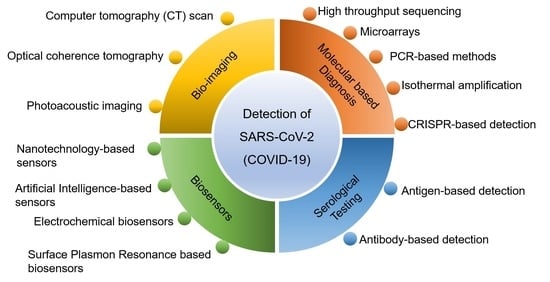
Advancements in Testing Strategies for COVID-19
Abstract
:1. Introduction
2. Detection Techniques of SARS-CoV-2
2.1. High Throughput Sequencing
2.2. Imaging
2.3. Microarrays
2.4. Molecular Assay-Based Diagnosis
2.4.1. PCR-Based Methods
Isothermal Amplification
RT-Loop-Mediated Isothermal Amplification (LAMP)
One-Pot Enzyme-Free Isothermal Amplification
RNA Auto Cycling
2.5. CRISPR Based Detection
2.6. Limitations of Molecular Diagnosis
2.7. Signal Readout Methods
2.8. Serological Testing
2.8.1. Antigen-Based Detection
2.8.2. Antibody-Based Detection
2.9. Challenges in Immunoassay-Based Diagnostics
3. Biosensors
3.1. Biosensor for Genome Based Detection
3.2. Biosensors for Surface Protein of SARS-CoV-2
3.2.1. Lateral Flow Immunoassays
3.2.2. Lateral Flow Assays
3.2.3. Microfluidic-Based Immunosensor
3.2.4. FET-Based Biosensor
3.3. Biosensors for the Detection of Antibodies
3.4. Miscellaneous Biosensor Technologies
3.4.1. Multiplexed Paper-Based Colorimetric Sensor
3.4.2. Nanotechnology-Based Sensors
3.4.3. Aptamer-Based Detection
3.4.4. Artificial Intelligence-Based Sensors
3.4.5. Electrochemical Biosensors
3.4.6. Surface Plasmon Resonance-Based Biosensor
3.4.7. Localized Surface Plasmon Resonance
3.4.8. Biosensors Designed for Alternative Target of SARS-CoV-2
4. An Overview of the Commercially Available Kits
5. Impacts of Mutants on Diagnosis
6. Summary and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peiris, J.S.M. Coronaviruses. In Medical Microbiology (Eighteenth Edition); Greenwood, D., Barer, M., Slack, R., Irving, W., Eds.; Churchill Livingstone: Edinburgh, UK, 2012; pp. 587–593. ISBN 978-0-7020-4089-4. [Google Scholar]
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.C.K.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016, 24, 490–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Wit, E.; Van Doremalen, N.; Falzarano, D.; Munster, V.J.; Singh, P.; Hanson, P.S.; Morris, C.M. SARS and MERS: Recent Insights into Emerging Coronaviruses. Nat. Rev. Microbiol. 2016, 14, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Azhar, E.I.; El-Kafrawy, S.A.; Farraj, S.A.; Hassan, A.M.; Al-Saeed, M.S.; Hashem, A.M.; Madani, T.A. Evidence for Camel-to-Human Transmission of MERS Coronavirus. N. Engl. J. Med. 2014, 370, 2499–2505. [Google Scholar] [CrossRef]
- Guan, Y.; Zheng, B.J.; He, Y.Q.; Liu, X.L.; Zhuang, Z.X.; Cheung, C.L.; Luo, S.W.; Li, P.H.; Zhang, L.J.; Guan, Y.J.; et al. Isolation and Characterization of Viruses Related to the SARS Coronavirus from Animals in Southern China. Science 2003, 302, 276–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Hoek, L.; Pyrc, K.; Jebbink, M.F.; Vermeulen-Oost, W.; Berkhout, R.J.M.; Wolthers, K.C.; Wertheim-van Dillen, P.M.E.; Kaandorp, J.; Spaargaren, J.; Berkhout, B. Identification of a New Human Coronavirus. Nat. Med. 2004, 10, 368–373. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A New Coronavirus Associated with Human Respiratory Disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [Green Version]
- Neuman, B.W.; Adair, B.D.; Yoshioka, C.; Quispe, J.D.; Orca, G.; Kuhn, P.; Milligan, R.A.; Yeager, M.; Buchmeier, M.J. Supramolecular Architecture of Severe Acute Respiratory Syndrome Coronavirus Revealed by Electron Cryomicroscopy. J. Virol. 2006, 80, 7918–7928. [Google Scholar] [CrossRef] [Green Version]
- Woo, P.C.Y.; Lau, S.K.P.; Chu, C.; Chan, K.; Tsoi, H.; Huang, Y.; Wong, B.H.L.; Poon, R.W.S.; Cai, J.J.; Luk, W.; et al. Characterization and Complete Genome Sequence of a Novel Coronavirus, Coronavirus HKU1, from Patients with Pneumonia. J. Virol. 2005, 79, 884–895. [Google Scholar] [CrossRef] [Green Version]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The Proximal Origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef] [Green Version]
- Letko, M.; Marzi, A.; Munster, V. Functional Assessment of Cell Entry and Receptor Usage for SARS-CoV-2 and Other Lineage B Betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Liu, Q.; Guo, D. Emerging Coronaviruses: Genome Structure, Replication, and Pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, J.F.-W.W.; Yuan, S.; Kok, K.-H.H.; To, K.K.-W.W.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.-Y.Y.; Poon, R.W.-S.S.; et al. A Familial Cluster of Pneumonia Associated with the 2019 Novel Coronavirus Indicating Person-to-Person Transmission: A Study of a Family Cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Schoeman, D.; Fielding, B.C. Coronavirus Envelope Protein: Current Knowledge. Virol. J. 2019, 16, 69. [Google Scholar] [CrossRef] [Green Version]
- Rota, P.A.; Oberste, M.S.; Monroe, S.S.; Nix, W.A.; Campagnoli, R.; Icenogle, J.P.; Peñaranda, S.; Bankamp, B.; Maher, K.; Chen, M.; et al. Characterization of a Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. Science 2003, 300, 1394–1399. [Google Scholar] [CrossRef] [Green Version]
- Weekly Epidemiological Update on COVID-19–21 September 2021. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---21-september-2021 (accessed on 30 September 2021).
- Barber, A.; Griffin, J.; Casey, M.; Collins, Á.B.; Lane, E.A.; Ten Bosch, Q.; De Jong, M.; Evoy, D.M.; Byrne, A.W.; McAloon, C.G.; et al. The Basic Reproduction Number of SARS-CoV-2: A Scoping Review of Available Evidence. medRxiv 2020. [Google Scholar] [CrossRef]
- Petersen, E.; Koopmans, M.; Go, U.; Hamer, D.H.; Petrosillo, N.; Castelli, F.; Storgaard, M.; Al Khalili, S.; Simonsen, L. Comparing SARS-CoV-2 with SARS-CoV and Influenza Pandemics. Lancet Infect. Dis. 2020, 20, e238–e244. [Google Scholar] [CrossRef]
- Lv, H.; Wu, N.C.; Tsang, O.T.-Y.; Yuan, M.; Perera, R.A.P.M.; Leung, W.S.; So, R.T.Y.; Chun Chan, J.M.; Yip, G.K.; Hong Chik, T.S.; et al. Cross-Reactive Antibody Response between SARS-CoV-2 and SARS-CoV Infections; Cold Spring Harbor Laboratory: New York, NY, USA, 2020; Volume 31. [Google Scholar]
- Perlman, S.; Netland, J. Coronaviruses Post-SARS: Update on Replication and Pathogenesis. Nat. Rev. Microbiol. 2009, 7, 439–450. [Google Scholar] [CrossRef] [Green Version]
- Fehr, A.R.; Perlman, S. Coronaviruses: An Overview of Their Replication and Pathogenesis. In Coronaviruses: Methods and Protocols; Maier, H.J., Bickerton, E., Britton, P., Eds.; Springer: New York, NY, USA, 2015; Volume 1282, pp. 1–23. ISBN 978-1-4939-2438-7. [Google Scholar]
- Coronavirus Incubation Period: How Long and When Most Contagious. Available online: https://www.webmd.com/lung/coronavirus-incubation-period#1 (accessed on 18 January 2022).
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Z.; Xiao, Y.; Kang, L.; Ma, W.; Shi, L.; Zhang, L.; Zhou, Z.; Yang, J.; Zhong, J.; Yang, D.; et al. Genomic Diversity of Severe Acute Respiratory Syndrome–Coronavirus 2 in Patients With Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 71, 713–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.-L.; Tseng, W.-P.; Lin, C.-H.; Lee, T.-F.; Chung, M.-Y.; Huang, C.-H.; Chen, S.-Y.; Hsueh, P.-R.; Chen, S.-C. Four Point-of-Care Lateral Flow Immunoassays for Diagnosis of COVID-19 and for Assessing Dynamics of Antibody Responses to SARS-CoV-2. J. Infect. 2020, 81, 435–442. [Google Scholar] [CrossRef] [PubMed]
- MacMullan, M.A.; Ibrayeva, A.; Trettner, K.; Deming, L.; Das, S.; Tran, F.; Moreno, J.R.; Casian, J.G.; Chellamuthu, P.; Kraft, J.; et al. ELISA Detection of SARS-CoV-2 Antibodies in Saliva. Sci. Rep. 2020, 10, 20818. [Google Scholar] [CrossRef] [PubMed]
- Mason, M.G.; Botella, J.R. Rapid (30-Second), Equipment-Free Purification of Nucleic Acids Using Easy-to-Make Dipsticks. Nat. Protoc. 2020, 15, 3663–3677. [Google Scholar] [CrossRef]
- Zhen, W.; Smith, E.; Manji, R.; Schron, D.; Berry, G.J.; McAdam, A.J. Clinical Evaluation of Three Sample-to-Answer Platforms for Detection of SARS-CoV-2. J. Clin. Microbiol. 2020, 58, e00783-20. [Google Scholar] [CrossRef] [Green Version]
- Abbott RealTime SARS-CoV-2 Assay (EUA), Abbott Molecular. Available online: https://www.molecular.abbott/us/en/products/infectious-disease/RealTime-SARS-CoV-2-Assay (accessed on 19 September 2021).
- Altona DIAGNOSTICS RealStar ® Instructions for Use RealStar ® SARS-CoV-2 RT-PCR Kit 1.0 03/2020 EN. Available online: https://altona-diagnostics.com/en/products/reagents-140/reagents/realstar-real-time-pcr-reagents/realstar-sars-cov-2-rt-pcr-kit-ruo.html (accessed on 20 January 2022).
- CareStart COVID-19 MDx RT-PCR – Access Bio. Available online: https://accessbiodiagnostics.net/covid-19-mdx-rt-pcr/ (accessed on 7 January 2022).
- SARS-CoV-2 Assay Kit | Applied BioCode. Available online: https://www.apbiocode.com/sars-cov-2.htm (accessed on 10 January 2022).
- Cepheid | Xpert® Xpress SARS-CoV-2 - FDA Emergency Use Authorization. Available online: https://www.cepheid.com/en/coronavirus (accessed on 20 May 2022).
- Respiratory Pathogen Panel Test | BioFire Diagnostics. Available online: https://www.biofiredx.com/products/the-filmarray-panels/filmarrayrp/ (accessed on 20 January 2022).
- Amaral, C.; Antunes, W.; Moe, E.; Duarte, A.G.; Lima, L.M.P.; Santos, C.; Gomes, I.L.; Afonso, G.S.; Vieira, R.; Teles, H.S.S.; et al. A Molecular Test Based on RT-LAMP for Rapid, Sensitive and Inexpensive Colorimetric Detection of SARS-CoV-2 in Clinical Samples. Sci. Rep. 2021, 11, 16430. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR–Cas12-Based Detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Yi, Y.; Luo, X.; Xiong, N.; Liu, Y.; Li, S.; Sun, R.; Wang, Y.; Hu, B.; Chen, W.; et al. Development and Clinical Application of a Rapid IgM-IgG Combined Antibody Test for SARS-CoV-2 Infection Diagnosis. J. Med. Virol. 2020, 92, 1518–1524. [Google Scholar] [CrossRef]
- Roda, A.; Cavalera, S.; Di Nardo, F.; Calabria, D.; Rosati, S.; Simoni, P.; Colitti, B.; Baggiani, C.; Roda, M.; Anfossi, L. Dual Lateral Flow Optical/Chemiluminescence Immunosensors for the Rapid Detection of Salivary and Serum IgA in Patients with COVID-19 Disease. Biosens. Bioelectron. 2021, 172, 112765. [Google Scholar] [CrossRef]
- Theel, E.S.; Harring, J.; Hilgart, H.; Granger, D.; McAdam, A.J. Performance Characteristics of Four High-Throughput Immunoassays for Detection of IgG Antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020, 58, e01243-20. [Google Scholar] [CrossRef] [PubMed]
- Human SARS-CoV-2 Spike (Trimer) IgM ELISA Kit – Invitrogen. Available online: https://www.thermofisher.com/elisa/product/Human-SARS-CoV-2-Spike-Trimer-IgM-ELISA-Kit/BMS2324 (accessed on 20 May 2022).
- Kudriashova, I.B.; Kirnos, M.D.; Vaniushin, B.F. DNA-Methylase Activities from Animal Mitochondria and Nuclei: Different Specificity of DNA Methylation. Biokhimiia 1976, 41, 1968–1977. [Google Scholar] [PubMed]
- Jing, M.; Bond, R.; Robertson, L.J.; Moore, J.; Kowalczyk, A.; Price, R.; Burns, W.; Nesbit, M.A.; McLaughlin, J.; Moore, T. User Experience Analysis of AbC-19 Rapid Test via Lateral Flow Immunoassays for Self-Administrated SARS-CoV-2 Antibody Testing. Sci. Rep. 2021, 11, 14026. [Google Scholar] [CrossRef]
- COVID-19 Total Antibody ELISA Kit. Available online: https://www.biopanda.co.uk/php/products/elisa/covid19tabelisa.php (accessed on 20 February 2022).
- SARS-CoV-2 Antigen Rapid Test - Results in 15 Minutes | Kiweno. Available online: https://kiweno.com/en/sars-cov2-antigen-test/ (accessed on 20 February 2022).
- Mertens, P.; De Vos, N.; Martiny, D.; Jassoy, C.; Mirazimi, A.; Cuypers, L.; den Wijngaert, S.; Monteil, V.; Melin, P.; Stoffels, K.; et al. Development and Potential Usefulness of the COVID-19 Ag Respi-Strip Diagnostic Assay in a Pandemic Context. Front. Med. 2020, 7, 225. [Google Scholar] [CrossRef]
- Use of SARS-CoV-2 Antigen-Detection Rapid Diagnostic Tests for COVID-19 Self-Testing. Available online: https://www.who.int/publications/i/item/WHO-2019-nCoV-Ag-RDTs-Self_testing-2022.1 (accessed on 21 February 2022).
- Rashid, Z.Z.; Othman, S.N.; Samat, M.N.A.; Ali, U.K.; Wong, K.K. Diagnostic Performance of COVID-19 Serology Assays. Malays. J. Pathol. 2020, 42, 13–21. [Google Scholar]
- Yüce, M.; Filiztekin, E.; Özkaya, K.G. COVID-19 Diagnosis—A Review of Current Methods. Biosens. Bioelectron. 2021, 172, 112752. [Google Scholar] [CrossRef]
- Edward_holmes Novel 2019 Coronavirus Genome - SARS-CoV-2 Coronavirus – Virological. Available online: https://virological.org/t/novel-2019-coronavirus-genome/319 (accessed on 20 March 2021).
- Hu, B.; Ge, X.; Wang, L.-F.; Shi, Z. Bat Origin of Human Coronaviruses. Virol. J. 2015, 12, 221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mostafa, H.H.; Fissel, J.A.; Fanelli, B.; Bergman, Y.; Gniazdowski, V.; Dadlani, M.; Carroll, K.C.; Colwell, R.R.; Simner, P.J. Metagenomic Next-Generation Sequencing of Nasopharyngeal Specimens Collected from Confirmed and Suspect COVID-19 Patients. MBio 2020, 11, e01969-20. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, L.; Deng, X.; Liang, R.; Su, M.; He, C.; Hu, L.; Su, Y.; Ren, J.; Yu, F.; et al. Recent Advances in the Detection of Respiratory Virus Infection in Humans. J. Med. Virol. 2020, 92, 408–417. [Google Scholar] [CrossRef]
- Harper, H.; Burridge, A.; Winfield, M.; Finn, A.; Davidson, A.; Matthews, D.; Hutchings, S.; Vipond, B.; Jain, N.; The COVID-19 Genomics UK (COG-UK) Consortium; et al. Detecting SARS-CoV-2 Variants with SNP Genotyping. PLoS ONE 2021, 16, e0243185. [Google Scholar] [CrossRef]
- Pan, Y.; Guan, H.; Zhou, S.; Wang, Y.; Li, Q.; Zhu, T.; Hu, Q.; Xia, L. Initial CT Findings and Temporal Changes in Patients with the Novel Coronavirus Pneumonia (2019-NCoV): A Study of 63 Patients in Wuhan, China. Eur. Radiol. 2020, 30, 3306–3309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, M.; Bernheim, A.; Mei, X.; Zhang, N.; Huang, M.; Zeng, X.; Cui, J.; Xu, W.; Yang, Y.; Fayad, Z.A.; et al. CT Imaging Features of 2019 Novel Coronavirus (2019-NCoV). Radiology 2020, 295, 202–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ooi, G.C.; Khong, P.L.; Müller, N.L.; Yiu, W.C.; Zhou, L.J.; Ho, J.C.M.; Lam, B.; Nicolaou, S.; Tsang, K.W.T. Severe Acute Respiratory Syndrome: Temporal Lung Changes at Thin-Section CT in 30 Patients. Radiology 2004, 230, 836–844. [Google Scholar] [CrossRef] [PubMed]
- Wijmans, L.; Mooij-Kalverda, K.A.; Bos, L.; Goorsenberg, A.; Smit, M.; Van Den Berk, I.; De Bruin, D.; Bonta, P.; Schultz, M.; Annema, J. Optical Coherence Tomography (OCT) in Patients with Acute Respiratory Failure on the ICU. Eur. Respir. J. 2019, 54, PA3172. [Google Scholar] [CrossRef]
- Steinberg, I.; Huland, D.M.; Vermesh, O.; Frostig, H.E.; Tummers, W.S.; Gambhir, S.S. Photoacoustic Clinical Imaging. Photoacoustics 2019, 14, 77–98. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Ma, W.; Wu, Q.; Zhang, B.; Song, Y.; Guo, Q.; Xiao, W.; Wang, Y.; Zheng, W. Design and Application of 60mer Oligonucleotide Microarray in SARS Coronavirus Detection. Chin. Sci. Bull. 2003, 48, 1165–1169. [Google Scholar] [CrossRef] [Green Version]
- de Kleber, S.L.L.; Volker, H.; Nicolas, R.; Marcus, P.; Felix, D.J.; Sabue, M.; Leo, P.; Sigrid, B.; Jan, H.B.; Laurent, K.; et al. Generic Detection of Coronaviruses and Differentiation at the Prototype Strain Level by Reverse Transcription-PCR and Nonfluorescent Low-Density Microarray. J. Clin. Microbiol. 2007, 45, 1049–1052. [Google Scholar] [CrossRef] [Green Version]
- Hardick, J.; Metzgar, D.; Risen, L.; Myers, C.; Balansay, M.; Malcom, T.; Rothman, R.; Gaydos, C. Initial Performance Evaluation of a Spotted Array Mobile Analysis Platform (MAP) for the Detection of Influenza A/B, RSV, and MERS Coronavirus. Diagn. Microbiol. Infect. Dis. 2018, 91, 245–247. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 Novel Coronavirus (2019-NCoV) by Real-Time RT-PCR. Euro Surveill. 2020, 25, 2000045. [Google Scholar] [CrossRef] [Green Version]
- Adachi, D.; Johnson, G.; Draker, R.; Ayers, M.; Mazzulli, T.; Talbot, P.J.; Tellier, R. Comprehensive Detection and Identification of Human Coronaviruses, Including the SARS-Associated Coronavirus, with a Single RT-PCR Assay. J. Virol. Methods 2004, 122, 29–36. [Google Scholar] [CrossRef]
- Setianingsih, T.Y.; Wiyatno, A.; Hartono, T.S.; Hindawati, E.; Dewantari, A.K.; Myint, K.S.; Lisdawati, V.; Safari, D. Detection of Multiple Viral Sequences in the Respiratory Tract Samples of Suspected Middle East Respiratory Syndrome Coronavirus Patients in Jakarta, Indonesia 2015–2016. Int. J. Infect. Dis. 2019, 86, 102–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corman, V.M.; Eckerle, I.; Bleicker, T.; Zaki, A.; Landt, O.; Eschbach-Bludau, M.; van Boheemen, S.; Gopal, R.; Ballhause, M.; Bestebroer, T.M.; et al. Detection of a Novel Human Coronavirus by Real-Time Reverse-Transcription Polymerase Chain Reaction. Eurosurveillance 2012, 17, 20285. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Whitaker, B.; Sakthivel, S.K.K.; Kamili, S.; Rose, L.E.; Lowe, L.; Mohareb, E.; Elassal, E.M.; Al-sanouri, T.; Haddadin, A.; et al. Real-Time Reverse Transcription-PCR Assay Panel for Middle East Respiratory Syndrome Coronavirus. J. Clin. Microbiol. 2014, 52, 67–75. [Google Scholar] [CrossRef] [Green Version]
- Bartholomew, R.A.; Hutchison, J.R.; Straub, T.M.; Call, D.R. PCR, Real-Time PCR, Digital PCR, and Isothermal Amplification. In Manual of Environmental Microbiology; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2016; pp. 2.3.2-1–2.3.2-13. ISBN 9781683670742. [Google Scholar]
- Lu, X.; Wang, L.; Sakthivel, S.K.; Whitaker, B.; Murray, J.; Kamili, S.; Lynch, B.; Malapati, L.; Burke, S.A.; Harcourt, J.; et al. US CDC Real-Time Reverse Transcription PCR Panel for Detection of Severe Acute Respiratory Syndrome Coronavirus 2 - Volume 26, Number 8—August 2020 - Emerging Infectious Diseases Journal – CDC. Available online: https://wwwnc.cdc.gov/eid/article/26/8/20-1246_article (accessed on 26 September 2021).
- Pyrc, K.; Milewska, A.; Potempa, J. Development of Loop-Mediated Isothermal Amplification Assay for Detection of Human Coronavirus-NL63. J. Virol. Methods 2011, 175, 133–136. [Google Scholar] [CrossRef] [PubMed]
- Poon, L.L.M.; Leung, C.S.W.; Tashiro, M.; Chan, K.H.; Wong, B.W.Y.; Yuen, K.Y.; Guan, Y.; Peiris, J.S.M. Rapid Detection of the Severe Acute Respiratory Syndrome (SARS) Coronavirus by a Loop-Mediated Isothermal Amplification Assay. Clin. Chem. 2004, 50, 1050–1052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, M.; Su, F.; Zhang, R.; Jiang, X.; Xiao, H.; Yan, X.; Yang, C.; Fan, X.; Wu, G. Rapid Point-of-Care Testing for SARS-CoV-2 Virus Nucleic Acid Detection by an Isothermal and Nonenzymatic Signal Amplification System Coupled with a Lateral Flow Immunoassay Strip. Sens. Actuators B Chem. 2021, 342, 129899. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Han, X.; Zheng, C. Evolution of CT Manifestations in a Patient Recovered from 2019 Novel Coronavirus (2019-NCoV) Pneumonia in Wuhan, China. Radiology 2020, 295, 20. [Google Scholar] [CrossRef]
- Park, T.J.; Lee, S.J.; Kim, D.-K.; Heo, N.S.; Park, J.Y.; Lee, S.Y. Development of Label-Free Optical Diagnosis for Sensitive Detection of Influenza Virus with Genetically Engineered Fusion Protein. Talanta 2012, 89, 246–252. [Google Scholar] [CrossRef]
- Njiru, Z.K. Loop-Mediated Isothermal Amplification Technology: Towards Point of Care Diagnostics. PLoS Negl. Trop. Dis. 2012, 6, e1572. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Dang, X.; Wang, Q.; Xu, M.; Zhao, Q.; Zhou, Y.; Zhao, H.; Wang, L.; Xu, Y.; Wang, J.; et al. Rapid Detection of SARS-CoV-2 Using Reverse Transcription RT-LAMP Method. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Mohammadniaei, M.; Yoon, J.; Choi, H.K.; Placide, V.; Bharate, B.G.; Lee, T.; Choi, J.-W. Multifunctional Nanobiohybrid Material Composed of Ag@Bi2Se3/RNA Three-Way Junction/MiRNA/Retinoic Acid for Neuroblastoma Differentiation. ACS Appl. Mater. Interfaces 2019, 11, 8779–8788. [Google Scholar] [CrossRef] [PubMed]
- Park, G.-S.; Ku, K.; Baek, S.-H.; Kim, S.-J.; Il Kim, S.; Kim, B.-T.; Maeng, J.-S. Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assays Targeting Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). J. Mol. Diagn. 2020, 22, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Shirato, K.; Semba, S.; El-Kafrawy, S.A.; Hassan, A.M.; Tolah, A.M.; Takayama, I.; Kageyama, T.; Notomi, T.; Kamitani, W.; Matsuyama, S.; et al. Development of Fluorescent Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP) Using Quenching Probes for the Detection of the Middle East Respiratory Syndrome Coronavirus. J. Virol. Methods 2018, 258, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Lai, M.; Wilson, R.; Glidle, A.; Reboud, J.; Cooper, J.M. Branched Hybridization Chain Reaction—Using Highly Dimensional DNA Nanostructures for Label-Free, Reagent-Less, Multiplexed Molecular Diagnostics. Microsystems Nanoeng. 2019, 5, 37. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Thachuk, C.; Ellington, A.D.; Winfree, E.; Soloveichik, D. Effective Design Principles for Leakless Strand Displacement Systems. Proc. Natl. Acad. Sci. USA 2018, 115, E12182–E12191. [Google Scholar] [CrossRef] [Green Version]
- Yin, P.; Choi, H.M.T.; Calvert, C.R.; Pierce, N.A. Programming Biomolecular Self-Assembly Pathways. Nature 2008, 451, 318–322. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Chen, X.; Ellington, A.D. Adapting Enzyme-Free DNA Circuits to the Detection of Loop-Mediated Isothermal Amplification Reactions. Anal. Chem. 2012, 84, 8371–8377. [Google Scholar] [CrossRef] [Green Version]
- Jiao, J.; Duan, C.; Xue, L.; Liu, Y.; Sun, W.; Xiang, Y. DNA Nanoscaffold-Based SARS-CoV-2 Detection for COVID-19 Diagnosis. Biosens. Bioelectron. 2020, 167, 112479. [Google Scholar] [CrossRef]
- Bhadra, S.; Riedel, T.E.; Lakhotia, S.; Tran, N.D.; Ellington, A.D. High-Surety Isothermal Amplification and Detection of SARS-CoV-2, Including with Crude Enzymes. bioRxiv 2020. [Google Scholar] [CrossRef]
- Do, J.Y.; Jeong, J.Y.; Hong, C.A. Catalytic Hairpin DNA Assembly-Based Chemiluminescent Assay for the Detection of Short SARS-CoV-2 Target CDNA. Talanta 2021, 233, 122505. [Google Scholar] [CrossRef]
- Christensen, U.B.; Wamberg, M.; El-Essawy, F.A.G.; Ismail, A.E.; Nielsen, C.B.; Filichev, V.V.; Jessen, C.H.; Petersen, M.; Pedersen, E.B. Intercalating Nucleic Acids: The Influence of Linker Length and Intercalator Type on Their Duplex Stabilities. Nucleosides. Nucleotides Nucleic Acids 2004, 23, 207–225. [Google Scholar] [CrossRef] [PubMed]
- Shirato, K.; Yano, T.; Senba, S.; Akachi, S.; Kobayashi, T.; Nishinaka, T.; Notomi, T.; Matsuyama, S. Detection of Middle East Respiratory Syndrome Coronavirus Using Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP). Virol. J. 2014, 11, 139. [Google Scholar] [CrossRef] [Green Version]
- Mautner, L.; Baillie, C.-K.; Herold, H.M.; Volkwein, W.; Guertler, P.; Eberle, U.; Ackermann, N.; Sing, A.; Pavlovic, M.; Goerlich, O.; et al. Rapid Point-of-Care Detection of SARS-CoV-2 Using Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP). Virol. J. 2020, 17, 160. [Google Scholar] [CrossRef] [PubMed]
- Rohaim, M.A.; Clayton, E.; Sahin, I.; Vilela, J.; Khalifa, M.E.; Al-Natour, M.Q.; Bayoumi, M.; Poirier, A.C.; Branavan, M.; Tharmakulasingam, M.; et al. Artificial Intelligence-Assisted Loop Mediated Isothermal Amplification (AI-LAMP) for Rapid Detection of SARS-CoV-2. Viruses 2020, 12, 972. [Google Scholar] [CrossRef] [PubMed]
- Mohammadniaei, M.; Zhang, M.; Ashley, J.; Christensen, U.B.; Friis-Hansen, L.J.; Gregersen, R.; Lisby, J.G.; Benfield, T.L.; Nielsen, F.E.; Henning Rasmussen, J.; et al. A Non-Enzymatic, Isothermal Strand Displacement and Amplification Assay for Rapid Detection of SARS-CoV-2 RNA. Nat. Commun. 2021, 12, 5089. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Liu, X.; Ye, S.; Liu, J.; Zheng, W.; Dong, X.; Yin, X. Colorimetric Isothermal Nucleic Acid Detection of SARS-CoV-2 with Dye Combination. Heliyon 2021, 7, e06886. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.; Xu, Y.; Zhang, X.; Ding, C.; Huang, R.; Zhang, Z.; Lv, J.; Xie, X.; Chen, Y.; Li, Y.; et al. CRISPR/Cas9-Mediated Gene Editing in Human Tripronuclear Zygotes. Protein Cell 2015, 6, 363–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, X.; Liu, Y.; Chemparathy, A.; Pande, T.; La Russa, M.; Qi, L.S. A Comprehensive Analysis and Resource to Use CRISPR-Cas13 for Broad-Spectrum Targeting of RNA Viruses. Cell Reports Med. 2021, 2, 100245. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Singh, J.; Streithorst, J.; Granados, A.; Sotomayor-Gonzalez, A.; Zorn, K.; Gopez, A.; et al. Rapid Detection of 2019 Novel Coronavirus SARS-CoV-2 Using a CRISPR-Based DETECTR Lateral Flow Assay. medRxiv Prepr. Serv. Health Sci. 2020. [Google Scholar] [CrossRef]
- Ding, X.; Yin, K.; Li, Z.; Liu, C. All-in-One Dual CRISPR-Cas12a (AIOD-CRISPR) Assay: A Case for Rapid, Ultrasensitive and Visual Detection of Novel Coronavirus SARS-CoV-2 and HIV Virus. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kushawah, G.; Hernandez-Huertas, L.; Abugattas-Nuñez del Prado, J.; Martinez-Morales, J.R.; DeVore, M.L.; Hassan, H.; Moreno-Sanchez, I.; Tomas-Gallardo, L.; Diaz-Moscoso, A.; Monges, D.E.; et al. CRISPR-Cas13d Induces Efficient MRNA Knockdown in Animal Embryos. Dev. Cell 2020, 54, 805–817.e7. [Google Scholar] [CrossRef]
- Rauch, J.N.; Valois, E.; Solley, S.C.; Braig, F.; Lach, R.S.; Audouard, M.; Ponce-Rojas, J.C.; Costello, M.S.; Baxter, N.J.; Kosik, K.S.; et al. A Scalable, Easy-to-Deploy, Protocol for Cas13-Based Detection of SARS-CoV-2 Genetic Material. bioRxiv 2020, 59, e02402-20. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, M.I.; Makhawi, A.M.; Kraft, C.S. SHERLOCK and DETECTR: CRISPR-Cas Systems as Potential Rapid Diagnostic Tools for Emerging Infectious Diseases. J. Clin. Microbiol. 2021, 59, e00745-20. [Google Scholar] [CrossRef] [PubMed]
- Santiago-Frangos, A.; Hall, L.N.; Nemudraia, A.; Nemudryi, A.; Krishna, P.; Wiegand, T.; Wilkinson, R.A.; Snyder, D.T.; Hedges, J.F.; Cicha, C.; et al. Intrinsic Signal Amplification by Type III CRISPR-Cas Systems Provides a Sequence-Specific SARS-CoV-2 Diagnostic. Cell Rep. Med. 2021, 2, 100319. [Google Scholar] [CrossRef] [PubMed]
- Fozouni, P.; Son, S.; Díaz de León Derby, M.; Knott, G.J.; Gray, C.N.; D’Ambrosio, M.V.; Zhao, C.; Switz, N.A.; Kumar, G.R.; Stephens, S.I.; et al. Amplification-Free Detection of SARS-CoV-2 with CRISPR-Cas13a and Mobile Phone Microscopy. Cell 2021, 184, 323–333.e9. [Google Scholar] [CrossRef]
- Guo, X.; Guo, Z.; Duan, C.; Chen, Z.; Wang, G.; Lu, Y.; Li, M.; Lu, J. Long-Term Persistence of IgG Antibodies in SARS-CoV Infected Healthcare Workers. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef] [Green Version]
- Green, D.A.; Zucker, J.; Westblade, L.F.; Whittier, S.; Rennert, H.; Velu, P.; Craney, A.; Cushing, M.; Liu, D.; Sobieszczyk, M.E.; et al. Clinical Performance of SARS-CoV-2 Molecular Tests. J. Clin. Microbiol. 2020, 58, e00995-20. [Google Scholar] [CrossRef]
- Tahamtan, A.; Ardebili, A. Real-Time RT-PCR in COVID-19 Detection: Issues Affecting the Results. Expert Rev. Mol. Diagn. 2020, 20, 453–454. [Google Scholar] [CrossRef] [Green Version]
- Loeffelholz, M.J.; Pong, D.L.; Pyles, R.B.; Xiong, Y.; Miller, A.L.; Bufton, K.K.; Chonmaitree, T. Comparison of the FilmArray Respiratory Panel and Prodesse Real-Time PCR Assays for Detection of Respiratory Pathogens. J. Clin. Microbiol. 2011, 49, 4083–4088. [Google Scholar] [CrossRef] [Green Version]
- CRISPR-Based Portable COVID Tests. Nat. Biotechnol. 2021, 39, 1031. [CrossRef] [PubMed]
- Chandrasekaran, A.R. DNA Nanobiosensors: An Outlook on Signal Readout Strategies. J. Nanomater. 2017, 2017, 1–9. [Google Scholar] [CrossRef]
- Rajendran, V.K.; Bakthavathsalam, P.; Bergquist, P.L.; Sunna, A. A Portable Nucleic Acid Detection System Using Natural Convection Combined with a Smartphone. Biosens. Bioelectron. 2019, 134, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Paradiso, A.V.; De Summa, S.; Loconsole, D.; Procacci, V.; Sallustio, A.; Centrone, F.; Silvestris, N.; Cafagna, V.; De Palma, G.; Tufaro, A.; et al. Rapid Serological Assays and SARS-CoV-2 Real-Time Polymerase Chain Reaction Assays for the Detection of SARS-CoV-2: Comparative Study. J. Med. Internet. Res. 2020, 22, e19152. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Tan, Z.; Zhao, K.; Zou, W.; Wang, H.; Gao, H.; Sun, S.; Bu, D.; Chai, W.; Li, Y. The Effect of N-Glycosylation of SARS-CoV-2 Spike Protein on the Virus Interaction with the Host Cell ACE2 Receptor. iScience 2021, 24, 103272. [Google Scholar] [CrossRef] [PubMed]
- Robson, B. COVID-19 Coronavirus Spike Protein Analysis for Synthetic Vaccines, a Peptidomimetic Antagonist, and Therapeutic Drugs, and Analysis of a Proposed Achilles’ Heel Conserved Region to Minimize Probability of Escape Mutations and Drug Resistance. Comput. Biol. Med. 2020, 121, 103749. [Google Scholar] [CrossRef] [PubMed]
- Jalkanen, P.; Pasternack, A.; Maljanen, S.; Melén, K.; Kolehmainen, P.; Huttunen, M.; Lundberg, R.; Tripathi, L.; Khan, H.; Ritvos, M.A.; et al. A Combination of N and S Antigens With IgA and IgG Measurement Strengthens the Accuracy of SARS-CoV-2 Serodiagnostics. J. Infect. Dis. 2021, 224, 218–228. [Google Scholar] [CrossRef]
- Singh, P.; Chakraborty, R.; Marwal, R.; Radhakrishan, V.S.; Bhaskar, A.K.; Vashisht, H.; Dhar, M.S.; Pradhan, S.; Ranjan, G.; Imran, M.; et al. A Rapid and Sensitive Method to Detect SARS-CoV-2 Virus Using Targeted-Mass Spectrometry. J. Proteins Proteomics 2020, 11, 159–165. [Google Scholar] [CrossRef]
- Dogan, M.; Kozhaya, L.; Placek, L.; Gunter, C.; Yigit, M.; Hardy, R.; Plassmeyer, M.; Coatney, P.; Lillard, K.; Bukhari, Z.; et al. SARS-CoV-2 Specific Antibody and Neutralization Assays Reveal the Wide Range of the Humoral Immune Response to Virus. Commun. Biol. 2021, 4, 129. [Google Scholar] [CrossRef]
- Frank, S.A. Specificity and Cross-Reactivity. In Immunology and Evolution of Infectious Disease; Princeton University Press: Princeton, NJ, USA, 2002. [Google Scholar]
- Zhou, Y.; Chen, Y.; Liu, W.; Fang, H.; Li, X.; Hou, L.; Liu, Y.; Lai, W.; Huang, X.; Xiong, Y. Development of a Rapid and Sensitive Quantum Dot Nanobead-Based Double-Antigen Sandwich Lateral Flow Immunoassay and Its Clinical Performance for the Detection of SARS-CoV-2 Total Antibodies. Sens. Actuators B Chem. 2021, 343, 130139. [Google Scholar] [CrossRef]
- Jung, C.; Levy, C.; Varon, E.; Biscardi, S.; Batard, C.; Wollner, A.; Deberdt, P.; Sellam, A.; Hau, I.; Cohen, R. Diagnostic Accuracy of SARS-CoV-2 Antigen Detection Test in Children: A Real-Life Study. Front. Pediatr. 2021, 9, 727. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.P.; Che, X.-Y.; Yuen, K.-Y.; Wong, B.H.L.; Cheng, V.C.C.; Woo, G.K.S.; Hung, I.F.N.; Poon, R.W.S.; Chan, K.-H.; Peiris, J.S.M.; et al. SARS Coronavirus Detection Methods. Emerg. Infect. Dis. J. 2005, 11, 1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, L.; Xie, X.; Gong, Q.; Feng, R.; Guo, X.; Su, B.; Chen, L. Transcriptional Difference between SARS-COV-2 and Other Human Coronaviruses Revealed by Sub-Genomic RNA Profiling. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Cheng, S.; Chen, R.; Ke, J.; Liu, Y.; Li, Y.; Feng, W.; Li, F.; Liu, C.; Geva, E.; et al. An Isothermal Amplification Reactor with an Integrated Isolation Membrane for Point-of-Care Detection of Infectious Diseases. Analyst 2020, 12, 2069–2076. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Cheng, S.; Chen, R.; Ke, J.; Liu, Y.; Li, Y.; Feng, W.; Li, F. Near-Infrared Lanthanide-Doped Nanoparticles for a Low Interference Lateral Flow Immunoassay Test. ACS Appl. Mater. Interfaces 2020, 12, 4358–4365. [Google Scholar] [CrossRef]
- Charles, A.; Janeway, J.; Travers, P.; Walport, M.; Shlomchik, M.J. The Distribution and Functions of Immunoglobulin Isotypes. In Immunobiology: The Immune System in Health and Disease; Garland Science: New York, NY, USA, 2001. [Google Scholar]
- Long, Q.-X.; Liu, B.-Z.; Deng, H.-J.; Wu, G.-C.; Deng, K.; Chen, Y.-K.; Liao, P.; Qiu, J.-F.; Lin, Y.; Cai, X.-F.; et al. Antibody Responses to SARS-CoV-2 in Patients with COVID-19. Nat. Med. 2020, 26, 845–848. [Google Scholar] [CrossRef]
- To, K.K.-W.; Tsang, O.T.-Y.; Leung, W.-S.; Tam, A.R.; Wu, T.-C.; Lung, D.C.; Yip, C.C.-Y.; Cai, J.-P.; Chan, J.M.-C.; Chik, T.S.-H.; et al. Temporal Profiles of Viral Load in Posterior Oropharyngeal Saliva Samples and Serum Antibody Responses during Infection by SARS-CoV-2: An Observational Cohort Study. Lancet Infect. Dis. 2020, 20, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to Biosensors. Essays Biochem. 2016, 60, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Land, K.J.; Boeras, D.I.; Chen, X.-S.; Ramsay, A.R.; Peeling, R.W. REASSURED Diagnostics to Inform Disease Control Strategies, Strengthen Health Systems and Improve Patient Outcomes. Nat. Microbiol. 2019, 4, 46–54. [Google Scholar] [CrossRef]
- Tripathy, S.; Singh, S.G. Label-Free Electrochemical Detection of DNA Hybridization: A Method for COVID-19 Diagnosis. Trans. Indian Natl. Acad. Eng. 2020, 5, 205–209. [Google Scholar] [CrossRef]
- Qiu, G.; Gai, Z.; Tao, Y.; Schmitt, J.; Kullak-Ublick, G.A.; Wang, J. Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano 2020, 14, 5268–5277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moitra, P.; Alafeef, M.; Dighe, K.; Frieman, M.B.; Pan, D. Selective Naked-Eye Detection of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles. ACS Nano 2020, 14, 7617–7627. [Google Scholar] [CrossRef] [PubMed]
- Mavrikou, S.; Moschopoulou, G.; Tsekouras, V.; Kintzios, S. Development of a Portable, Ultra-Rapid and Ultra-Sensitive Cell-Based Biosensor for the Direct Detection of the SARS-CoV-2 S1 Spike Protein Antigen. Sensors 2020, 20, 3121. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Ding, L.; Zhou, J.; Chen, S.; Chen, F.; Zhao, C.; Xu, J.; Hu, W.; Ji, J.; Xu, H.; et al. One-Step Rapid Quantification of SARS-CoV-2 Virus Particles via Low-Cost Nanoplasmonic Sensors in Generic Microplate Reader and Point-of-Care Device. Biosens. Bioelectron. 2021, 171, 112685. [Google Scholar] [CrossRef]
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.-H.; Choi, M.; Ku, K.B.; Lee, C.-S.; Jun, S.; Park, D.; Kim, H.G.; et al. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; Ke, X.; Gu, Y.; Liu, H.; Sun, X. Whole Genome Identification of Potential G-Quadruplexes and Analysis of the G-Quadruplex Binding Domain for SARS-CoV-2. Front. Genet. 2020, 1430. [Google Scholar] [CrossRef]
- Parisi, O.I.; Dattilo, M.; Patitucci, F.; Malivindi, R.; Pezzi, V.; Perrotta, I.; Ruffo, M.; Amone, F.; Puoci, F. ‘Monoclonal-Type’ Plastic Antibodies for SARS-CoV-2 Based on Molecularly Imprinted Polymers. bioRxiv 2020. [Google Scholar] [CrossRef]
- Mahari, S.; Roberts, A.; Shahdeo, D.; Gandhi, S. ECovSens-Ultrasensitive Novel In-House Built Printed Circuit Board Based Electrochemical Device for Rapid Detection of NCovid-19 Antigen, a Spike Protein Domain 1 of SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Vadlamani, B.S.; Uppal, T.; Verma, S.C.; Misra, M. Functionalized TiO2 Nanotube-Based Electrochemical Biosensor for Rapid Detection of SARS-CoV-2. Sensors 2020, 20, 5871. [Google Scholar] [CrossRef]
- Lee, J.-H.; Choi, M.; Jung, Y.; Lee, S.K.; Lee, C.-S.; Kim, J.; Kim, J.; Kim, N.H.; Kim, B.-T.; Kim, H.G. A Novel Rapid Detection for SARS-CoV-2 Spike 1 Antigens Using Human Angiotensin Converting Enzyme 2 (ACE2). Biosens. Bioelectron. 2021, 171, 112715. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, Z.; Zhai, X.; Li, Y.; Lin, L.; Zhao, H.; Bian, L.; Li, P.; Yu, L.; Wu, Y.; et al. Rapid and Sensitive Detection of Anti-SARS-CoV-2 IgG, Using Lanthanide-Doped Nanoparticles-Based Lateral Flow Immunoassay. Anal. Chem. 2020, 92, 7226–7231. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Li, Y.; Liu, J.; Guo, L.; Wang, Z.; Xu, X.; Song, S.; Hao, C.; Liu, L.; Xin, M.; et al. Rapid, Ultrasensitive and Highly Specific Biosensor for the Diagnosis of SARS-CoV-2 in Clinical Blood Samples. Mater. Chem. Front. 2020, 4, 2000–2005. [Google Scholar] [CrossRef]
- Cady, N.C.; Tokranova, N.; Minor, A.; Nikvand, N.; Strle, K.; Lee, W.T.; Page, W.; Guignon, E.; Pilar, A.; Gibson, G.N. Multiplexed Detection and Quantification of Human Antibody Response to COVID-19 Infection Using a Plasmon Enhanced Biosensor Platform. Biosens. Bioelectron. 2021, 171, 112679. [Google Scholar] [CrossRef] [PubMed]
- Funari, R.; Chu, K.-Y.; Shen, A.Q. Detection of Antibodies against SARS-CoV-2 Spike Protein by Gold Nanospikes in an Opto-Microfluidic Chip. Biosens. Bioelectron. 2020, 169, 112578. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Manzano, J.; Malpartida-Cardenas, K.; Moser, N.; Pennisi, I.; Cavuto, M.; Miglietta, L.; Moniri, A.; Penn, R.; Satta, G.; Randell, P.; et al. Handheld Point-of-Care System for Rapid Detection of SARS-CoV-2 Extracted RNA in under 20 Min. ACS Cent. Sci. 2021, 7, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Abbas, N.; Shin, S. A Rapid Diagnosis of SARS-CoV-2 Using DNA Hydrogel Formation on Microfluidic Pores. Biosens. Bioelectron. 2021, 177, 113005. [Google Scholar] [CrossRef]
- Ramachandran, A.; Huyke, D.A.; Sharma, E.; Sahoo, M.K.; Huang, C.; Banaei, N.; Pinsky, B.A.; Santiago, J.G. Electric Field-Driven Microfluidics for Rapid CRISPR-Based Diagnostics and Its Application to Detection of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 29518–29525. [Google Scholar] [CrossRef]
- Wang, D.; He, S.; Wang, X.; Yan, Y.; Liu, J.; Wu, S.; Liu, S.; Lei, Y.; Chen, M.; Li, L.; et al. Rapid Lateral Flow Immunoassay for the Fluorescence Detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1150–1158. [Google Scholar] [CrossRef]
- Huang, X.; Aguilar, Z.P.; Xu, H.; Lai, W.; Xiong, Y. Membrane-Based Lateral Flow Immunochromatographic Strip with Nanoparticles as Reporters for Detection: A Review. Biosens. Bioelectron. 2016, 75, 166–180. [Google Scholar] [CrossRef]
- Wang, X.; Wu, X.; Lu, Z.; Tao, X. Comparative Study of Time-Resolved Fluorescent Nanobeads, Quantum Dot Nanobeads and Quantum Dots as Labels in Fluorescence Immunochromatography for Detection of Aflatoxin B1 in Grains. Biomolecules 2020, 10, 575. [Google Scholar] [CrossRef] [Green Version]
