Point-of-Care Diagnostics for Farm Animal Diseases: From Biosensors to Integrated Lab-on-Chip Devices
Abstract
:1. Introduction
2. Categories of POC Devices
2.1. Paper-Based POC Devices
2.1.1. Dipstick and Strip Tests
2.1.2. Lateral Flow Assays (LFAs)
2.2. Microfluidic POC Devices
2.2.1. Micro Total Analysis Systems (μTAS)—Lab-on-Chip (LOC) Devices
2.2.2. Microfluidic Paper-Based Analytical Devices (μPAD)
2.2.3. Applications of Microfluidic Technologies
3. Biosensors in Animal Production
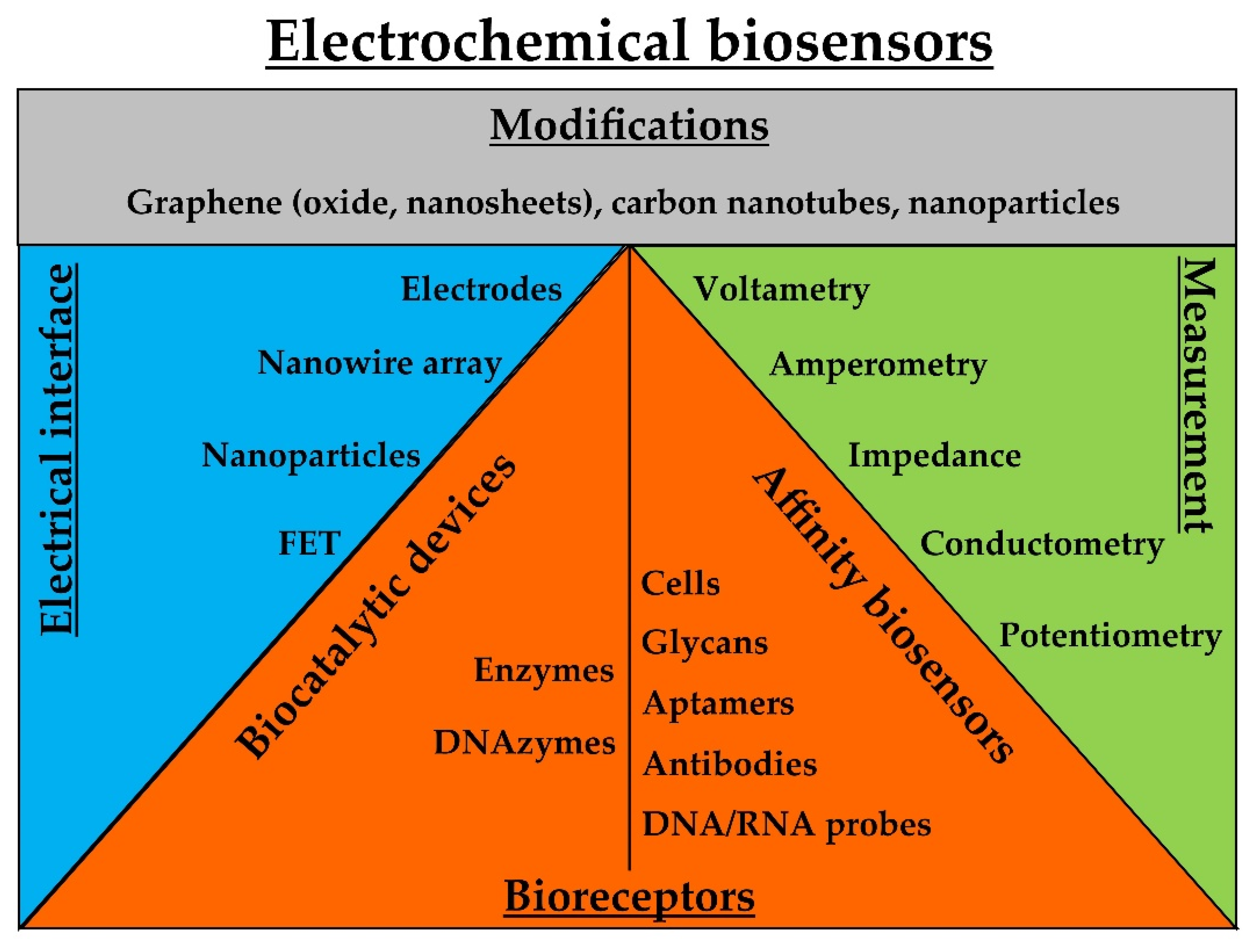
3.1. Electrochemical Biosensors
3.2. Optical Biosensors
3.3. Piezoelectric Biosensors
3.4. Magnetic Biosensors
3.5. Other Approaches in Signal Transduction
4. POC Tests and Devices for Mastitis and Animal Diseases
4.1. LFAs
4.2. Lab-on-Chip (LOC) Devices
5. Regulation of POC Tests for Animal Diseases
6. Challenges of Veterinary POC Testing
7. Future Perspectives
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gouel, C.; Guimbard, H. Nutrition Transition and the Structure of Global Food Demand. Am. J. Agric. Econ. 2018, 101, 383–403. [Google Scholar] [CrossRef] [Green Version]
- Tian, X.; Engel, B.A.; Qian, H.; Hua, E.; Sun, S.; Wang, Y. Will reaching the maximum achievable yield potential meet future global food demand? J. Clean. Prod. 2021, 294, 126285. [Google Scholar] [CrossRef]
- Henchion, M.; Moloney, A.; Hyland, J.; Zimmermann, J.; McCarthy, S. Review: Trends for meat, milk and egg consumption for the next decades and the role played by livestock systems in the global production of proteins. Animal 2021, 15, 100287. [Google Scholar] [CrossRef] [PubMed]
- VanderWaal, K.; Deen, J. Global trends in infectious diseases of swine. Proc. Natl. Acad. Sci. USA 2018, 115, 11495–11500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgan, N.; Prakash, A.; Jutzi, S. Influencia de las enfermedades animales en los mercados agropecuarios internacionales. Rev. Sci. Tech. 2006, 25, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Perry, B.D.; Grace, D.; Sones, K. Current drivers and future directions of global livestock disease dynamics. Proc. Natl. Acad. Sci. USA 2011, 110, 20871–20877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foudeh, A.M.; Didar, T.F.; Veres, T.; Tabrizian, M. Microfluidic designs and techniques using lab-on-a-chip devices for pathogen detection for point-of-care diagnostics. Lab Chip 2012, 12, 3249–3266. [Google Scholar] [CrossRef]
- Manessis, G.; Gelasakis, A.; Bossis, I. The challenge of introducing Point of Care Diagnostics in Farm Animal Health Management. Biomed. J. Sci. Tech. Res. 2019, 14, 001–004. [Google Scholar] [CrossRef]
- Cummins, B.M.; Ligler, F.S.; Walker, G.M. Point-of-care diagnostics for niche applications. Biotechnol. Adv. 2016, 34, 161–176. [Google Scholar] [CrossRef] [Green Version]
- Drain, P.K.; Hyle, E.P.; Noubary, F.; Freedberg, K.A.; Wilson, D.; Bishai, W.R.; Rodriguez, W.; Bassett, I.V. Diagnostic point-of-care tests in resource-limited settings. Lancet Infect. Dis. 2014, 14, 239–249. [Google Scholar] [CrossRef] [Green Version]
- Soin, N.; Fishlock, S.; Kelsey, C.; Smith, S. Triboelectric Effect Enabled Self-Powered, Point-of-Care Diagnostics: Opportunities for Developing ASSURED and REASSURED Devices. Micromachines 2021, 12, 337. [Google Scholar] [CrossRef]
- Sharma, S.; Zapatero-Rodríguez, J.; Estrela, P.; O’Kennedy, R.J. Point-of-Care Diagnostics in Low Resource Settings: Present Status and Future Role of Microfluidics. Biosensors 2015, 5, 577–601. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.T.; Lantigua, D.; Meka, A.; Taing, S.; Pandher, M.; Camci-Unal, G. Paper-Based Sensors: Emerging Themes and Applications. Sensors 2018, 18, 2838. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Wang, S.; Wang, L.; Li, F.; Pingguan-Murphy, B.; Lu, T.J.; Xu, F. Advances in paper-based point-of-care diagnostics. Biosens. Bioelectron. 2014, 54, 585–597. [Google Scholar] [CrossRef]
