The Structure of Bilirubin Oxidase from Bacillus pumilus Reveals a Unique Disulfide Bond for Site-Specific Direct Electron Transfer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cloning, Expression, and Purification of BpBOD
2.3. Crystallization, Data Collection, and Structure Determination
2.4. Bilirubin Oxidase Structure Analysis
2.5. Bilirubin Oxidase-Based Cathode Fabrication
2.6. Fluorescence Intensity of the BpBOD-Pyrene-Maleimide-TCEP Mixture
2.7. Estimation of the BpBOD Deposited in Electrochemically Active Orientation on the Electrode
2.8. Electron Transfer Rate (Ket) Calculations
2.9. Effect of TCEP and PyMal on BpBOD Activity
3. Results and Discussion
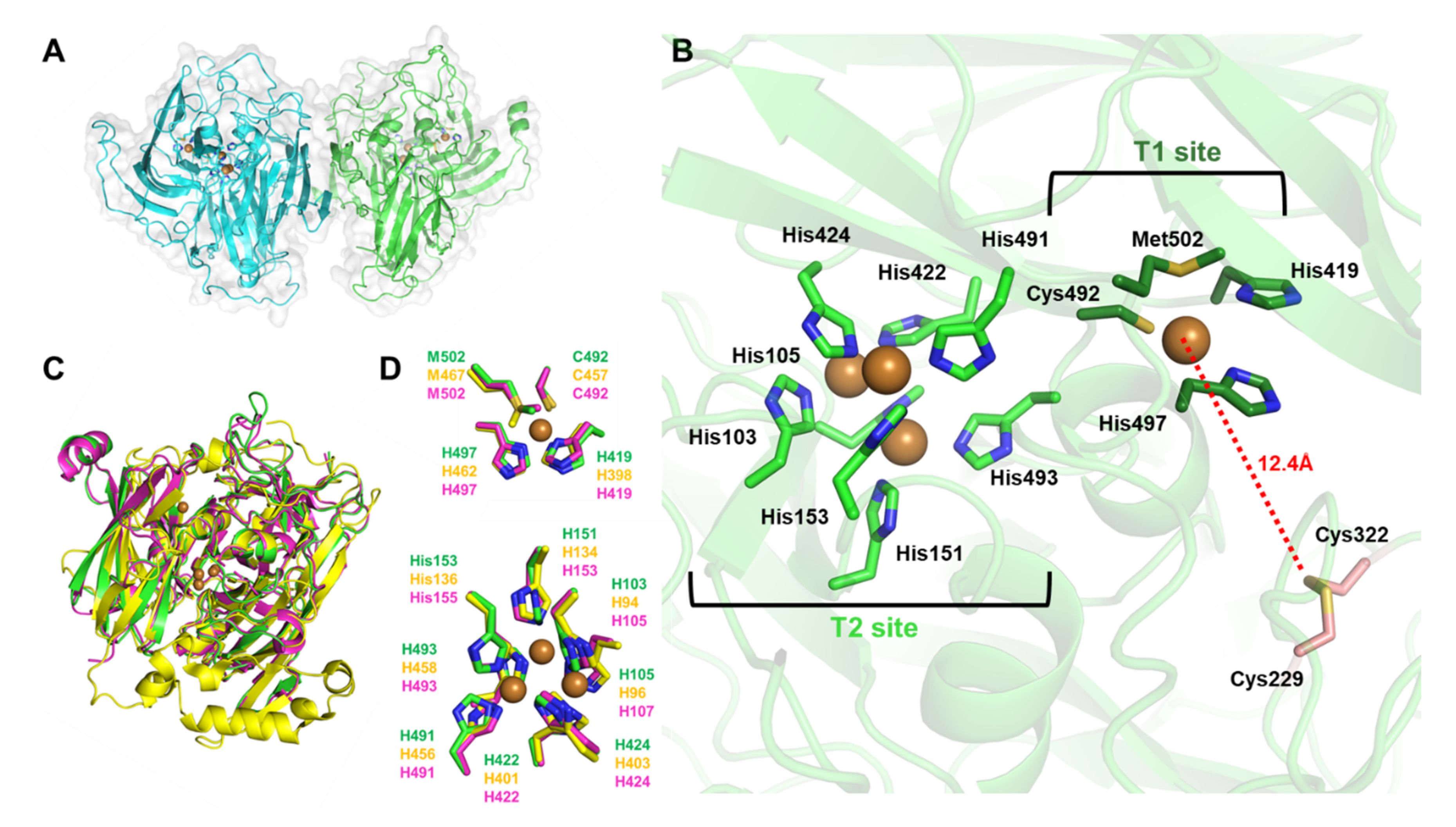
3.1. BpBOD Structure Revealed a Unique Disulfide Bond
| Protein [PDB Code] | BpBOD [7Z5P] |
|---|---|
| Data collection | |
| Synchrotron | DESY, Hamburg |
| Beamline | P14 |
| Wavelength | 0.97625 |
| Space group | C 1 2 1 |
| Resolution range * | 59.22–2.991 (3.098–2.991) |
| Unit cell dimensions a, b, c (Å) α, β, γ (°) | 198.96, 63.51, 115.96 90, 124.567, 90 |
| Total reflections * | 166,046 (10,488) |
| Unique reflections * | 24,048 (2229) |
| Completeness (%) * | 98.12 (92.57) |
| Mean I/sigma(I) * | 7.4 (1.5) |
| Wilson B-factor | 78.94 |
| R-merge *,† | 0.188 (1.47) |
| R-meas * | 0.204 (1.60) |
| CC1/2 * | 99.5 (71.9) |
| Refinement | |
| Resolution (Å) | 59.22–3.5 |
| Reflections used in refinement * | 23,986 (2219) |
| Reflections used for R-free * | 2398 (221) |
| R/Rfree‡,* | 0.254/0.267 (0.491/0.479) |
| Number of non-hydrogen atoms | 8244 |
| Macromolecules | 8236 |
| Ligands | 8 |
| Protein residues | 1010 |
| RMSD bonds length (Å) | 0.009 |
| RMSD bond angles (°) | 1.45 |
| Ramachandran favored (%) | 90.18 |
| Ramachandran allowed (%) | 8.62 |
| Ramachandran outliers (%) | 1.20 |
| Average B-factor (Å2) | 90.64 |
| Macromolecules | 90.63 |
| Ligands | 101.28 |
3.2. BpBOD Site-Specific Direct Electron Transfer
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Messerschmidt, A. Multi-Copper Oxidases; World Scientific: Singapore, 1997; ISBN 978-981-02-2711-1. [Google Scholar]
- Rosenzweig, A.C.; Sazinsky, M.H. Structural Insights into Dioxygen-Activating Copper Enzymes. Curr. Opin. Struct. Biol. 2006, 16, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Kjaergaard, C.H.; Durand, F.; Tasca, F.; Qayyum, M.F.; Kauffmann, B.; Gounel, S.; Suraniti, E.; Hodgson, K.O.; Hedman, B.; Mano, N.; et al. Spectroscopic and Crystallographic Characterization of “Alternative Resting” and “Resting Oxidized” Enzyme Forms of Bilirubin Oxidase: Implications for Activity and Electrochemical Behavior of Multicopper Oxidases. J. Am. Chem. Soc. 2012, 134, 5548–5551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, T.J.; Dalton, H. Chapter 6 Biocatalysis by Methane Monooxygenase and Its Implications for the Petroleum Industry. In Studies in Surface Science and Catalysis; Vazquez-Duhalt, R., Quintero-Ramirez, R., Eds.; Petroleum Biotechnology; Elsevier: Amsterdam, The Netherlands, 2004; Volume 151, pp. 177–192. [Google Scholar]
- Lawton, T.J.; Rosenzweig, A.C. Biocatalysts for Methane Conversion: Big Progress on Breaking a Small Substrate. Curr. Opin. Chem. Biol. 2016, 35, 142–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldfeder, M.; Kanteev, M.; Isaschar-Ovdat, S.; Adir, N.; Fishman, A. Determination of Tyrosinase Substrate-Binding Modes Reveals Mechanistic Differences between Type-3 Copper Proteins. Nat. Commun. 2014, 5, 4505. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, K.U.; Ali, A.S.; Ali, S.A.; Naaz, I. Microbial Tyrosinases: Promising Enzymes for Pharmaceutical, Food Bioprocessing, and Environmental Industry. Biochem. Res. Int. 2014, 2014, e854687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mano, N.; de Poulpiquet, A. O2 Reduction in Enzymatic Biofuel Cells. Chem. Rev. 2018, 118, 2392–2468. [Google Scholar] [CrossRef] [Green Version]
- Lalaoui, N.; Rousselot-Pailley, P.; Robert, V.; Mekmouche, Y.; Villalonga, R.; Holzinger, M.; Cosnier, S.; Tron, T.; Le Goff, A. Direct Electron Transfer between a Site-Specific Pyrene-Modified Laccase and Carbon Nanotube/Gold Nanoparticle Supramolecular Assemblies for Bioelectrocatalytic Dioxygen Reduction. ACS Catal. 2016, 6, 1894–1900. [Google Scholar] [CrossRef]
- Batra, B.; Lata, S.; Sunny; Rana, J.S.; Pundir, C.S. Construction of an Amperometric Bilirubin Biosensor Based on Covalent Immobilization of Bilirubin Oxidase onto Zirconia Coated Silica Nanoparticles/Chitosan Hybrid Film. Biosens. Bioelectron. 2013, 44, 64–69. [Google Scholar] [CrossRef]
- Calabrese Barton, S.; Gallaway, J.; Atanassov, P. Enzymatic Biofuel Cells for Implantable and Microscale Devices. Chem. Rev. 2004, 104, 4867–4886. [Google Scholar] [CrossRef]
- Mukha, D.; Cohen, Y.; Yehezkeli, O. Bismuth Vanadate/Bilirubin Oxidase Photo(Bio)Electrochemical Cells for Unbiased, Light-Triggered Electrical Power Generation. ChemSusChem 2020, 13, 2684–2692. [Google Scholar] [CrossRef]
- Mano, N.; Edembe, L. Bilirubin Oxidases in Bioelectrochemistry: Features and Recent Findings. Biosens. Bioelectron. 2013, 50, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Gounel, S.; Rouhana, J.; Stines-Chaumeil, C.; Cadet, M.; Mano, N. Increasing the Catalytic Activity of Bilirubin Oxidase from Bacillus Pumilus: Importance of Host Strain and Chaperones Proteins. J. Biotechnol. 2016, 230, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Herzallh, N.S.; Cohen, Y.; Cohen, R.; Chmelnik, O.; Shoham, Y.; Yehezkeli, O. Cellulose to Electricity Conversion by an Enzymatic Biofuel Cell. Sustain. Energy Fuels 2021, 5, 4580–4586. [Google Scholar] [CrossRef]
- Cohen, R.; Bitton, R.E.; Herzallh, N.S.; Cohen, Y.; Yehezkeli, O. Utilization of FAD-Glucose Dehydrogenase from T. Emersonii for Amperometric Biosensing and Biofuel Cell Devices. Anal. Chem. 2021, 93, 11585–11591. [Google Scholar] [CrossRef]
- Durand, F.; Kjaergaard, C.H.; Suraniti, E.; Gounel, S.; Hadt, R.G.; Solomon, E.I.; Mano, N. Bilirubin Oxidase from Bacillus Pumilus: A Promising Enzyme for the Elaboration of Efficient Cathodes in Biofuel Cells. Biosens. Bioelectron. 2012, 35, 140–146. [Google Scholar] [CrossRef] [Green Version]
- Gentil, S.; Carrière, M.; Cosnier, S.; Gounel, S.; Mano, N.; Le Goff, A. Direct Electrochemistry of Bilirubin Oxidase from Magnaporthe Orizae on Covalently-Functionalized MWCNT for the Design of High-Performance Oxygen-Reducing Biocathodes. Chem.–A Eur. J. 2018, 24, 8404–8408. [Google Scholar] [CrossRef]
- Reiss, R.; Ihssen, J.; Thöny-Meyer, L. Bacillus Pumiluslaccase: A Heat Stable Enzyme with a Wide Substrate Spectrum. BMC Biotechnol. 2011, 11, 9. [Google Scholar] [CrossRef] [Green Version]
- dos Santos, L.; Climent, V.; Blanford, C.F.; Armstrong, F.A. Mechanistic Studies of the ‘Blue’ Cu Enzyme, Bilirubin Oxidase, as a Highly Efficient Electrocatalyst for the Oxygen Reduction Reaction. Phys. Chem. Chem. Phys. 2010, 12, 13962–13974. [Google Scholar] [CrossRef]
- Tasca, F.; Farias, D.; Castro, C.; Acuna-Rougier, C.; Antiochia, R. Bilirubin Oxidase from Myrothecium Verrucaria Physically Absorbed on Graphite Electrodes. Insights into the Alternative Resting Form and the Sources of Activity Loss. PLoS ONE 2015, 10, e0132181. [Google Scholar] [CrossRef]
- Shleev, S.; El Kasmi, A.; Ruzgas, T.; Gorton, L. Direct Heterogeneous Electron Transfer Reactions of Bilirubin Oxidase at a Spectrographic Graphite Electrode. Electrochem. Commun. 2004, 6, 934–939. [Google Scholar] [CrossRef]
- Dagys, M.; Laurynėnas, A.; Ratautas, D.; Kulys, J.; Vidžiūnaitė, R.; Talaikis, M.; Niaura, G.; Marcinkevičienė, L.; Meškys, R.; Shleev, S. Oxygen Electroreduction Catalysed by Laccase Wired to Gold Nanoparticles via the Trinuclear Copper Cluster. Energy Environ. Sci. 2017, 10, 498–502. [Google Scholar] [CrossRef]
- Milton, R.D.; Giroud, F.; Thumser, A.E.; Minteer, S.D.; Slade, R.C.T. Bilirubin Oxidase Bioelectrocatalytic Cathodes: The Impact of Hydrogen Peroxide. Chem. Commun. 2014, 50, 94–96. [Google Scholar] [CrossRef] [PubMed]
- Bourourou, M.; Elouarzaki, K.; Lalaoui, N.; Agnès, C.; Le Goff, A.; Holzinger, M.; Maaref, A.; Cosnier, S. Supramolecular Immobilization of Laccase on Carbon Nanotube Electrodes Functionalized with (Methylpyrenylaminomethyl)Anthraquinone for Direct Electron Reduction of Oxygen. Chem.–A Eur. J. 2013, 19, 9371–9375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lalaoui, N.; Le Goff, A.; Holzinger, M.; Cosnier, S. Fully Oriented Bilirubin Oxidase on Porphyrin-Functionalized Carbon Nanotube Electrodes for Electrocatalytic Oxygen Reduction. Chem.–A Eur. J. 2015, 21, 16868–16873. [Google Scholar] [CrossRef]
- Yan, Y.-M.; Yehezkeli, O.; Willner, I. Integrated, Electrically Contacted NAD(P)+-Dependent Enzyme–Carbon Nanotube Electrodes for Biosensors and Biofuel Cell Applications. Chem.–A Eur. J. 2007, 13, 10168–10175. [Google Scholar] [CrossRef]
- Yan, Y.; Zheng, W.; Su, L.; Mao, L. Carbon-Nanotube-Based Glucose/O2 Biofuel Cells. Adv. Mater. 2006, 18, 2639–2643. [Google Scholar] [CrossRef]
- Schubert, K.; Goebel, G.; Lisdat, F. Bilirubin Oxidase Bound to Multi-Walled Carbon Nanotube-Modified Gold. Electrochim. Acta 2009, 54, 3033–3038. [Google Scholar] [CrossRef]
- Gross, A.J.; Chen, X.; Giroud, F.; Travelet, C.; Borsali, R.; Cosnier, S. Redox-Active Glyconanoparticles as Electron Shuttles for Mediated Electron Transfer with Bilirubin Oxidase in Solution. J. Am. Chem. Soc. 2017, 139, 16076–16079. [Google Scholar] [CrossRef]
- Elouarzaki, K.; Cheng, D.; Fisher, A.C.; Lee, J.-M. Coupling Orientation and Mediation Strategies for Efficient Electron Transfer in Hybrid Biofuel Cells. Nat. Energy 2018, 3, 574. [Google Scholar] [CrossRef]
- Mizutani, K.; Toyoda, M.; Sagara, K.; Takahashi, N.; Sato, A.; Kamitaka, Y.; Tsujimura, S.; Nakanishi, Y.; Sugiura, T.; Yamaguchi, S.; et al. X-Ray Analysis of Bilirubin Oxidase from Myrothecium Verrucaria at 2.3 Å Resolution Using a Twinned Crystal. Acta Cryst. F 2010, 66, 765–770. [Google Scholar] [CrossRef] [Green Version]
- Cracknell, J.A.; McNamara, T.P.; Lowe, E.D.; Blanford, C.F. Bilirubin Oxidase from Myrothecium Verrucaria: X-Ray Determination of the Complete Crystal Structure and a Rational Surface Modification for Enhanced Electrocatalytic O2reduction. Dalton Trans. 2011, 40, 6668–6675. [Google Scholar] [CrossRef] [PubMed]
- Kovaľ, T.; Švecová, L.; Østergaard, L.H.; Skalova, T.; Dušková, J.; Hašek, J.; Kolenko, P.; Fejfarová, K.; Stránský, J.; Trundová, M.; et al. Trp-His Covalent Adduct in Bilirubin Oxidase Is Crucial for Effective Bilirubin Binding but Has a Minor Role in Electron Transfer. Sci. Rep. 2019, 9, 13700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akter, M.; Tokiwa, T.; Shoji, M.; Nishikawa, K.; Shigeta, Y.; Sakurai, T.; Higuchi, Y.; Kataoka, K.; Shibata, N. Redox Potential-Dependent Formation of an Unusual His-Trp Bond in Bilirubin Oxidase. Chemistry 2018, 24, 18052–18058. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. XDS. Acta Crystallogr. Sect. D 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, P. Scaling and Assessment of Data Quality. Acta Crystallogr. Sect. D 2006, 62, 72–82. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A Comprehensive Python-Based System for Macromolecular Structure Solution. Acta Crystallogr. Sect. D 2010, 66, 213–221. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Cowtan, K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr. Sect. D 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [Green Version]
- Delano, W.L. The PyMOL Molecular Graphics System. 2002. Available online: http://www.pymol.org (accessed on 17 November 2021).
- Krissinel, E.; Henrick, K. Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Ashkenazy, H.; Erez, E.; Martz, E.; Pupko, T.; Ben-Tal, N. ConSurf 2010: Calculating Evolutionary Conservation in Sequence and Structure of Proteins and Nucleic Acids. Nucleic Acids Res. 2010, 38, W529–W533. [Google Scholar] [CrossRef] [Green Version]
- Mazurenko, I.; Monsalve, K.; Rouhana, J.; Parent, P.; Laffon, C.; Goff, A.L.; Szunerits, S.; Boukherroub, R.; Giudici-Orticoni, M.-T.; Mano, N.; et al. How the Intricate Interactions between Carbon Nanotubes and Two Bilirubin Oxidases Control Direct and Mediated O2 Reduction. ACS Appl. Mater. Interfaces 2016, 8, 23074–23085. [Google Scholar] [CrossRef]
- Yan, X.; Ma, S.; Tang, J.; Tanner, D.; Ulstrup, J.; Xiao, X.; Zhang, J. Direct Electron Transfer of Fructose Dehydrogenase Immobilized on Thiol-Gold Electrodes. Electrochim. Acta 2021, 392, 138946. [Google Scholar] [CrossRef]
- Cohen, R.; Cohen, Y.; Mukha, D.; Yehezkeli, O. Oxygen Insensitive Amperometric Glucose Biosensor Based on FAD Dependent Glucose Dehydrogenase Co-Entrapped with DCPIP or DCNQ in a Polydopamine Layer. Electrochim. Acta 2021, 367, 137477. [Google Scholar] [CrossRef]
- Xie, T.; Liu, Z.; Liu, Q.; Wang, G. Structural Insight into the Oxidation of Sinapic Acid by CotA Laccase. J. Struct. Biol. 2015, 190, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Komori, H.; Higuchi, Y. Structural Insights into the O2 Reduction Mechanism of Multicopper Oxidase. J. Biochem. 2015, 158, 293–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Lolage, F.A.; Bartlett, P.N.; Gounel, S.; Staigre, P.; Mano, N. Site-Directed Immobilization of Bilirubin Oxidase for Electrocatalytic Oxygen Reduction. ACS Catal. 2019, 9, 2068–2078. [Google Scholar] [CrossRef]
- Giroud, F.; Minteer, S.D. Anthracene-Modified Pyrenes Immobilized on Carbon Nanotubes for Direct Electroreduction of O2 by Laccase. Electrochem. Commun. 2013, 34, 157–160. [Google Scholar] [CrossRef]
- Hickey, D.P.; Lim, K.; Cai, R.; Patterson, A.R.; Yuan, M.; Sahin, S.; Abdellaoui, S.; Minteer, S.D. Pyrene Hydrogel for Promoting Direct Bioelectrochemistry: ATP-Independent Electroenzymatic Reduction of N2. Chem. Sci. 2018, 9, 5172–5177. [Google Scholar] [CrossRef] [Green Version]
- Blanchard, P.-Y.; Buzzetti, P.H.M.; Davies, B.; Nedellec, Y.; Girotto, E.M.; Gross, A.J.; Le Goff, A.; Nishina, Y.; Cosnier, S.; Holzinger, M. Electrosynthesis of Pyrenediones on Carbon Nanotube Electrodes for Efficient Electron Transfer with FAD-Dependent Glucose Dehydrogenase in Biofuel Cell Anodes. ChemElectroChem 2019, 6, 5242–5247. [Google Scholar] [CrossRef]
- Szczesny, J.; Marković, N.; Conzuelo, F.; Zacarias, S.; Pereira, I.A.C.; Lubitz, W.; Plumeré, N.; Schuhmann, W.; Ruff, A. A Gas Breathing Hydrogen/Air Biofuel Cell Comprising a Redox Polymer/Hydrogenase-Based Bioanode. Nat. Commun. 2018, 9, 4715. [Google Scholar] [CrossRef]
- Dombkowski, A.A.; Sultana, K.Z.; Craig, D.B. Protein Disulfide Engineering. FEBS Lett. 2014, 588, 206–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bulaj, G. Formation of Disulfide Bonds in Proteins and Peptides. Biotechnol. Adv. 2005, 23, 87–92. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gihaz, S.; Herzallh, N.S.; Cohen, Y.; Bachar, O.; Fishman, A.; Yehezkeli, O. The Structure of Bilirubin Oxidase from Bacillus pumilus Reveals a Unique Disulfide Bond for Site-Specific Direct Electron Transfer. Biosensors 2022, 12, 258. https://doi.org/10.3390/bios12050258
Gihaz S, Herzallh NS, Cohen Y, Bachar O, Fishman A, Yehezkeli O. The Structure of Bilirubin Oxidase from Bacillus pumilus Reveals a Unique Disulfide Bond for Site-Specific Direct Electron Transfer. Biosensors. 2022; 12(5):258. https://doi.org/10.3390/bios12050258
Chicago/Turabian StyleGihaz, Shalev, Nidaa Shrara Herzallh, Yifat Cohen, Oren Bachar, Ayelet Fishman, and Omer Yehezkeli. 2022. "The Structure of Bilirubin Oxidase from Bacillus pumilus Reveals a Unique Disulfide Bond for Site-Specific Direct Electron Transfer" Biosensors 12, no. 5: 258. https://doi.org/10.3390/bios12050258
APA StyleGihaz, S., Herzallh, N. S., Cohen, Y., Bachar, O., Fishman, A., & Yehezkeli, O. (2022). The Structure of Bilirubin Oxidase from Bacillus pumilus Reveals a Unique Disulfide Bond for Site-Specific Direct Electron Transfer. Biosensors, 12(5), 258. https://doi.org/10.3390/bios12050258