- Vaitukaitis, J.L.; Braunstein, G.D.; Ross, G.T. A Radioimmunoassay Which Specifically Measures Human Chorionic Gonadotropin in the Presence of Human Luteinizing Hormone. Am. J. Obstet. Gynecol. 1972, 113, 751–758. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, X.; Han, L.; Chen, T.; Wang, L.; Li, H.; Li, S.; He, L.; Fu, X.; Chen, S.; et al. Multiplex Reverse Transcription Loop-Mediated Isothermal Amplification Combined with Nanoparticle-Based Lateral Flow Biosensor for the Diagnosis of COVID-19. Biosens. Bioelectron. 2020, 166, 112437. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Cui, J.; Huang, L.; Du, B.; Chen, L.; Xue, G.; Li, S.; Zhang, W.; Zhao, L.; Sun, Y.; et al. Rapid and Visual Detection of 2019 Novel Coronavirus (SARS-CoV-2) by a Reverse Transcription Loop-Mediated Isothermal Amplification Assay. Clin. Microbiol. Infect. 2020, 26, 773–779. [Google Scholar] [CrossRef]
- Xiong, E.; Jiang, L.; Tian, T.; Hu, M.; Yue, H.; Huang, M.; Lin, W.; Jiang, Y.; Zhu, D.; Zhou, X. Simultaneous Dual-Gene Diagnosis of SARS-CoV-2 Based on CRISPR/Cas9-Mediated Lateral Flow Assay. Angew. Chem. Int. Ed. 2021, 60, 5307–5315. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zheng, T.; Wang, H.; Chen, W.; Huang, X.; Liang, J.; Qiu, L.; Han, D.; Tan, W. Rapid One-Pot Detection of SARS-CoV-2 Based on a Lateral Flow Assay in Clinical Samples. Anal. Chem. 2021, 93, 3325–3330. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Boswell, S.A.; Chidley, C.; Lu, Z.; Pettit, M.E.; Gaudio, B.L.; Fajnzylber, J.M.; Ingram, R.T.; Ward, R.H.; Li, J.Z.; et al. An Enhanced Isothermal Amplification Assay for Viral Detection. Nat. Commun. 2020, 11, 5920. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Sui, Z.; Huang, Z.; Xie, J.; Wen, K.; Zhang, Y.; Huang, W.; Mi, W.; Peng, K.; Dai, X.; et al. Point-of-Care Test System for Detection of Immunoglobulin-G and -M against Nucleocapsid Protein and Spike Glycoprotein of SARS-CoV-2. Sens. Actuators B Chem. 2021, 331, 129415. [Google Scholar] [CrossRef] [PubMed]
- Bayin, Q.; Huang, L.; Ren, C.; Fu, Y.; Ma, X.; Guo, J. Anti-SARS-CoV-2 IgG and IgM Detection with a GMR Based LFIA System. Talanta 2021, 227, 122207. [Google Scholar] [CrossRef]
- Grant, B.D.; Anderson, C.E.; Williford, J.R.; Alonzo, L.F.; Glukhova, V.A.; Boyle, D.S.; Weigl, B.H.; Nichols, K.P. SARS-CoV-2 Coronavirus Nucleocapsid Antigen-Detecting Half-Strip Lateral Flow Assay Toward the Development of Point of Care Tests Using Commercially Available Reagents. Anal. Chem. 2020, 92, 11305–11309. [Google Scholar] [CrossRef]
- Acquah, C.; Jeevanandam, J.; Tan, K.X.; Danquah, M.K. Engineered Aptamers for Enhanced COVID-19 Theranostics. Cell. Mol. Bioeng. 2021, 14, 209–221. [Google Scholar] [CrossRef]
- Zhang, L.; Fang, X.; Liu, X.; Ou, H.; Zhang, H.; Wang, J.; Li, Q.; Cheng, H.; Zhang, W.; Luo, Z. Discovery of Sandwich Type COVID-19 Nucleocapsid Protein DNA Aptamers. Chem. Commun. 2020, 56, 10235–10238. [Google Scholar] [CrossRef] [PubMed]
- Devi, A.; Chaitanya, N.S.N. Designing of Peptide Aptamer Targeting the Receptor-Binding Domain of Spike Protein of SARS-CoV-2: An in Silico Study. Mol. Divers. 2022, 26, 157–169. [Google Scholar] [CrossRef]
- Li, J.; Lillehoj, P.B. Microfluidic Magneto Immunosensor for Rapid, High Sensitivity Measurements of SARS-CoV-2 Nucleocapsid Protein in Serum. ACS Sensors 2021, 6, 1270–1278. [Google Scholar] [CrossRef] [PubMed]
- Nehra, A.; Pal Singh, K. Current Trends in Nanomaterial Embedded Field Effect Transistor-Based Biosensor. Biosens. Bioelectron. 2015, 74, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Janissen, R.; Sahoo, P.K.; Santos, C.A.; da Silva, A.M.; von Zuben, A.A.G.; Souto, D.E.P.; Costa, A.D.T.; Celedon, P.; Zanchin, N.I.T.; Almeida, D.B.; et al. InP Nanowire Biosensor with Tailored Biofunctionalization: Ultrasensitive and Highly Selective Disease Biomarker Detection. Nano Lett. 2017, 17, 5938–5949. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Ren, R.; Pu, H.; Guo, X.; Chang, J.; Zhou, G.; Mao, S.; Kron, M.; Chen, J. Field-Effect Transistor Biosensor for Rapid Detection of Ebola Antigen. Sci. Rep. 2017, 7, 10974. [Google Scholar] [CrossRef] [Green Version]
- Geim, A.K.; Novoselov, K.S. The Rise of Graphene. In Nanoscience and Technology; Co-Published with Macmillan Publishers Ltd.: London, UK, 2009; pp. 11–19. ISBN 978-981-4282-68-0. [Google Scholar]
- Lin, Q.; Wen, D.; Wu, J.; Liu, L.; Wu, W.; Fang, X.; Kong, J. Microfluidic Immunoassays for Sensitive and Simultaneous Detection of IgG/IgM/Antigen of SARS-CoV-2 within 15 Min. Anal. Chem. 2020, 92, 9454–9458. [Google Scholar] [CrossRef]
- Laksanasopin, T.; Guo, T.W.; Nayak, S.; Sridhara, A.A.; Xie, S.; Olowookere, O.O.; Cadinu, P.; Meng, F.; Chee, N.H.; Kim, J.; et al. A Smartphone Dongle for Diagnosis of Infectious Diseases at the Point of Care. Sci. Transl. Med. 2015, 7, 273re1. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Cordero, J.L.; Maerkl, S.J. Mechanically Induced Trapping of Molecular Interactions and Its Applications. J. Lab. Autom. 2015, 21, 356–367. [Google Scholar] [CrossRef] [Green Version]
- Swank, Z.; Michielin, G.; Yip, H.M.; Cohen, P.; Andrey, D.O.; Vuilleumier, N.; Kaiser, L.; Eckerle, I.; Meyer, B.; Maerkl, S.J. A High-Throughput Microfluidic Nanoimmunoassay for Detecting Anti–SARS-CoV-2 Antibodies in Serum or Ultralow-Volume Blood Samples. Proc. Natl. Acad. Sci. USA 2021, 118, e2025289118. [Google Scholar] [CrossRef]
- Mettakoonpitak, J.; Boehle, K.; Nantaphol, S.; Teengam, P.; Adkins, J.A.; Srisa-Art, M.; Henry, C.S. Electrochemistry on Paper-Based Analytical Devices: A Review. Electroanalysis 2016, 28, 1420–1436. [Google Scholar] [CrossRef]
- Yetisen, A.K.; Akram, M.S.; Lowe, C.R. Paper-Based Microfluidic Point-of-Care Diagnostic Devices. Lab Chip 2013, 13, 2210–2251. [Google Scholar] [CrossRef] [PubMed]
- Teengam, P.; Siangproh, W.; Tuantranont, A.; Vilaivan, T.; Chailapakul, O.; Henry, C.S. Multiplex Paper-Based Colorimetric DNA Sensor Using Pyrrolidinyl Peptide Nucleic Acid-Induced AgNPs Aggregation for Detecting MERS-CoV, MTB, and HPV Oligonucleotides. Anal. Chem. 2017, 89, 5428–5435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, G.-R.; Ki, H.; Kim, M.-G. Automated, Universal, and Mass-Producible Paper-Based Lateral Flow Biosensing Platform for High-Performance Point-of-Care Testing. ACS Appl. Mater. Interfaces 2020, 12, 1885–1894. [Google Scholar] [CrossRef]
- Nguyen, V.-T.; Song, S.; Park, S.; Joo, C. Recent Advances in High-Sensitivity Detection Methods for Paper-Based Lateral-Flow Assay. Biosens. Bioelectron. 2020, 152, 112015. [Google Scholar] [CrossRef]
- Pilavaki, E.; Demosthenous, A. Optimized Lateral Flow Immunoassay Reader for the Detection of Infectious Diseases in Developing Countries. Sensors 2017, 17, 2673. [Google Scholar] [CrossRef] [Green Version]
- Tabish, T.A.; Hamblin, M.R. Multivalent Nanomedicines to Treat COVID-19: A Slow Train Coming. Nano Today 2020, 35, 100962. [Google Scholar] [CrossRef]
- Farooq, T.; Adeel, M.; He, Z.; Umar, M.; Shakoor, N.; da Silva, W.; Elmer, W.; White, J.C.; Rui, Y. Nanotechnology and Plant Viruses: An Emerging Disease Management Approach for Resistant Pathogens. ACS Nano 2021, 15, 6030–6037. [Google Scholar] [CrossRef]
- Amin, M.T.; Alazba, A.A.; Manzoor, U. A Review of Removal of Pollutants from Water/Wastewater Using Different Types of Nanomaterials. Adv. Mater. Sci. Eng. 2014, 2014, 825910. [Google Scholar] [CrossRef] [Green Version]
- Xiang, J.; Yan, M.; Li, H.; Liu, T.; Lin, C.; Huang, S.; Shen, C. Evaluation of Enzyme-Linked Immunoassay and Colloidal Gold-Immunochromatographic Assay Kit for Detection of Novel Coronavirus (SARS-Cov-2) Causing an Outbreak of Pneumonia (COVID-19). medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Zodrow, K.; Brunet, L.; Mahendra, S.; Li, D.; Zhang, A.; Li, Q.; Alvarez, P.J.J. Polysulfone Ultrafiltration Membranes Impregnated with Silver Nanoparticles Show Improved Biofouling Resistance and Virus Removal. Water Res. 2009, 43, 715–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Huang, M.; Liu, Y.; Meng, L.; Ma, M. Functionalized Electrospun Nanofiber Membranes for Water Treatment: A Review. Sci. Total Environ. 2020, 739, 139944. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhou, Q.; Li, S.; Yan, H.; Chang, B.; Wang, Y.; Dong, S. Rapid and Visual Detection of SARS-CoV-2 Using Multiplex Reverse Transcription Loop-Mediated Isothermal Amplification Linked With Gold Nanoparticle-Based Lateral Flow Biosensor. Front. Cell. Infect. Microbiol. 2021, 11, 656. [Google Scholar] [CrossRef] [PubMed]
- Karakuş, E.; Erdemir, E.; Demirbilek, N.; Liv, L. Colorimetric and Electrochemical Detection of SARS-CoV-2 Spike Antigen with a Gold Nanoparticle-Based Biosensor. Anal. Chim. Acta 2021, 1182, 338939. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Wang, C.; Yang, X.; Zheng, S.; Cheng, X.; Liu, Z.; Zhao, B.; Xiao, R. Rapid Field Determination of SARS-CoV-2 by a Colorimetric and Fluorescent Dual-Functional Lateral Flow Immunoassay Biosensor. Sens. Actuators B Chem. 2022, 351, 130897. [Google Scholar] [CrossRef]
- Wandtke, T.; Woźniak, J.; Kopiński, P. Aptamers in Diagnostics and Treatment of Viral Infections. Viruses 2015, 7, 751–780. [Google Scholar] [CrossRef] [Green Version]
- Ahn, D.-G.; Jeon, I.-J.; Kim, J.D.; Song, M.-S.; Han, S.-R.; Lee, S.-W.; Jung, H.; Oh, J.-W. RNA Aptamer-Based Sensitive Detection of SARS Coronavirus Nucleocapsid Protein. Analyst 2009, 134, 1896–1901. [Google Scholar] [CrossRef]
- Shu, Y.; Shu, D.; Haque, F.; Guo, P. Fabrication of PRNA Nanoparticles to Deliver Therapeutic RNAs and Bioactive Compounds into Tumor Cells. Nat. Protoc. 2013, 8, 1635–1659. [Google Scholar] [CrossRef] [Green Version]
- Dey, L.; Chakraborty, S.; Mukhopadhyay, A. Machine Learning Techniques for Sequence-Based Prediction of Viral–Host Interactions between SARS-CoV-2 and Human Proteins. Biomed. J. 2020, 43, 438–450. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, H.; Ye, T.; Juhas, M. Deep Learning for Imaging and Detection of Microorganisms. Trends Microbiol. 2021, 29, 569–572. [Google Scholar] [CrossRef]
- Layqah, L.A.; Eissa, S. An Electrochemical Immunosensor for the Corona Virus Associated with the Middle East Respiratory Syndrome Using an Array of Gold Nanoparticle-Modified Carbon Electrodes. Microchim. Acta 2019, 186, 224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, B.E.; Ong, S.W.X.; Kalimuddin, S.; Low, J.G.; Tan, S.Y.; Loh, J.; Ng, O.-T.; Marimuthu, K.; Ang, L.W.; Mak, T.M.; et al. Epidemiologic Features and Clinical Course of Patients Infected With SARS-CoV-2 in Singapore. JAMA 2020, 323, 1488–1494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, H.; Shin, J.; Sim, J.; Cho, H.; Hong, S. Reusable Surface Plasmon Resonance Biosensor Chip for the Detection of H1N1 Influenza Virus. Biosens. Bioelectron. 2020, 168, 112561. [Google Scholar] [CrossRef] [PubMed]
- Estmer Nilsson, C.; Abbas, S.; Bennemo, M.; Larsson, A.; Hämäläinen, M.D.; Frostell-Karlsson, Å. A Novel Assay for Influenza Virus Quantification Using Surface Plasmon Resonance. Vaccine 2010, 28, 759–766. [Google Scholar] [CrossRef] [PubMed]
- Garcia, B.H.; Goodman, R.M. Use of Surface Plasmon Resonance Imaging to Study Viral RNA:Protein Interactions. J. Virol. Methods 2008, 147, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Park, T.J.; Hyun, M.S.; Lee, H.J.; Lee, S.Y.; Ko, S. A Self-Assembled Fusion Protein-Based Surface Plasmon Resonance Biosensor for Rapid Diagnosis of Severe Acute Respiratory Syndrome. Talanta 2009, 79, 295–301. [Google Scholar] [CrossRef]
- Zhang, H.; Harpster, M.H.; Wilson, W.C.; Johnson, P.A. Surface-Enhanced Raman Scattering Detection of DNAs Derived from Virus Genomes Using Au-Coated Paramagnetic Nanoparticles. Langmuir 2012, 28, 4030–4037. [Google Scholar] [CrossRef]
- Singh, R.; Thakur, P.; Thakur, A.; Kumar, H.; Chawla, P.; Rohit, J.V.; Kaushik, R.; Kumar, N. Colorimetric Sensing Approaches of Surface-Modified Gold and Silver Nanoparticles for Detection of Residual Pesticides: A Review. Int. J. Environ. Anal. Chem. 2021, 101, 3006–3022. [Google Scholar] [CrossRef]
- Cao, J.; Galbraith, E.K.; Sun, T.; Grattan, K.T.V. Comparison of Surface Plasmon Resonance and Localized Surface Plasmon Resonance-Based Optical Fibre Sensors. J. Phys. Conf. Ser. 2011, 307, 12050. [Google Scholar] [CrossRef]
- Takemura, K.; Adegoke, O.; Takahashi, N.; Kato, T.; Li, T.-C.; Kitamoto, N.; Tanaka, T.; Suzuki, T.; Park, E.Y. Versatility of a Localized Surface Plasmon Resonance-Based Gold Nanoparticle-Alloyed Quantum Dot Nanobiosensor for Immunofluorescence Detection of Viruses. Biosens. Bioelectron. 2017, 89, 998–1005. [Google Scholar] [CrossRef]
- Inci, F.; Tokel, O.; Wang, S.; Gurkan, U.A.; Tasoglu, S.; Kuritzkes, D.R.; Demirci, U. Nanoplasmonic Quantitative Detection of Intact Viruses from Unprocessed Whole Blood. ACS Nano 2013, 7, 4733–4745. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Rong, Z.; Wang, J.; Xiao, R.; Wang, S. A Fluorescent Aptasensor for H5N1 Influenza Virus Detection Based-on the Core–Shell Nanoparticles Metal-Enhanced Fluorescence (MEF). Biosens. Bioelectron. 2015, 66, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Jiang, Y.-Z.; Wu, L.-L.; Wu, Z.; Bi, Y.; Wong, G.; Qiu, X.; Chen, J.; Pang, D.-W.; Zhang, Z.-L. Dual-Signal Readout Nanospheres for Rapid Point-of-Care Detection of Ebola Virus Glycoprotein. Anal. Chem. 2017, 89, 13105–13111. [Google Scholar] [CrossRef] [PubMed]
- Driskell, J.D.; Shanmukh, S.; Liu, Y.-J.; Hennigan, S.; Jones, L.; Zhao, Y.-P.; Dluhy, R.A.; Krause, D.C.; Tripp, R.A. Infectious Agent Detection With SERS-Active Silver Nanorod Arrays Prepared by Oblique Angle Deposition. IEEE Sens. J. 2008, 8, 863–870. [Google Scholar] [CrossRef]
- Shanmukh, S.; Jones, L.; Driskell, J.; Zhao, Y.; Dluhy, R.; Tripp, R.A. Rapid and Sensitive Detection of Respiratory Virus Molecular Signatures Using a Silver Nanorod Array SERS Substrate. Nano Lett. 2006, 6, 2630–2636. [Google Scholar] [CrossRef]
- Driskell, J.D.; Zhu, Y.; Kirkwood, C.D.; Zhao, Y.; Dluhy, R.A.; Tripp, R.A. Rapid and Sensitive Detection of Rotavirus Molecular Signatures Using Surface Enhanced Raman Spectroscopy. PLoS ONE 2010, 5, e10222. [Google Scholar] [CrossRef]
- Sivashanmugan, K.; Liao, J.-D.; You, J.-W.; Wu, C.-L. Focused-Ion-Beam-Fabricated Au/Ag Multilayered Nanorod Array as SERS-Active Substrate for Virus Strain Detection. Sens. Actuators B Chem. 2013, 181, 361–367. [Google Scholar] [CrossRef]
- Sanjuán, R. Collective Infectious Units in Viruses. Trends Microbiol. 2017, 25, 402–412. [Google Scholar] [CrossRef] [Green Version]
- Saviñon-Flores, F.; Méndez, E.; López-Castaños, M.; Carabarin-Lima, A.; López-Castaños, K.A.; González-Fuentes, M.A.; Méndez-Albores, A. A Review on SERS-Based Detection of Human Virus Infections: Influenza and Coronavirus. Biosensors 2021, 11, 66. [Google Scholar] [CrossRef]
- Lukose, J.; Chidangil, S.; George, S.D. Optical Technologies for the Detection of Viruses like COVID-19: Progress and Prospects. Biosens. Bioelectron. 2021, 178, 113004. [Google Scholar] [CrossRef]
- Xu, S.; Ji, X.; Xu, W.; Li, X.; Wang, L.; Bai, Y.; Zhao, B.; Ozaki, Y. Immunoassay Using Probe-Labelling Immunogold Nanoparticles with Silver Staining Enhancement via Surface-Enhanced Raman Scattering. Analyst 2004, 129, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from SARS-CoV-2 and Discovery of Its Inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef] [Green Version]
- Tai, W.; He, L.; Zhang, X.; Pu, J.; Voronin, D.; Jiang, S.; Zhou, Y.; Du, L. Characterization of the Receptor-Binding Domain (RBD) of 2019 Novel Coronavirus: Implication for Development of RBD Protein as a Viral Attachment Inhibitor and Vaccine. Cell. Mol. Immunol. 2020, 17, 613–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ñique, A.M.; Coronado-Marquina, F.; Mendez Rico, J.A.; García Mendoza, M.P.; Rojas-Serrano, N.; Simas, P.V.M.; Cabezas Sanchez, C.; Drexler, J.F. A Faster and Less Costly Alternative for RNA Extraction of SARS-CoV-2 Using Proteinase k Treatment Followed by Thermal Shock. PLoS ONE 2021, 16, e0248885. [Google Scholar] [CrossRef] [PubMed]
- Smyrlaki, I.; Ekman, M.; Lentini, A.; Rufino de Sousa, N.; Papanicolaou, N.; Vondracek, M.; Aarum, J.; Safari, H.; Muradrasoli, S.; Rothfuchs, A.G.; et al. Massive and Rapid COVID-19 Testing Is Feasible by Extraction-Free SARS-CoV-2 RT-PCR. Nat. Commun. 2020, 11, 4812. [Google Scholar] [CrossRef]
- Gerber, P.P.; Duncan, L.M.; Greenwood, E.J.D.; Marelli, S.; Naamati, A.; Teixeira-Silva, A.; Crozier, T.W.M.; Gabaev, I.; Zhan, J.R.; Mulroney, T.E.; et al. A Protease-Activatable Luminescent Biosensor and Reporter Cell Line for Authentic SARS-CoV-2 Infection. PLoS Pathog. 2022, 18, 1–23. [Google Scholar] [CrossRef]
- COVID-19 Tests and Collection Kits Authorized by the FDA: Infographic | FDA. Available online: https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/covid-19-tests-and-collection-kits-authorized-fda-infographic (accessed on 19 September 2021).
- Afzal, A. Molecular Diagnostic Technologies for COVID-19: Limitations and Challenges. J. Adv. Res. 2020, 26, 149–159. [Google Scholar] [CrossRef]
- Antigen-Detection in the Diagnosis of SARS-CoV-2 Infection. Available online: https://www.who.int/publications/i/item/antigen-detection-in-the-diagnosis-of-sars-cov-2infection-using-rapid-immunoassays (accessed on 21 May 2022).
- Shen, L.; Zhang, Q.; Luo, X.; Xiao, H.; Gu, M.; Cao, L.; Zhao, F.; Chen, Z. A Rapid Lateral Flow Immunoassay Strip for Detection of SARS-CoV-2 Antigen Using Latex Microspheres. J. Clin. Lab. Anal. 2021, 35, e24091. [Google Scholar] [CrossRef]
- Han, W.; Shin, J.H. Low-Cost, Open-Source 3D Printed Antibody Dispenser for Development and Small-Scale Production of Lateral Flow Assay Strips. HardwareX 2021, 9, e00188. [Google Scholar] [CrossRef]
- Pretti, M.A.M.; Galvani, R.G.; Scherer, N.M.; Farias, A.S.; Boroni, M. In Silico Analysis of Mutant Epitopes in New SARS-CoV-2 Lineages Suggest Global Enhanced CD8+ T Cell Reactivity and Also Signs of Immune Response Escape. Infect. Genet. Evol. 2022, 99, 105236. [Google Scholar] [CrossRef]
- Zhang, L.; Jackson, C.B.; Mou, H.; Ojha, A.; Rangarajan, E.S.; Izard, T.; Farzan, M.; Choe, H. The D614G Mutation in the SARS-CoV-2 Spike Protein Reduces S1 Shedding and Increases Infectivity. bioRxiv 2020. [Google Scholar] [CrossRef]
- Lubinski, B.; Fernandes, M.H.V.; Frazier, L.; Tang, T.; Daniel, S.; Diel, D.G.; Jaimes, J.A.; Whittaker, G.R. Functional Evaluation of the P681H Mutation on the Proteolytic Activation the SARS-CoV-2 Variant B.1.1.7 (Alpha) Spike. bioRxiv 2021. [Google Scholar] [CrossRef] [PubMed]
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.D.; Dingens, A.S.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C.; et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 2020, 182, 1295–1310. [Google Scholar] [CrossRef]
- Gobeil, S.M.-C.; Janowska, K.; McDowell, S.; Mansouri, K.; Parks, R.; Stalls, V.; Kopp, M.F.; Manne, K.; Li, D.; Wiehe, K.; et al. Effect of Natural Mutations of SARS-CoV-2 on Spike Structure, Conformation, and Antigenicity. Science 2021, 373, eabi6226. [Google Scholar] [CrossRef] [PubMed]
- Madhi, S.A.; Baillie, V.; Cutland, C.L.; Voysey, M.; Koen, A.L.; Fairlie, L.; Padayachee, S.D.; Dheda, K.; Barnabas, S.L.; Bhorat, Q.E.; et al. Efficacy of the ChAdOx1 NCoV-19 Covid-19 Vaccine against the B.1.351 Variant. N. Engl. J. Med. 2021, 384, 1885–1898. [Google Scholar] [CrossRef]
- Cele, S.; Gazy, I.; Jackson, L.; Hwa, S.-H.; Tegally, H.; Lustig, G.; Giandhari, J.; Pillay, S.; Wilkinson, E.; Naidoo, Y.; et al. Escape of SARS-CoV-2 501Y.V2 from Neutralization by Convalescent Plasma. Nature 2021, 593, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Lopez Bernal, J.; Andrews, N.; Gower, C.; Gallagher, E.; Simmons, R.; Thelwall, S.; Stowe, J.; Tessier, E.; Groves, N.; Dabrera, G.; et al. Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N. Engl. J. Med. 2021, 385, 585–594. [Google Scholar] [CrossRef]
- Sheikh, A.; McMenamin, J.; Taylor, B.; Robertson, C. SARS-CoV-2 Delta VOC in Scotland: Demographics, Risk of Hospital Admission, and Vaccine Effectiveness. Lancet 2021, 397, 2461–2462. [Google Scholar] [CrossRef]
- Jangra, S.; Ye, C.; Rathnasinghe, R.; Stadlbauer, D.; PVI Study Group; Krammer, F.; Simon, V.; Martinez-Sobrido, L.; Garćia-Sastre, A.; Schotsaert, M. The E484K Mutation in the SARS-CoV-2 Spike Protein Reduces but Does Not Abolish Neutralizing Activity of Human Convalescent and Post-Vaccination Sera. medRxiv 2021. [Google Scholar] [CrossRef]
- Baum, A.; Fulton, B.O.; Wloga, E.; Copin, R.; Pascal, K.E.; Russo, V.; Giordano, S.; Lanza, K.; Negron, N.; Ni, M.; et al. Antibody Cocktail to SARS-CoV-2 Spike Protein Prevents Rapid Mutational Escape Seen with Individual Antibodies. Science 2020, 369, 1014–1018. [Google Scholar] [CrossRef]
- Jangra, S.; Ye, C.; Rathnasinghe, R.; Stadlbauer, D.; Alshammary, H.; Amoako, A.A.; Awawda, M.H.; Beach, K.F.; Bermúdez-González, M.C.; Chernet, R.L.; et al. SARS-CoV-2 Spike E484K Mutation Reduces Antibody Neutralisation. Lancet Microbe 2021, 2, e283–e284. [Google Scholar] [CrossRef]
- Cov-Lineages. Available online: https://cov-lineages.org/ (accessed on 21 September 2021).
- O’Toole, Á.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J.T.; Colquhoun, R.; Ruis, C.; Abu-Dahab, K.; Taylor, B.; et al. Assignment of Epidemiological Lineages in an Emerging Pandemic Using the Pangolin Tool. Virus Evol. 2021, 7, veab064. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, Á.; Kraemer, M.U.G.; Hill, V.; Pybus, O.G.; Watts, A.; Bogoch, I.I.; Khan, K.; Messina, J.P.; Tegally, H.; Lessells, R.R.; et al. Tracking the International Spread of SARS-CoV-2 Lineages B.1.1.7 and B.1.351/501Y-V2. Wellcome Open Res. 2021, 6. [Google Scholar] [CrossRef]
- Rambaut, A.; Holmes, E.C.; O’Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A Dynamic Nomenclature Proposal for SARS-CoV-2 Lineages to Assist Genomic Epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef] [PubMed]
- Davies, N.G.; Jarvis, C.I.; van Zandvoort, K.; Clifford, S.; Sun, F.Y.; Funk, S.; Medley, G.; Jafari, Y.; Meakin, S.R.; Lowe, R.; et al. Increased Mortality in Community-Tested Cases of SARS-CoV-2 Lineage B.1.1.7. Nature 2021, 593, 270–274. [Google Scholar] [CrossRef]
- Davies, N.G.; Abbott, S.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.D.; Pearson, C.A.B.; Russell, T.W.; Tully, D.C.; Washburne, A.D.; et al. Estimated Transmissibility and Impact of SARS-CoV-2 Lineage B.1.1.7 in England. Science 2021, 372, eabg3055. [Google Scholar] [CrossRef]
- Funk, T.; Pharris, A.; Spiteri, G.; Bundle, N.; Melidou, A.; Carr, M.; Gonzalez, G.; Garcia-Leon, A.; Crispie, F.; O’Connor, L.; et al. Characteristics of SARS-CoV-2 Variants of Concern B.1.1.7, B.1.351 or P.1: Data from Seven EU/EEA Countries, Weeks 38/2020 to 10/2021. Eurosurveillance 2021, 26, 2100348. [Google Scholar] [CrossRef]
- Pereira, F.; Tosta, S.; Lima, M.M.; de Oliveira da Silva, L.; Nardy, V.B.; Gómez, M.K.A.; Lima, J.G.; Fonseca, V.; de Oliveira, T.; Lourenço, J.; et al. Genomic Surveillance Activities Unveil the Introduction of the SARS-CoV-2 B.1.525 Variant of Interest in Brazil: Case Report. J. Med. Virol. 2021, 93, 5523–5526. [Google Scholar] [CrossRef]
- Ozer, E.A.; Simons, L.M.; Adewumi, O.M.; Fowotade, A.A.; Omoruyi, E.C.; Adeniji, J.A.; Dean, T.J.; Zayas, J.; Bhimalli, P.P.; Ash, M.K.; et al. Coincident Rapid Expansion of Two SARS-CoV-2 Lineages with Enhanced Infectivity in Nigeria. medRxiv 2021. [Google Scholar] [CrossRef]
- Nonaka, C.K.V.; Gräf, T.; de Lorenzo Barcia, C.A.; Costa, V.F.; de Oliveira, J.L.; da Hora Passos, R.; Bastos, I.N.; de Santana, M.C.B.; Santos, I.M.; de Sousa, K.A.F.; et al. SARS-CoV-2 Variant of Concern P.1 (Gamma) Infection in Young and Middle-Aged Patients Admitted to the Intensive Care Units of a Single Hospital in Salvador, Northeast Brazil, February 2021. Int. J. Infect. Dis. 2021, 111, 47–54. [Google Scholar] [CrossRef]
- Bugembe, D.L.; Phan, M.V.T.; Ssewanyana, I.; Semanda, P.; Nansumba, H.; Dhaala, B.; Nabadda, S.; O’Toole, Á.N.; Rambaut, A.; Kaleebu, P.; et al. A SARS-CoV-2 Lineage A Variant (A.23.1) with Altered Spike Has Emerged and Is Dominating the Current Uganda Epidemic. medRxiv 2021. [Google Scholar] [CrossRef]
- Elaissari, A.; Holt, L.; Meunier, F.; Voisset, C.; Pichot, C.; Mandrand, B.; Mabilat, C. Hydrophilic and Cationic Latex Particles for the Specific Extraction of Nucleic Acids. J. Biomater. Sci. Polym. Ed. 1999, 10, 403–420. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Deng, C.; Liu, Y.; Zhao, X.; Tang, Y.; Liu, R.; Xia, Q.; Yan, W.; Ge, G. Optimization of Influencing Factors of Nucleic Acid Adsorption onto Silica-Coated Magnetic Particles: Application to Viral Nucleic Acid Extraction from Serum. J. Chromatogr. A 2014, 1325, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Dighe, K.; Moitra, P.; Alafeef, M.; Gunaseelan, N.; Pan, D. A Rapid RNA Extraction-Free Lateral Flow Assay for Molecular Point-of-Care Detection of SARS-CoV-2 Augmented by Chemical Probes. Biosens. Bioelectron. 2022, 200, 113900. [Google Scholar] [CrossRef] [PubMed]
- Bruno, A.; de Mora, D.; Freire-Paspuel, B.; Rodriguez, A.S.; Paredes-Espinosa, M.B.; Olmedo, M.; Sanchez, M.; Romero, J.; Paez, M.; Gonzalez, M.; et al. Analytical and Clinical Evaluation of a Heat Shock SARS-CoV-2 Detection Method without RNA Extraction for N and E Genes RT-QPCR. Int. J. Infect. Dis. 2021, 109, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Chomczynski, P.; Sacchi, N. The Single-Step Method of RNA Isolation by Acid Guanidinium Thiocyanate–Phenol–Chloroform Extraction: Twenty-Something Years On. Nat. Protoc. 2006, 1, 581–585. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-Step Method of RNA Isolation by Acid Guanidinium Thiocyanate-Phenol-Chloroform Extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- He, H.; Li, R.; Chen, Y.; Pan, P.; Tong, W.; Dong, X.; Chen, Y.; Yu, D. Integrated DNA and RNA Extraction Using Magnetic Beads from Viral Pathogens Causing Acute Respiratory Infections. Sci. Rep. 2017, 7, 45199. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Long, L.; Zhang, D.; Yuan, T.; Cui, S.; Yang, P.; Wang, Q.; Ren, S. Potential False-Negative Nucleic Acid Testing Results for Severe Acute Respiratory Syndrome Coronavirus 2 from Thermal Inactivation of Samples with Low Viral Loads. Clin. Chem. 2020, 66, 794–801. [Google Scholar] [CrossRef] [Green Version]
- Nyan, D.-C.C.; Ulitzky, L.E.; Cehan, N.; Williamson, P.; Winkelman, V.; Rios, M.; Taylor, D.R. Rapid Detection of Hepatitis B Virus in Blood Plasma by a Specific and Sensitive Loop-Mediated Isothermal Amplification Assay. Clin. Infect. Dis. 2014, 59, 16–23. [Google Scholar] [CrossRef] [Green Version]
- Pan, L.; Mu, M.; Yang, P.; Sun, Y.; Wang, R.; Yan, J.; Li, P.; Hu, B.; Wang, J.; Hu, C.; et al. Clinical Characteristics of COVID-19 Patients With Digestive Symptoms in Hubei, China: A Descriptive, Cross-Sectional, Multicenter Study. Off. J. Am. Coll. Gastroenterol. | ACG 2020, 115. [Google Scholar] [CrossRef] [PubMed]
- Perumal, N.; Jain, R.; Shrivastava, R.; Lalwani, J.; Chaurasia, D. Stability of SARS-CoV-2 RNA in Viral Lysis Buffer Stored at Different Temperatures. J. Lab. Physicians 2020, 12, 268–270. [Google Scholar] [CrossRef]
- Wozniak, A.; Cerda, A.; Ibarra-Henríquez, C.; Sebastian, V.; Armijo, G.; Lamig, L.; Miranda, C.; Lagos, M.; Solari, S.; Guzmán, A.M.; et al. A Simple RNA Preparation Method for SARS-CoV-2 Detection by RT-QPCR. Sci. Rep. 2020, 10, 16608. [Google Scholar] [CrossRef] [PubMed]
- Barza, R.; Patel, P.; Sabatini, L.; Singh, K. Use of a Simplified Sample Processing Step without RNA Extraction for Direct SARS-CoV-2 RT-PCR Detection. J. Clin. Virol. 2020, 132, 104587. [Google Scholar] [CrossRef]
- Ranoa, D.R.E.; Holland, R.L.; Alnaji, F.G.; Green, K.J.; Wang, L.; Brooke, C.B.; Burke, M.D.; Fan, T.M.; Hergenrother, P.J. Saliva-Based Molecular Testing for SARS-CoV-2 That Bypasses RNA Extraction. bioRxiv 2020. [Google Scholar] [CrossRef]
- Mauriz, E. Clinical Applications of Visual Plasmonic Colorimetric Sensing. Sensors 2020, 20, 6214. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M. Biosphere 2’s Lessons about Living on Earth and in Space. Sp. Sci. Technol. 2021, 2021, 8067539. [Google Scholar] [CrossRef]
- Cinelli, I.; Russomano, T. Advances in Space Medicine Applied to Pandemics on Earth. Sp. Sci. Technol. 2021, 2021, 9821480. [Google Scholar] [CrossRef]
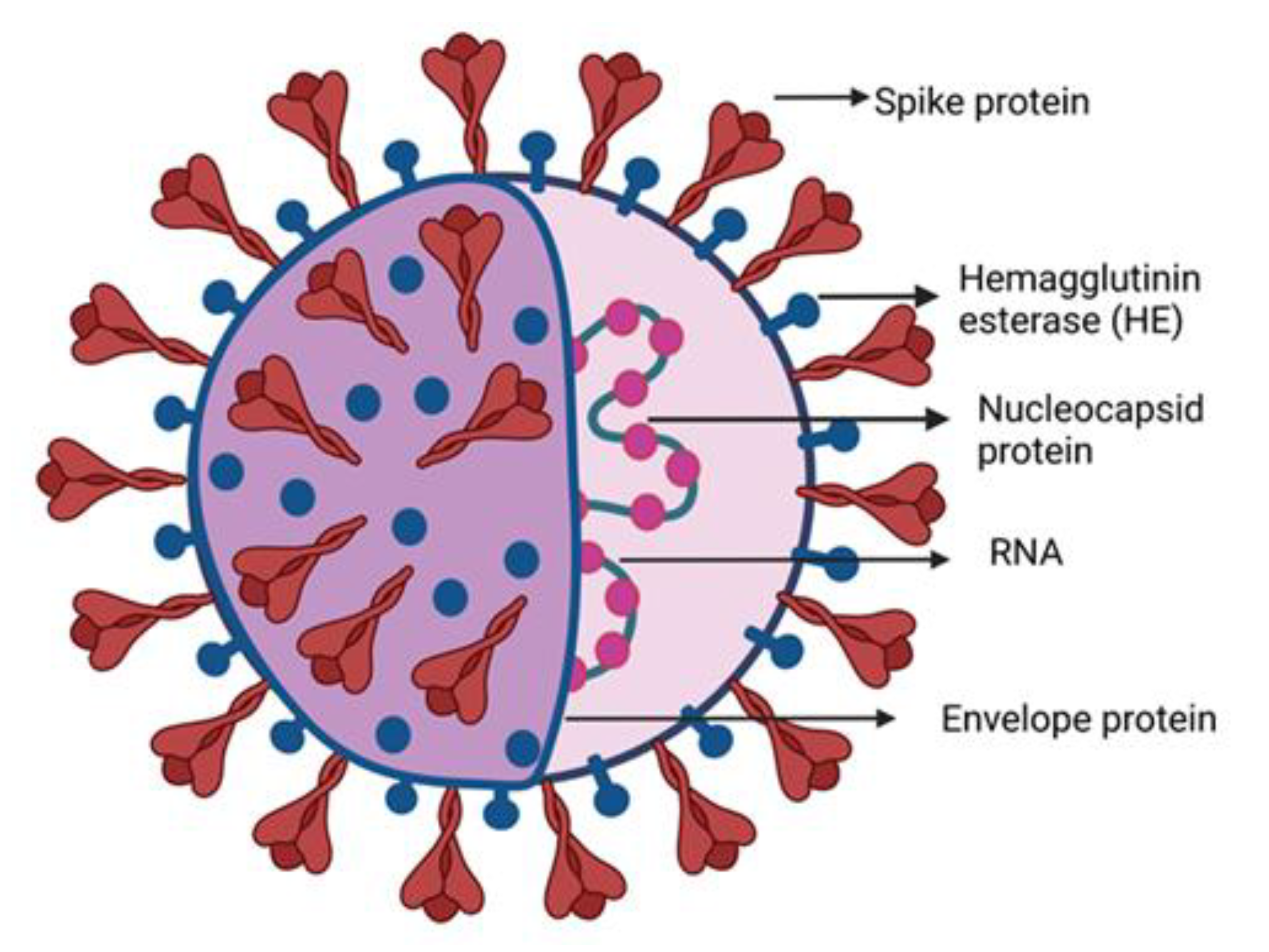

 ) to a quencher (
) to a quencher (  ). Mutant in the right graph showed a lower LOD (3-folds) than the wild in the left graph. (d) Colorimetric detection of SARS-CoV-2 by mutant N1 complex by a dye phenol red (a pH-sensitive dye) incubated at 60 °C for 30 min. (e) Visible fluorometric detection by the mutant N1 complex using calcein, incubated for 60 min at 60 °C [102].
). Mutant in the right graph showed a lower LOD (3-folds) than the wild in the left graph. (d) Colorimetric detection of SARS-CoV-2 by mutant N1 complex by a dye phenol red (a pH-sensitive dye) incubated at 60 °C for 30 min. (e) Visible fluorometric detection by the mutant N1 complex using calcein, incubated for 60 min at 60 °C [102].
 ) to a quencher (
) to a quencher (  ). Mutant in the right graph showed a lower LOD (3-folds) than the wild in the left graph. (d) Colorimetric detection of SARS-CoV-2 by mutant N1 complex by a dye phenol red (a pH-sensitive dye) incubated at 60 °C for 30 min. (e) Visible fluorometric detection by the mutant N1 complex using calcein, incubated for 60 min at 60 °C [102].
). Mutant in the right graph showed a lower LOD (3-folds) than the wild in the left graph. (d) Colorimetric detection of SARS-CoV-2 by mutant N1 complex by a dye phenol red (a pH-sensitive dye) incubated at 60 °C for 30 min. (e) Visible fluorometric detection by the mutant N1 complex using calcein, incubated for 60 min at 60 °C [102].



| Molecular Assay | RNA | Test Format | Turnaround Time (min.) | Target Analyte | Validated Samples |
| NAT reagent kit (Open source) | 60–120 <13 [32] 40–50 [33] 83 [34] 8 h (188 samples) [35] | N-gene,E-gene, S-gene, ORF1ab, RdRp gene, ORF1a, | Nasopharyngeal swab, Unknown, Oropharyngeal swab Nasal swab Bronchoalveolar lavage | ||
Cartridge based
| 36 [36], 45 [37], 30 [38] | Orf1ab, N-gene, RdRp | Nasopharyngeal, Oropharyngeal, Nasal swabs | ||
Dipstick
| <40 [39] | Orf1ab, N-gene | Nasopharyngeal | ||
| Immunoassay | Antibody(serological) | Cartridge-based processing | 15 [40] | IgG, Total Antibody, IgM | Serum, Plasma |
| Chemiluminescence | 15 [41] 29, 80, 120, 48 [42] | IgG, IgM, Total Antibody, Nucleocapsid protein, Unknown | Serum, Unknown samples | ||
Rapid diagnostic
| 15–20 60–90 [43] 10 [44] | IgG,N-protein, Total antibody Unknown | Serum Plasma Unknown saliva | ||
Reagent Kit
| 20 [45] <90 [46] | IgG, Total antibody,IgA,N-protein, Unknown | Serum Nasopharyngeal Unknown | ||
| Antigen | Cartridge-based process
immunoassay (CLIA) | 15 [47] | N-protein, S-protein (RBD) N-protein | Nasopharyngeal swab Saliva Nasal swab Unknown Oropharyngeal swab | |
Rapid diagnostic
| 15 [48] 20–30 [49,50] | N-protein S-protein (unknown type) S-protein RBD S-protein S1 S-protein S2 | Nasopharyngeal swabs Nasal swab Saliva Unknown Oropharyngeal swab Sputum | ||
| Reagent kit | 15 [51] | N-protein S-protein S1 S-protein (unknown type) | Nasopharyngeal swab Unknown Serum Nasal swab Oropharyngeal swab |
| Target | Biosensors | Principle | LOD/Detection Time/Cutoff Value/Sensitivity, Specificity | Drawbacks/Targets | References |
| Genome Based Detection | Label-free electrochemical biosensor | DNA hybridization of electrodeposited AuNPs immobilized with single-stranded nucleotide | - | RNA extraction Sensitive sample handling | [130] |
| Plasmonic biosensors (optical-LSPR) | Dual-functional plasmonic biosensor combining the plasmonic photothermal (PPT) effect and localized surface plasmon resonance (LSPR) sensing transduction | 0.22 pmol/L, 3 min | From multigene mixture | [131] | |
| Naked-eye colorimetric | Thiol-modified antisense oligonucleotide is used to cap AuNPs, which change color upon finding the target N-gene | 0.18 ng/μL, 10 min | N-gene | [132] | |
| Viral Particle Based Detection | Cell-based biosensor | Membrane-engineered mammalian cell containing antibody to bind with S-antigen | 1 fg/mL, 3 min | Not applicable for the detection of variants. | [133] |
| Nanoplasmonic sensors | Optical measurement of the SARS-CoV-2 particle | 370 vp/mL, 15 min | Restricted for the S-antigen | [134] | |
| Field effect-based transistor | Graphene-coated sheets with SARS-CoV-2 antibody | 2.42 × 102 copies/mL (in clinical samples) | s-antigen | [135] | |
| G-druplex-based biosensors | Whole genome | [136] | |||
| Molecularly imprinted polymers | Monoclonal-type, synthetic antibodies of SARS-CoV-2 | - | Only applied to the S-antigen | [137] | |
| eCovSens | Potentiostat-based sensor fluorine doped tin oxide + AuNPs immobilized with monoclonal antibody | 90 fmol/L, 10–30 s | S-antigen | [138] | |
| Electrochemical Biosensor | Functionalized TiO2 nanotube-based electrochemical | 0.7 nmol/L, 30 s | S-glycoprotein | [139] | |
| Lateral flow immunoassay | ACE2 enzyme binding captured antibody | 1.86 × 105 copies/mL | Spike antigen | [140] | |
| Antibody Based Detection | Lateral flow immunoassay | Lanthanide-doped Nanoparticles | 0.06666, 10 min | Anti-SARS-CoV-2-IgG | [141] |
| Immunochromatographic | 15 min, 85.2% and 100% | IgG-IgM combined | [142] | ||
| Immobilization on AuNPs | 15 min, 88.66% and 90.63% | IgG-IgM combined | [40] | ||
| Plasmon-enhanced biosensor | Grating Coupled Fluorescent Plasmonic (GC-FP) based on ELISA from dried blood spot samples | 30 min 100% and 100% | Multiplexed (IgG, IgM, IgA) | [143] | |
| Opto-microfluidic | A microfluidic device fabricated by the electrodeposition of Au-nanospikes linked with the optic probe to detect the target by using localized surface plasmon resonance | 0.5 pmol/L, 30 min | Antibodies against the spike protein | [144] |
| Variants (Notations, Lineages) | Identification Date and Countries and Total No. of Countries | Sequences (GISAID) | Notable Mutations Occur at S-Protein Notable Mutations (S-Protein) | Effect on Antigenicity | |||
| (Alpha) B.1.1.7(VOC-202012/01, 20/501Y.V1) | UK, 20 November 2020 | 168 | 1,133,025 | 23 | 8 | N501Y, [223] D614G [224], P681H [225] | N501Y effects on RBD [226] No effect on the serum neutralization [227] |
| (Beta) B.1.351, 501Y.V2; 20C/501Y.V2 | South Africa, 20 December | 110 | 10,095,100 | 21 | 9 | K417N, E484K, N501Y, D614G, A701V | K417T possibility escaping some monoclonal antibodies [228,229] |
| (Delta) B.1.617.2 | India | 141 | 10,095,100 | 12 | S:P681R S:L452R, | Yes, Increased [230,231] | |
| (Eta) B.1.525 | Nigeria | 80 | 9719 | 10 | aa:S:E484K aa:S:Q677H aa:S:F888L | Yes, reduce serum neutralization against IgG [232]. | |
| (Gamma) P.1, 501Y.V3, | Brazil and Japan, 20 December | 74 | 68,754 | 17 | 10 | aa:S:E484K aa:S:N501Y aa:S:K417T aa:S:D614G, aa:S:H655Y | aa:S:E484K escape mutation affects the nAbs [233]. |
| A.23.1 E484K | Ugenda Brazil UK | 48 | 1126 | 16 | 4 | aa:S:F157L aa:S:V367F aa:S:Q613H aa:S:P681R | Yes [234] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asghar, R.; Rasheed, M.; ul Hassan, J.; Rafique, M.; Khan, M.; Deng, Y. Advancements in Testing Strategies for COVID-19. Biosensors 2022, 12, 410. https://doi.org/10.3390/bios12060410
Asghar R, Rasheed M, ul Hassan J, Rafique M, Khan M, Deng Y. Advancements in Testing Strategies for COVID-19. Biosensors. 2022; 12(6):410. https://doi.org/10.3390/bios12060410
Chicago/Turabian StyleAsghar, Rabia, Madiha Rasheed, Jalees ul Hassan, Mohsin Rafique, Mashooq Khan, and Yulin Deng. 2022. "Advancements in Testing Strategies for COVID-19" Biosensors 12, no. 6: 410. https://doi.org/10.3390/bios12060410