- Morbioli, G.G.; Mazzu-Nascimento, T.; Stockton, A.M.; Carrilho, E. Technical aspects and challenges of colorimetric detection with microfluidic paper-based analytical devices (μPADs)—A review. Anal. Chim. Acta 2017, 970, 1–22. [Google Scholar] [CrossRef]
- Yetisen, A.K.; Akram, M.S.; Lowe, C.R. Paper-based microfluidic point-of-care diagnostic devices. Lab Chip 2013, 13, 2210–2251. [Google Scholar] [CrossRef]
- Ge, L.; Yu, J.; Ge, S.; Yan, M. Lab-on-paper-based devices using chemiluminescence and electrogenerated chemiluminescence detection. Anal. Bioanal. Chem. 2014, 406, 5613–5630. [Google Scholar] [CrossRef]
- Vashist, S.K.; Luppa, P.B.; Yeo, L.Y.; Ozcan, A.; Luong, J.H.T. Emerging Technologies for Next-Generation Point-of-Care Testing. Trends Biotechnol. 2015, 33, 692–705. [Google Scholar] [CrossRef]
- Bahadır, E.B.; Sezgintürk, M.K. Lateral flow assays: Principles, designs and labels. TrAC Trends Anal. Chem. 2016, 82, 286–306. [Google Scholar] [CrossRef]
- Mahato, K.; Srivastava, A.; Chandra, P. Paper based diagnostics for personalized health care: Emerging technologies and commercial aspects. Biosens. Bioelectron. 2017, 96, 246–259. [Google Scholar] [CrossRef]
- Quesada-González, D.; Merkoçi, A. Nanomaterial-based devices for point-of-care diagnostic applications. Chem. Soc. Rev. 2018, 47, 4697–4709. [Google Scholar] [CrossRef] [PubMed]
- Link, N.; Weber, W.; Fussenegger, M. A novel generic dipstick-based technology for rapid and precise detection of tetracycline, streptogramin and macrolide antibiotics in food samples. J. Biotechnol. 2007, 128, 668–680. [Google Scholar] [CrossRef] [PubMed]
- Brady, M.A.; Dennis, J.S.; Wagner-Mann, C. Evaluating the use of plasma hematocrit samples to detect ketones utilizing urine dipstick colorimetric methodology in diabetic dogs and cats. J. Veter. Emerg. Crit. Care 2003, 13, 15. [Google Scholar] [CrossRef]
- Carrier, J.; Stewart, S.; Godden, S.; Fetrow, J.; Rapnicki, P. Evaluation and Use of Three Cowside Tests for Detection of Subclinical Ketosis in Early Postpartum Cows. J. Dairy Sci. 2004, 87, 3725–3735. [Google Scholar] [CrossRef] [Green Version]
- Eltzov, E.; Guttel, S.; Kei, A.L.Y.; Sinawang, P.D.; Ionescu, R.E.; Marks, R.S. Lateral Flow Immunoassays—From Paper Strip to Smartphone Technology. Electroanalysis 2015, 27, 2116–2130. [Google Scholar] [CrossRef]
- Mohd Hanafiah, K.; Arifin, N.; Bustami, Y.; Noordin, R.; Garcia, M.; Anderson, D. Development of Multiplexed Infectious Disease Lateral Flow Assays: Challenges and Opportunities. Diagnostics 2017, 7, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.; Yang, J.; Zhang, G.; Wang, X.; Qiao, S.; Zhao, D.; Zhi, Y.; Li, X.; Xing, G.; Luo, J.; et al. Development of an immunochromatographic strip for the detection of antibodies against foot-and-mouth disease virus serotype O. J. Virol. Methods 2010, 165, 139–144. [Google Scholar] [CrossRef]
- Brüning-Richardson, A.; Akerblom, L.; Klingeborn, B.; Anderson, J. Improvement and development of rapid chromatographic strip-tests for the diagnosis of rinderpest and peste des petits ruminants viruses. J. Virol. Methods 2011, 174, 42–46. [Google Scholar] [CrossRef]
- Busin, V.; Wells, B.; Kersaudy-Kerhoas, M.; Shu, W.; Burgess, S.T. Opportunities and challenges for the application of microfluidic technologies in point-of-care veterinary diagnostics. Mol. Cell. Probes 2016, 30, 331–341. [Google Scholar] [CrossRef] [Green Version]
- McDonald, J.C.; Duffy, D.C.; Anderson, J.R.; Chiu, D.T.; Wu, H.; Schueller, O.J.A.; Whitesides, G.M. Fabrication of Microfluidic Systems in Poly(dimethylsiloxane). Electrophoresis 2000, 21, 27–40. [Google Scholar] [CrossRef]
- Teh, S.-Y.; Lin, R.; Hung, L.-H.; Lee, A.P. Droplet microfluidics. Lab Chip 2008, 8, 198–220. [Google Scholar] [CrossRef]
- Reyes, D.R.; Iossifidis, D.; Auroux, P.; Manz, A. Micro Total Analysis Systems 1. Introduction, Theory, and Technology. Anal. Chem. 2002, 74, 2623–2636. [Google Scholar] [CrossRef]
- Taberham, A.; Kraft, M.; Mowlem, M.; Morgan, H. The fabrication of lab-on-chip devices from fluoropolymers. J. Micromech. Microeng. 2008, 18, 064011. [Google Scholar] [CrossRef]
- Rossier, J.; Reymond, F.; Michel, P.E. Polymer microfluidic chips for electrochemical and biochemical analyses. Electrophoresis 2002, 23, 858–867. [Google Scholar] [CrossRef]
- Nasseri, B.; Soleimani, N.; Rabiee, N.; Kalbasi, A.; Karimi, M.; Hamblin, M.R. Point-of-care microfluidic devices for pathogen detection. Biosens. Bioelectron. 2018, 117, 112–128. [Google Scholar] [CrossRef]
- Dittrich, P.S.; Tachikawa, K.; Manz, A. Micro Total Analysis Systems. Latest Advancements and Trends. Anal. Chem. 2006, 78, 3887–3908. [Google Scholar] [CrossRef] [PubMed]
- Lisowski, P.; Zarzycki, P.K. Microfluidic Paper-Based Analytical Devices (μPADs) and Micro Total Analysis Systems (μTAS): Development, Applications and Future Trends. Chromatographia 2013, 76, 1201–1214. [Google Scholar] [CrossRef] [Green Version]
- Martinez, A.W.; Phillips, S.T.; Whitesides, G.M. Diagnostics for the Developing World: Microfluidic Paper-Based Analytical Devices. Anal. Chem. 2010, 82, 3–10. [Google Scholar] [CrossRef]
- Akyazi, T.; Basabe-Desmonts, L.; Benito-Lopez, F. Review on microfluidic paper-based analytical devices towards commercialisation. Anal. Chim. Acta 2018, 1001, 1–17. [Google Scholar] [CrossRef]
- Sher, M.; Zhuang, R.; Demirci, U.; Asghar, W. Paper-based analytical devices for clinical diagnosis: Recent advances in the fabrication techniques and sensing mechanisms. Expert Rev. Mol. Diagn. 2017, 17, 351–366. [Google Scholar] [CrossRef]
- Carrilho, E.; Martinez, A.W.; Whitesides, G.M. Understanding Wax Printing: A Simple Micropatterning Process for Paper-Based Microfluidics. Anal. Chem. 2009, 81, 7091–7095. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Si, J.; Li, Z. Fabrication techniques for microfluidic paper-based analytical devices and their applications for biological testing: A review. Biosens. Bioelectron. 2016, 77, 774–789. [Google Scholar] [CrossRef] [PubMed]
- Olmos, C.M.; Vaca, A.; Rosero, G.; Peñaherrera, A.; Perez, C.; Carneiro, I.D.S.; Vizuete, K.; Arroyo, C.; Debut, A.; Pérez, M.S.; et al. Epoxy resin mold and PDMS microfluidic devices through photopolymer flexographic printing plate. Sens. Actuators B Chem. 2019, 288, 742–748. [Google Scholar] [CrossRef]
- Yehia, A.M.; Farag, M.A.; Tantawy, M.A. A novel trimodal system on a paper-based microfluidic device for on-site detection of the date rape drug “ketamine”. Anal. Chim. Acta 2020, 1104, 95–104. [Google Scholar] [CrossRef]
- Qi, J.; Li, B.; Wang, X.; Zhang, Z.; Wang, Z.; Han, J.; Chen, L. Three-dimensional paper-based microfluidic chip device for multiplexed fluorescence detection of Cu2+ and Hg2+ ions based on ion imprinting technology. Sens. Actuators B Chem. 2017, 251, 224–233. [Google Scholar] [CrossRef]
- Caglayan, M.G.; Sheykhi, S.; Mosca, L.; Anzenbacher, P. Fluorescent zinc and copper complexes for detection of adrafinil in paper-based microfluidic devices. Chem. Commun. 2016, 52, 8279–8282. [Google Scholar] [CrossRef]
- Davaji, B.; Lee, C.H. A paper-based calorimetric microfluidics platform for bio-chemical sensing. Biosens. Bioelectron. 2014, 59, 120–126. [Google Scholar] [CrossRef]
- Delaney, J.L.; Hogan, C.F.; Tian, J.; Shen, W. Electrogenerated Chemiluminescence Detection in Paper-Based Microfluidic Sensors. Anal. Chem. 2011, 83, 1300–1306. [Google Scholar] [CrossRef]
- Jangid, A.R.; Strong, E.B.; Escamilla, E.; Lore, B.A.; Tod, N.J.; Thiel, R.; Martinez, A.W.; Martinez, N.W. Chronometric Quantitation of Analytes in Paper-Based Microfluidic Devices (MicroPADs) via Enzymatic Degradation of a Metastable Biomatrix. Inventions 2019, 4, 48. [Google Scholar] [CrossRef] [Green Version]
- Fu, L.-M.; Wang, Y.-N. Detection methods and applications of microfluidic paper-based analytical devices. TrAC Trends Anal. Chem. 2018, 107, 196–211. [Google Scholar] [CrossRef]
- Jung, W.; Han, J.; Choi, J.-W.; Ahn, C.H. Point-of-care testing (POCT) diagnostic systems using microfluidic lab-on-a-chip technologies. Microelectron. Eng. 2015, 132, 46–57. [Google Scholar] [CrossRef]
- Wadhwa, A.; Foote, R.S.; Shaw, R.W.; Eda, S. Bead-based microfluidic immunoassay for diagnosis of Johne’s disease. J. Immunol. Methods 2012, 382, 196–202. [Google Scholar] [CrossRef]
- Kimura, S.; Fukuda, J.; Tajima, A.; Suzuki, H. On-chip diagnosis of subclinical mastitis in cows by electrochemical measurement of neutrophil activity in milk. Lab Chip 2012, 12, 1309–1315. [Google Scholar] [CrossRef]
- Su, L.; Jia, W.; Hou, C.; Lei, Y. Microbial biosensors: A review. Biosens. Bioelectron. 2010, 26, 1788–1799. [Google Scholar] [CrossRef]
- Vidic, J.; Manzano, M.; Chang, C.-M.; Jaffrezic-Renault, N. Advanced biosensors for detection of pathogens related to livestock and poultry. Veter. Res. 2017, 48, 11. [Google Scholar] [CrossRef] [Green Version]
- Mohankumar, P.; Ajayan, J.; Mohanraj, T.; Yasodharan, R. Recent developments in biosensors for healthcare and biomedical applications: A review. Measurement 2021, 167, 108293. [Google Scholar] [CrossRef]
- Ronkainen, N.J.; Halsall, H.B.; Heineman, W.R. Electrochemical biosensors. Chem. Soc. Rev. 2010, 39, 1747–1763. [Google Scholar] [CrossRef]
- Hammond, J.L.; Formisano, N.; Estrela, P.; Carrara, S.; Tkac, J. Electrochemical biosensors and nanobiosensors. Essays Biochem. 2016, 60, 69–80. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Liu, S.; Dai, Z.; Bao, J.; Yang, X. Applications of Nanomaterials in Electrochemical Enzyme Biosensors. Sensors 2009, 9, 8547–8561. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Wang, Y.; Liu, S.; Yu, J.; Wang, H.; Wang, Y.; Huang, J. Label-free and highly sensitive electrochemical detection of E. coli based on rolling circle amplifications coupled peroxidase-mimicking DNAzyme amplification. Biosens. Bioelectron. 2016, 75, 315–319. [Google Scholar] [CrossRef]
- Veerapandian, M.; Hunter, R.; Neethirajan, S. Lipoxygenase-modified Ru-bpy/graphene oxide: Electrochemical biosensor for on-farm monitoring of non-esterified fatty acid. Biosens. Bioelectron. 2015, 78, 253–258. [Google Scholar] [CrossRef]
- Karash, S.; Wang, R.; Kelso, L.; Lu, H.; Huang, T.J.; Li, Y. Rapid detection of avian influenza virus H5N1 in chicken tracheal samples using an impedance aptasensor with gold nanoparticles for signal amplification. J. Virol. Methods 2016, 236, 147–156. [Google Scholar] [CrossRef] [Green Version]
- Arora, K.; Prabhakar, N.; Chand, A.S.; Malhotra, B.D. Escherichia coli Genosensor Based on Polyaniline. Anal. Chem. 2007, 79, 6152–6158. [Google Scholar] [CrossRef]
- Rahi, A.; Sattarahmady, N.; Heli, H. An ultrasensitive electrochemical genosensor for Brucella based on palladium nanoparticles. Anal. Biochem. 2016, 510, 11–17. [Google Scholar] [CrossRef] [Green Version]
- Tarasov, A.; Gray, D.; Tsai, M.-Y.; Shields, N.; Montrose, A.; Creedon, N.; Lovera, P.; O’Riordan, A.; Mooney, M.H.; Vogel, E.M. A potentiometric biosensor for rapid on-site disease diagnostics. Biosens. Bioelectron. 2016, 79, 669–678. [Google Scholar] [CrossRef] [Green Version]
- Hideshima, S.; Hinou, H.; Ebihara, D.; Sato, R.; Kuroiwa, S.; Nakanishi, T.; Nishimura, S.-I.; Osaka, T. Attomolar Detection of Influenza A Virus Hemagglutinin Human H1 and Avian H5 Using Glycan-Blotted Field Effect Transistor Biosensor. Anal. Chem. 2013, 85, 5641–5644. [Google Scholar] [CrossRef]
- Shao, Y.; Wang, J.; Wu, H.; Liu, J.; Aksay, I.A.; Lin, Y. Graphene Based Electrochemical Sensors and Biosensors: A Review. Electroanalysis 2010, 22, 1027–1036. [Google Scholar] [CrossRef]
- Wang, J. Carbon-Nanotube Based Electrochemical Biosensors: A Review. Electroanalysis 2005, 17, 7–14. [Google Scholar] [CrossRef]
- Tuteja, S.K.; Duffield, T.; Neethirajan, S. Graphene-based multiplexed disposable electrochemical biosensor for rapid on-farm monitoring of NEFA and βHBA dairy biomarkers. J. Mater. Chem. B 2017, 5, 6930–6940. [Google Scholar] [CrossRef]
- Bhattacharya, M.; Hong, S.; Lee, D.; Cui, T.; Goyal, S. Carbon nanotube based sensors for the detection of viruses. Sens. Actuators B Chem. 2011, 155, 67–74. [Google Scholar] [CrossRef]
- Lv, S.; Sheng, J.; Zhao, S.; Liu, M.; Chen, L. The detection of brucellosis antibody in whole serum based on the low-fouling electrochemical immunosensor fabricated with magnetic Fe3O4@Au@PEG@HA nanoparticles. Biosens. Bioelectron. 2018, 117, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Fei, J.; Dou, W.; Zhao, G. Amperometric immunoassay for the detection of Salmonella pullorum using a screen—Printed carbon electrode modified with gold nanoparticle-coated reduced graphene oxide and immunomagnetic beads. Mikrochim. Acta 2015, 183, 757–764. [Google Scholar] [CrossRef]
- Haun, J.B.; Yoon, T.-J.; Lee, H.; Weissleder, R. Magnetic nanoparticle biosensors. WIREs Nanomed. Nanobiotechnol. 2010, 2, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Jaffrezic-Renault, N.; Martelet, C.; Chevolot, Y.; Cloarec, J.-P. Biosensors and Bio-Bar Code Assays Based on Biofunctionalized Magnetic Microbeads. Sensors 2007, 7, 589–614. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Wang, J. Optical biosensors: An exhaustive and comprehensive review. Analyst 2020, 145, 1605–1628. [Google Scholar] [CrossRef] [PubMed]
- Borisov, S.M.; Wolfbeis, O.S. Optical Biosensors. Chem. Rev. 2008, 108, 423–461. [Google Scholar] [CrossRef]
- Petrakova, A.V.; Urusov, A.E.; Zherdev, A.; Dzantiev, B.B. Gold nanoparticles of different shape for bicolor lateral flow test. Anal. Biochem. 2018, 568, 7–13. [Google Scholar] [CrossRef]
- Fang, Z.; Wu, W.; Lu, X.; Zeng, L. Lateral flow biosensor for DNA extraction-free detection of salmonella based on aptamer mediated strand displacement amplification. Biosens. Bioelectron. 2014, 56, 192–197. [Google Scholar] [CrossRef]
- Kumar, N.; Bhatia, S.; Pateriya, A.K.; Sood, R.; Nagarajan, S.; Murugkar, H.V.; Kumar, S.; Singh, P.; Singh, V.P. Label-free peptide nucleic acid biosensor for visual detection of multiple strains of influenza A virus suitable for field applications. Anal. Chim. Acta 2019, 1093, 123–130. [Google Scholar] [CrossRef]
- Ali, M.M.; Brown, C.L.; Jahanshahi-Anbuhi, S.; Kannan, B.; Li, Y.; Filipe, C.D.M.; Brennan, J.D. A Printed Multicomponent Paper Sensor for Bacterial Detection. Sci. Rep. 2017, 7, 12335. [Google Scholar] [CrossRef]
- Peedel, D.; Rinken, T. Rapid biosensing of Staphylococcus aureus bacteria in milk. Anal. Methods 2014, 6, 2642–2647. [Google Scholar] [CrossRef]
- Lu, L.; Jun, S. Evaluation of a microwire sensor functionalized to detect Escherichia coli bacterial cells. Biosens. Bioelectron. 2012, 36, 257–261. [Google Scholar] [CrossRef]
- Wagner, A.M.; Knipe, J.M.; Orive, G.; Peppas, N.A. Quantum dots in biomedical applications. Acta Biomater. 2019, 94, 44–63. [Google Scholar] [CrossRef]
- Weng, X.; Chen, L.; Neethirajan, S.; Duffield, T. Development of quantum dots-based biosensor towards on-farm detection of subclinical ketosis. Biosens. Bioelectron. 2015, 72, 140–147. [Google Scholar] [CrossRef]
- Shao, K.; Zhang, C.; Ye, S.; Cai, K.; Wu, L.; Wang, B.; Zou, C.; Lu, Z.; Han, H. Near–infrared electrochemiluminesence biosensor for high sensitive detection of porcine reproductive and respiratory syndrome virus based on cyclodextrin-grafted porous Au/PtAu nanotube. Sens. Actuators B Chem. 2017, 240, 586–594. [Google Scholar] [CrossRef]
- Zhao, Y.; Tong, R.-J.; Xia, F.; Peng, Y. Current status of optical fiber biosensor based on surface plasmon resonance. Biosens. Bioelectron. 2019, 142, 111505. [Google Scholar] [CrossRef]
- Biagetti, M.; Cuccioloni, M.; Bonfili, L.; Cecarini, V.; Sebastiani, C.; Curcio, L.; Giammarioli, M.; De Mia, G.M.; Eleuteri, A.M.; Angeletti, M. Chimeric DNA/LNA-based biosensor for the rapid detection of African swine fever virus. Talanta 2018, 184, 35–41. [Google Scholar] [CrossRef]
- Åkerstedt, M.; Björck, L.; Waller, K.P.; Sternesjo, A. Biosensor assay for determination of haptoglobin in bovine milk. J. Dairy Res. 2006, 73, 299–305. [Google Scholar] [CrossRef] [Green Version]
- Fan, X.; White, I.M.; Shopova, S.I.; Zhu, H.; Suter, J.; Sun, Y. Sensitive optical biosensors for unlabeled targets: A review. Anal. Chim. Acta 2008, 620, 8–26. [Google Scholar] [CrossRef]
- Gómez-Gómez, M.; Sánchez, C.; Peransi, S.; Zurita, D.; Bellieres, L.; Recuero, S.; Rodrigo, M.; Simón, S.; Camarca, A.; Capo, A.; et al. Photonic Label-Free Biosensors for Fast and Multiplex Detection of Swine Viral Diseases. Sensors 2022, 22, 708. [Google Scholar] [CrossRef]
- Luo, B.; Wu, S.; Zou, W.; Zhang, Z.; Zhao, M.; Shi, S.; Liu, Y.; Xi, X.; Zeng, Z.; Liang, W.; et al. Label-free immunoassay for porcine circovirus type 2 based on excessively tilted fiber grating modified with staphylococcal protein A. Biosens. Bioelectron. 2016, 86, 1054–1060. [Google Scholar] [CrossRef] [Green Version]
- Qi, C.; Tian, X.-S.; Chen, S.; Yan, J.-H.; Cao, Z.; Tian, K.-G.; Gao, G.F.; Jin, G. Detection of avian influenza virus subtype H5 using a biosensor based on imaging ellipsometry. Biosens. Bioelectron. 2010, 25, 1530–1534. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.Y.; Penn, L.S.; Xi, J. Quartz crystal microbalance: Sensing cell-substrate adhesion and beyond. Biosens. Bioelectron. 2017, 99, 593–602. [Google Scholar] [CrossRef]
- Hewa, T.M.P.; Tannock, G.A.; Mainwaring, D.E.; Harrison, S.; Fecondo, J.V. The detection of influenza A and B viruses in clinical specimens using a quartz crystal microbalance. J. Virol. Methods 2009, 162, 14–21. [Google Scholar] [CrossRef]
- Wang, R.; Wang, L.; Callaway, Z.T.; Lu, H.; Huang, T.J.; Li, Y. A nanowell-based QCM aptasensor for rapid and sensitive detection of avian influenza virus. Sens. Actuators B Chem. 2016, 240, 934–940. [Google Scholar] [CrossRef] [Green Version]
- Bayramoglu, G.; Ozalp, C.; Oztekin, M.; Arica, M.Y. Rapid and label-free detection of Brucella melitensis in milk and milk products using an aptasensor. Talanta 2019, 200, 263–271. [Google Scholar] [CrossRef]
- Barnett, J.M.; Monnier, B.M.; Tyler, S.; West, D.; Ballantine-Dykes, H.; Regan, E.; Wraith, P.; Kiely, J.; Luxton, R. Initial trail results of a magnetic biosensor for the rapid detection of Porcine Reproductive and Respiratory Virus (PRRSV) infection. Sens. Bio-Sens. Res. 2019, 27, 100315. [Google Scholar] [CrossRef]
- Krishna, V.D.; Wu, K.; Perez, A.M.; Wang, J.-P. Giant Magnetoresistance-based Biosensor for Detection of Influenza A Virus. Front. Microbiol. 2016, 7, 400. [Google Scholar] [CrossRef] [Green Version]
- Chai, Y.; Li, S.; Horikawa, S.; Park, M.-K.; Vodyanoy, V.; Chin, B.A. Rapid and Sensitive Detection of Salmonella Typhimurium on Eggshells by Using Wireless Biosensors. J. Food Prot. 2012, 75, 631–636. [Google Scholar] [CrossRef]
- Fritz, J. Cantilever biosensors. Analyst 2008, 133, 855–863. [Google Scholar] [CrossRef]
- Jia, K.; Toury, T.; Ionescu, R.E. Fabrication of an atrazine acoustic immunosensor based on a drop-deposition procedure. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2012, 59, 2015–2021. [Google Scholar] [CrossRef] [PubMed]
- Grenvall, C.; Folkenberg, J.R.; Augustsson, P.; Laurell, T. Label-free somatic cell cytometry in raw milk using acoustophoresis. Cytom. Part A 2012, 81, 1076–1083. [Google Scholar] [CrossRef] [PubMed]
- Zarei, M. Portable biosensing devices for point-of-care diagnostics: Recent developments and applications. TrAC Trends Anal. Chem. 2017, 91, 26–41. [Google Scholar] [CrossRef]
- Tan, X.; Ding, S.-Q.; Hu, Y.-X.; Li, J.-J.; Zhou, J.-Y. Development of an immunosensor assay for detection of haptoglobin in mastitic milk. Veter. Clin. Pathol. 2012, 41, 575–581. [Google Scholar] [CrossRef]
- Tuteja, S.K.; Neethirajan, S. Exploration of two-dimensional bio-functionalized phosphorene nanosheets (black phosphorous) for label free haptoglobin electro-immunosensing applications. Nanotechnology 2018, 29, 135101. [Google Scholar] [CrossRef]
- Nidzworski, D.; Pranszke, P.; Grudniewska, M.; Krol, E.; Gromadzka, B. Universal biosensor for detection of influenza virus. Biosens. Bioelectron. 2014, 59, 239–242. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Wang, R.; Jiao, P.; Li, Y.; Li, Y.; Liao, M.; Yu, Y.; Wang, M. An impedance immunosensor based on low-cost microelectrodes and specific monoclonal antibodies for rapid detection of avian influenza virus H5N1 in chicken swabs. Biosens. Bioelectron. 2015, 67, 546–552. [Google Scholar] [CrossRef]
- Fu, Y.; Callaway, Z.; Lum, J.; Wang, R.; Lin, J.; Li, Y. Exploiting Enzyme Catalysis in Ultra-Low Ion Strength Media for Impedance Biosensing of Avian Influenza Virus Using a Bare Interdigitated Electrode. Anal. Chem. 2013, 86, 1965–1971. [Google Scholar] [CrossRef]
- Diouani, M.; Helali, S.; Hafaid, I.; Hassen, W.; Snoussi, M.; Ghram, A.; Jaffrezic-Renault, N.; Abdelghani, A. Miniaturized biosensor for avian influenza virus detection. Mater. Sci. Eng. C 2008, 28, 580–583. [Google Scholar] [CrossRef]
- Dong, S.; Zhao, R.; Zhu, J.; Lu, X.; Li, Y.; Qiu, S.; Jia, L.; Jiao, X.; Song, S.; Fan, C.; et al. Electrochemical DNA Biosensor Based on a Tetrahedral Nanostructure Probe for the Detection of Avian Influenza A (H7N9) Virus. ACS Appl. Mater. Interfaces 2015, 7, 8834–8842. [Google Scholar] [CrossRef]
- Krejcova, L.; Nejdl, L.; Rodrigo, M.A.M.; Zurek, M.; Matousek, M.; Hynek, D.; Zitka, O.; Kopel, P.; Adam, V.; Kizek, R. 3D printed chip for electrochemical detection of influenza virus labeled with CdS quantum dots. Biosens. Bioelectron. 2013, 54, 421–427. [Google Scholar] [CrossRef]
- Niamh, A.M.; Sayers, C.R.; O’Riordan, S.B.A.A. Novel Single Gold Nanowire-based Electrochemical Immunosensor for Rapid Detection of Bovine Viral Diarrhoea Antibodies in Serum. J. Biosens. Bioelectron. 2015, 6, 3. [Google Scholar] [CrossRef]
- Creedon, N.; Sayers, R.; O’sullivan, B.; Kennedy, E.; Lovera, P.; O’riordan, A. Label-Free Impedimetric Nanoband Sensor for Detection of Both Bovine Viral Diarrhoea Virus (BVDV) and Antibody (BVDAb) in Serum. 2021. Available online: https://chemrxiv.org/engage/chemrxiv/article-details/60c73f5c337d6ce8fae26511 (accessed on 9 May 2022).
- Ahmed, S.R.; Mogus, J.; Chand, R.; Nagy, E.; Neethirajan, S. Optoelectronic fowl adenovirus detection based on local electric field enhancement on graphene quantum dots and gold nanobundle hybrid. Biosens. Bioelectron. 2018, 103, 45–53. [Google Scholar] [CrossRef]
- Huan, T.N.; Ha, V.T.T.; Hung, L.Q.; Yoon, M.-Y.; Han, S.-H.; Chung, H. Square wave voltammetric detection of Anthrax utilizing a peptide for selective recognition of a protein biomarker. Biosens. Bioelectron. 2009, 25, 469–474. [Google Scholar] [CrossRef]
- Wang, H.; Yuan, R.; Chai, Y.; Cao, Y.; Gan, X.; Chen, Y.; Wang, Y. An ultrasensitive peroxydisulfate electrochemiluminescence immunosensor for Streptococcus suis serotype 2 based on l-cysteine combined with mimicking bi-enzyme synergetic catalysis to in situ generate coreactant. Biosens. Bioelectron. 2012, 43, 63–68. [Google Scholar] [CrossRef]
- El Ichi, S.; Leon, F.; Vossier, L.; Marchandin, H.; Errachid, A.; Coste, J.; Jaffrezic-Renault, N.; Fournier-Wirth, C. Microconductometric immunosensor for label-free and sensitive detection of Gram-negative bacteria. Biosens. Bioelectron. 2014, 54, 378–384. [Google Scholar] [CrossRef]
- Liébana, S.; Lermo, A.; Campoy, S.; Cortés, M.P.; Alegret, S.; Pividori, M.I. Rapid detection of Salmonella in milk by electrochemical magneto-immunosensing. Biosens. Bioelectron. 2009, 25, 510–513. [Google Scholar] [CrossRef]
- Wu, H.; Zuo, Y.; Cui, C.; Yang, W.; Ma, H.; Wang, X. Rapid Quantitative Detection of Brucella melitensis by a Label-Free Impedance Immunosensor Based on a Gold Nanoparticle-Modified Screen-Printed Carbon Electrode. Sensors 2013, 13, 8551–8563. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.; Ansari, Z.A.; Seo, H.K.; Ansari, S.G. Synthesis and Application of Cu-Doped Nickel and Zirconium Oxide Nanoparticles as Brucella abortus Electrochemical Device Development. Sens. Lett. 2018, 16, 267–276. [Google Scholar] [CrossRef]
- Silva, M.G.; Helali, S.; Esseghaier, C.; Suarez, C.E.; Oliva, A.; Abdelghani, A. An impedance spectroscopy method for the detection and evaluation of Babesia bovis antibodies in cattle. Sens. Actuators B Chem. 2008, 135, 206–213. [Google Scholar] [CrossRef]
- Tseng, Y.-T.; Wang, C.-H.; Chang, C.-P.; Lee, G.-B. Integrated microfluidic system for rapid detection of influenza H1N1 virus using a sandwich-based aptamer assay. Biosens. Bioelectron. 2016, 82, 105–111. [Google Scholar] [CrossRef]
- Su, L.-C.; Chang, C.-M.; Tseng, Y.-L.; Chang, Y.-F.; Li, Y.-C.; Chang, Y.-S.; Chou, C. Rapid and Highly Sensitive Method for Influenza A (H1N1) Virus Detection. Anal. Chem. 2012, 84, 3914–3920. [Google Scholar] [CrossRef]
- Ahmed, S.R.; Nagy, É.; Neethirajan, S. Self-assembled star-shaped chiroplasmonic gold nanoparticles for an ultrasensitive chiro-immunosensor for viruses. RSC Adv. 2017, 7, 40849–40857. [Google Scholar] [CrossRef] [Green Version]
- Weng, X.; Neethirajan, S. Immunosensor Based on Antibody-Functionalized MoS2 for Rapid Detection of Avian Coronavirus on Cotton Thread. IEEE Sens. J. 2018, 18, 4358–4363. [Google Scholar] [CrossRef]
- Lu, T.; Ma, Q.; Yan, W.; Wang, Y.; Zhang, Y.; Zhao, L.; Chen, H. Selection of an aptamer against Muscovy duck parvovirus for highly sensitive rapid visual detection by label-free aptasensor. Talanta 2018, 176, 214–220. [Google Scholar] [CrossRef]
- Stringer, R.C.; Schommer, S.; Hoehn, D.; Grant, S.A. Development of an optical biosensor using gold nanoparticles and quantum dots for the detection of Porcine Reproductive and Respiratory Syndrome Virus. Sens. Actuators B Chem. 2008, 134, 427–431. [Google Scholar] [CrossRef]
- Chen, L.; Ye, S.; Cai, K.; Zhang, C.; Zhou, G.; He, Z.; Han, H. An aqueous platinum nanotube based fluorescent immuno-assay for porcine reproductive and respiratory syndrome virus detection. Talanta 2015, 144, 324–328. [Google Scholar] [CrossRef]
- Heinze, B.C.; Song, J.-Y.; Lee, C.-H.; Najam, A.; Yoon, J.-Y. Microfluidic immunosensor for rapid and sensitive detection of bovine viral diarrhea virus. Sens. Actuators B Chem. 2009, 138, 491–496. [Google Scholar] [CrossRef]
- Jain, B.; Lambe, U.; Tewari, A.; Kadian, S.K.; Prasad, M. Development of a rapid test for detection of foot-and-mouth disease virus specific antibodies using gold nanoparticles. Virusdisease 2018, 29, 192–198. [Google Scholar] [CrossRef]
- Lopez, C.A.; Daaboul, G.G.; Vedula, R.S.; Özkumur, E.; Bergstein, D.A.; Geisbert, T.W.; Fawcett, H.E.; Goldberg, B.B.; Connor, J.; Ünlü, M.S. Label-free multiplexed virus detection using spectral reflectance imaging. Biosens. Bioelectron. 2011, 26, 3432–3437. [Google Scholar] [CrossRef] [Green Version]
- McCutcheon, K.; Bandara, A.B.; Zuo, Z.; Heflin, J.R.; Inzana, T.J. The Application of a Nanomaterial Optical Fiber Biosensor Assay for Identification of Brucella Nomenspecies. Biosensors 2019, 9, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ko, S.; Grant, S.A. A novel FRET-based optical fiber biosensor for rapid detection of Salmonella typhimurium. Biosens. Bioelectron. 2006, 21, 1283–1290. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Caterer, N.R.; Xu, W.; Goolia, M. Development of a multiplex lateral flow strip test for foot-and-mouth disease virus detection using monoclonal antibodies. J. Virol. Methods 2015, 221, 119–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, M.; Zhmendak, D.; Mioulet, V.; King, D.P.; Burman, A.; Nfon, C.K. Combining a Universal Capture Ligand and Pan-Serotype Monoclonal Antibody to Develop a Pan-Serotype Lateral Flow Strip Test for Foot-and-Mouth Disease Virus Detection. Viruses 2022, 14, 785. [Google Scholar] [CrossRef]
- Agrawal, A.; Varshney, R.; Gattani, A.; Kirthika, P.; Khan, M.H.; Singh, R.; Kodape, S.; