Nucleic Acids Detection for Mycobacterium tuberculosis Based on Gold Nanoparticles Counting and Rolling-Circle Amplification
Abstract
:1. Introduction
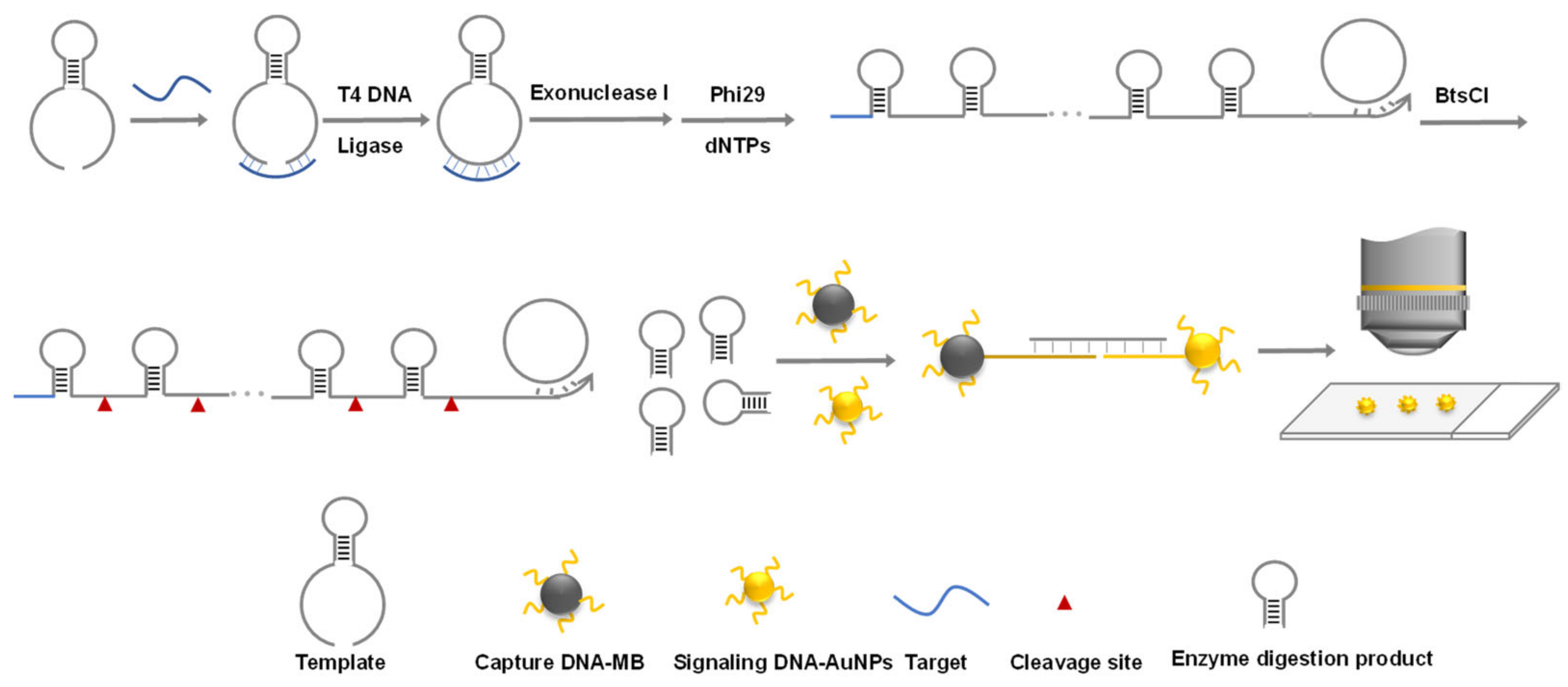
2. Materials and Methods
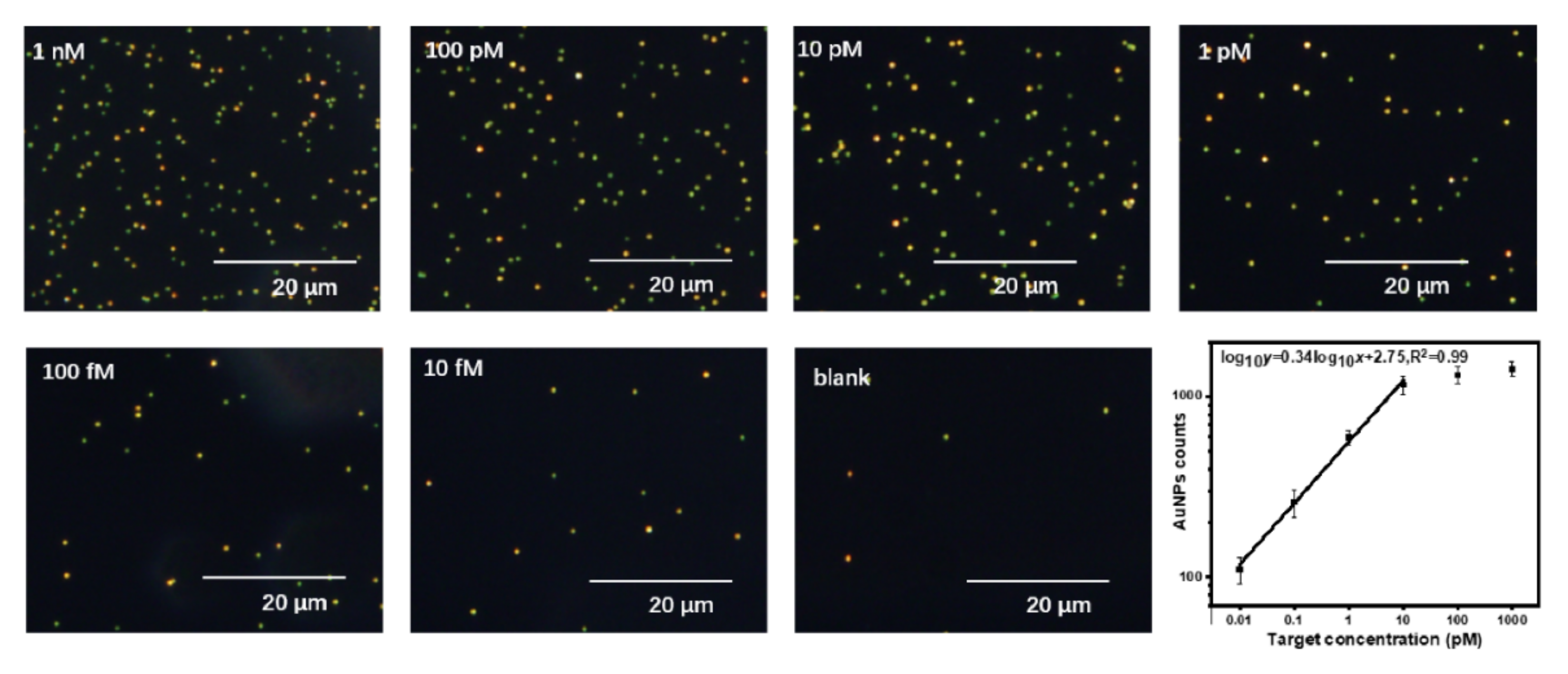
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gagneux, S. Ecology and evolution of Mycobacterium tuberculosis. Nat. Rev. Microbiol. 2018, 16, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Gengenbacher, M.; Kaufmann, S.H.E. Mycobacterium tuberculosis: Success through dormancy. FEMS Microbiol. Rev. 2012, 36, 514–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Getahun, H.; Matteelli, A.; Chaisson, R.E.; Raviglione, M. Latent Mycobacterium tuberculosis Infection. N. Engl. J. Med. 2015, 372, 2127–2135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordillo-Marroquin, C.; Gomez-Velasco, A.; Sanchez-Perez, H.J.; Pryg, K.; Shinners, J.; Murray, N.; Munoz-Jimenez, S.G.; Bencomo-Alerm, A.; Gomez-Bustamante, A.; Jonapa-Gomez, L.; et al. Magnetic Nanoparticle-Based Biosensing Assay Quantitatively Enhances Acid-Fast Bacilli Count in Paucibacillary Pulmonary Tuberculosis. Biosensors 2018, 8, 128. [Google Scholar] [CrossRef] [Green Version]
- Arias-Guillen, M.; Sanchez Menendez, M.M.; Alperi, M.; Riestra-Menendez, S.; Gonzalez Budino, M.T.; Maria Garcia-Clemente, M.; Martinez-Gonzalez, S.; Isabel Enriquez, A.; Alonso-Arias, R.; Palacios Gutierrez, J.J.; et al. High rates of tuberculin skin test positivity due to methotrexate therapy: False positive results. In Seminars in Arthritis and Rheumatism; W. B. Saunders: Philadelphia, PA, USA, 2018; Volume 48, pp. 538–546. [Google Scholar]
- Zhou, G.; Luo, Q.; Luo, S.; Teng, Z.; Ji, Z.; Yang, J.; Wang, F.; Wen, S.; Ding, Z.; Li, L.; et al. Interferon-gamma release assays or tuberculin skin test for detection and management of latent tuberculosis infection: A systematic review and meta-analysis. Lancet Infect. Dis. 2020, 20, 1457–1469. [Google Scholar] [CrossRef]
- Luetkemeyer, A.F.; Firnhaber, C.; Kendall, M.A.; Wu, X.; Mazurek, G.H.; Benator, D.A.; Arduino, R.; Fernandez, M.; Guy, E.; Johnson, P.; et al. Evaluation of Xpert MTB/RIF Versus AFB Smear and Culture to Identify Pulmonary Tuberculosis in Patients with Suspected Tuberculosis From Low and Higher Prevalence Settings. Clin. Infect. Dis. 2016, 62, 1081–1088. [Google Scholar] [CrossRef] [Green Version]
- Ng, B.Y.C.; Wee, E.J.H.; West, N.P.; Trau, M. Rapid DNA detection of Mycobacterium tuberculosis-towards single cell sensitivity in point-of-care diagnosis. Sci. Rep. 2015, 5, 15027. [Google Scholar] [CrossRef]
- Boyle, D.S.; McNerney, R.; Low, H.T.; Leader, B.T.; Perez-Osorio, A.C.; Meyer, J.C.; O’Sullivan, D.M.; Brooks, D.G.; Piepenburg, O.; Forrest, M.S. Rapid Detection of Mycobacterium tuberculosis by Recombinase Polymerase Amplification. PLoS ONE 2014, 9, e103091. [Google Scholar]
- Correa, R.A.M.S.; da Cruz, F.S.; Santos, C.C.; Pimenta, T.C.; Franco, D.L.; Ferreira, L.F. Optimization and Application of Electrochemical Transducer for Detection of Specific Oligonucleotide Sequence for Mycobacterium tuberculosis. Biosensors 2018, 8, 84. [Google Scholar] [CrossRef] [Green Version]
- Boehme, C.C.; Nabeta, P.; Hillemann, D.; Nicol, M.P.; Shenai, S.; Krapp, F.; Allen, J.; Tahirli, R.; Blakemore, R.; Rustomjee, R.; et al. Rapid Molecular Detection of Tuberculosis and Rifampin Resistance. N. Engl. J. Med. 2010, 363, 1005–1015. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.W.; Tang, X.F.; Ming, Z.H.; Ren, L.D.; Zhang, W.; Xiao, X.J. Short-DNA Specific Blocker PCR for Efficient and Simple Enrichment of Cell Free Fetal DNAs with Short Lengths. Chin. J. Chem. 2021, 39, 2101–2106. [Google Scholar] [CrossRef]
- Yang, Z.Y.; Chen, W.; Wang, J.Y.; Shi, M.H.; Zhang, R.L.; Dai, S.B.; Wu, T.B.; Zhao, M.P. Programmable One-Pot Enzymatic Reaction for Direct Fluorescence Detection of Ultralow-Abundance Mutations in the DNA Duplex. Anal. Chem. 2021, 93, 7086–7093. [Google Scholar] [CrossRef]
- Rosi, N.L.; Mirkin, C.A. Nanostructures in biodiagnostics. Chem. Rev. 2005, 105, 1547–1562. [Google Scholar] [CrossRef]
- Yang, T.; Luo, Z.; Tian, Y.; Qian, C.; Duan, Y. Design strategies of AuNPs-based nucleic acid colorimetric biosensors. TrAC-Trend Anal. Chem. 2020, 124, 115795. [Google Scholar] [CrossRef]
- Qi, F.; Han, Y.; Ye, Z.; Liu, H.; Wei, L.; Xiao, L. Color-Coded Single-Particle Pyrophosphate Assay with Dark-Field Optical Microscopy. Anal. Chem. 2018, 90, 11146–11153. [Google Scholar] [CrossRef]
- Bhusal, N.; Shrestha, S.; Pote, N.; Alocilja, E.C. Nanoparticle-Based Biosensing of Tuberculosis, an Affordable and Practical Alternative to Current Methods. Biosensor 2018, 9, 1. [Google Scholar] [CrossRef] [Green Version]
- Poon, C.-Y.; Wei, L.; Xu, Y.; Chen, B.; Xiao, L.; Li, H.-W. Quantification of Cancer Biomarkers in Serum Using Scattering-Based Quantitative Single Particle Intensity Measurement with a Dark-Field Microscope. Anal. Chem. 2016, 88, 8849–8856. [Google Scholar] [CrossRef]
- Kim, S.; Park, J.-E.; Hwang, W.; Seo, J.; Lee, Y.-K.; Hwang, J.-H.; Nam, J.-M. Optokinetically Encoded Nanoprobe-Based Multiplexing Strategy for MicroRNA Profiling. J. Am. Chem. Soc. 2017, 139, 3558–3566. [Google Scholar] [CrossRef]
- Li, T.; Wu, X.; Liu, F.; Li, N. Analytical methods based on the light-scattering of plasmonic nanoparticles at the single particle level with dark-field microscopy imaging. Analyst 2017, 142, 248–256. [Google Scholar] [CrossRef]
- Shen, J.; Liang, L.; Xiao, M.; Xie, X.; Wang, F.; Li, Q.; Ge, Z.; Li, J.; Shi, J.; Wang, L.; et al. Fractal Nanoplasmonic Labels for Supermultiplex Imaging in Single Cells. J. Am. Chem. Soc. 2019, 141, 11938–11946. [Google Scholar] [CrossRef]
- Wang, F.; Li, Y.; Han, Y.; Ye, Z.; Wei, L.; Luo, H.-B.; Xiao, L. Single-Particle Enzyme Activity Assay with Spectral-Resolved Dark-Field Optical Microscopy. Anal. Chem. 2019, 91, 6329–6339. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Li, T.; Tao, G.; Lin, R.; Pei, X.; Liu, F.; Li, N. A universal and enzyme-free immunoassay platform for biomarker detection based on gold nanoparticle enumeration with a dark-field microscope. Analyst 2017, 142, 4201–4205. [Google Scholar] [CrossRef] [PubMed]
- Pei, X.; Lai, T.; Tao, G.; Hong, H.; Liu, F.; Li, N. Ultraspecific Multiplexed Detection of Low-Abundance Single-Nucleotide Variants by Combining a Masking Tactic with Fluorescent Nanoparticle Counting. Anal. Chem. 2018, 90, 4226–4233. [Google Scholar] [CrossRef] [PubMed]
- Pei, X.; Yin, H.; Lai, T.; Zhang, J.; Liu, F.; Xu, X.; Li, N. Multiplexed Detection of Attomoles of Nucleic Acids Using Fluorescent Nanoparticle Counting Platform. Anal. Chem. 2018, 90, 1376–1383. [Google Scholar] [CrossRef]
- Pei, X.; Huang, H.; Chen, Y.; Li, C.; Liu, F.; Li, N. Modulating fluorescence anisotropy of dye-labeled DNA without involving mass amplification. Talanta 2016, 154, 567–573. [Google Scholar] [CrossRef]
- Fu, S.N.; Li, N.; Li, J.J.; Deng, Y.N.; Xu, L.D.; Yu, C.Y.; Su, X. Engineering high-robustness DNA molecular circuits by utilizing nucleases. Nanoscale 2020, 12, 6964–6970. [Google Scholar] [CrossRef]
- Li, T.; Xu, X.; Zhang, G.; Lin, R.; Chen, Y.; Li, C.; Liu, F.; Li, N. Nonamplification Sandwich Assay Platform for Sensitive Nucleic Acid Detection Based on AuNPs Enumeration with the Dark-Field Microscope. Anal. Chem. 2016, 88, 4188–4191. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Chen, Y.; Wei, H.; Xia, B.; Liu, F.; Li, N. Counting Bacteria Using Functionalized Gold Nanoparticles as the Light-Scattering Reporter. Anal. Chem. 2012, 84, 9721–9728. [Google Scholar] [CrossRef]
- Tian, W.M.; Li, P.J.; He, W.L.; Liu, C.H.; Li, Z.P. Rolling circle extension-actuated loop-mediated isothermal amplification (RCA-LAMP) for ultrasensitive detection of microRNAs. Biosens.Bioelectron. 2019, 128, 17–22. [Google Scholar] [CrossRef]
- Wei, J.; Wang, H.; Wu, Q.; Gong, X.; Ma, K.; Liu, X.; Wang, F. A Smart, Autocatalytic, DNAzyme Biocircuit for in Vivo, Amplified, MicroRNA Imaging. Angew. Chem. Int. Ed. 2020, 59, 5965–5971. [Google Scholar] [CrossRef]
- Liling, H.; Wei, W.; Xueqing, S.; Shuliu, W.; Qian, L.; Faliang, A.; Shijia, W. A fluorescent DNA hydrogel aptasensor based on the self-assembly of rolling circle amplification products for sensitive detection of ochratoxin A. J. Agric. Food Chem. 2020, 68, 369–375. [Google Scholar]
- Xu, X.; Li, T.; Xu, Z.; Wei, H.; Lin, R.; Xia, B.; Liu, F.; Li, N. Automatic Enumeration of Gold Nanomaterials at the Single-Particle Level. Anal. Chem. 2015, 87, 2576–2581. [Google Scholar] [CrossRef]
- Bastus, N.G.; Comenge, J.; Puntes, V. Kinetically Controlled Seeded Growth Synthesis of Citrate-Stabilized Gold Nanoparticles of up to 200 nm: Size Focusing versus Ostwald Ripening. Langmuir 2011, 27, 11098–11105. [Google Scholar] [CrossRef]
- Liu, J.; Lu, Y. Preparation of aptamer-linked gold nanoparticle purple aggregates for colorimetric sensing of analytes. Nat. Protoc. 2006, 1, 246–252. [Google Scholar] [CrossRef]
- Zhou, W.; Sun, J.; Li, X. Low-Cost Quantitative Photothermal Genetic Detection of Pathogens on a Paper Hybrid Device Using a Thermometer. Anal. Chem. 2020, 92, 14830–14837. [Google Scholar] [CrossRef]
- Hu, O.; Li, Z.; He, Q.; Tong, Y.; Tan, Y.; Chen, Z. Fluorescence Biosensor for One-Step Simultaneous Detection of Mycobacterium tuberculosis Multidrug-Resistant Genes Using nanoCoTPyP and Double Quantum Dots. Anal. Chem. 2022, 94, 7918–7927. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; He, X.; Ju, L.; Liu, X.; Li, F.; Cui, H. A label-free method for the detection of specific DNA sequences using gold nanoparticles bifunctionalized with a chemiluminescent reagent and a catalyst as signal reporters. Anal. Bioanal. Chem. 2016, 408, 8747–8754. [Google Scholar] [CrossRef]
- Wang, Y. TB-QUICK: CRISPR-Cas12b-assisted rapid and sensitive detection of Mycobacterium tuberculosis. J. Infect. 2021, 83, 54–60. [Google Scholar]
- Zhang, Y.; Wang, Y.; Xu, L.; Lou, C.; Ouyang, Q.; Qian, L. Paired dCas9 design as a nucleic acid detection platform for pathogenic strains. Methods 2021, 203, 70–77. [Google Scholar] [CrossRef]
- Long, Q. A Highly Sensitive and Specific Detection Method for Mycobacterium tuberculosis Fluoroquinolone Resistance Mutations Utilizing the CRISPR-Cas13a System. Front. Microbiol. 2022, 13, 847373. [Google Scholar]



| Target | 100 pM | 1 pM |
|---|---|---|
| Genomic DNA of healthy donors | 86.2% ± 12.2% | 100.0% ± 7.9% |
| Genomic DNA of Klebsiella pneumoniae | 93.1% ± 2.4% | 105.6% ± 3.9% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pei, X.; Hong, H.; Liu, S.; Li, N. Nucleic Acids Detection for Mycobacterium tuberculosis Based on Gold Nanoparticles Counting and Rolling-Circle Amplification. Biosensors 2022, 12, 448. https://doi.org/10.3390/bios12070448
Pei X, Hong H, Liu S, Li N. Nucleic Acids Detection for Mycobacterium tuberculosis Based on Gold Nanoparticles Counting and Rolling-Circle Amplification. Biosensors. 2022; 12(7):448. https://doi.org/10.3390/bios12070448
Chicago/Turabian StylePei, Xiaojing, Hu Hong, Sitong Liu, and Na Li. 2022. "Nucleic Acids Detection for Mycobacterium tuberculosis Based on Gold Nanoparticles Counting and Rolling-Circle Amplification" Biosensors 12, no. 7: 448. https://doi.org/10.3390/bios12070448
APA StylePei, X., Hong, H., Liu, S., & Li, N. (2022). Nucleic Acids Detection for Mycobacterium tuberculosis Based on Gold Nanoparticles Counting and Rolling-Circle Amplification. Biosensors, 12(7), 448. https://doi.org/10.3390/bios12070448






