiluminescent (ECL) Sensing of Mild Traumatic Brain Injury (mTBI) Biomarkers" Biosensors 12, no. 3: 172. https://doi.org/10.3390/bios12030172
APA StyleJović, M., Prim, D., Saini, E., & Pfeifer, M. E. (2022). Towards a Point-of-Care (POC) Diagnostic Platform for the Multiplex Electrochemiluminescent (ECL) Sensing of Mild Traumatic Brain Injury (mTBI) Biomarkers. Biosensors, 12(3), 172. https://doi.org/10.3390/bios12030172








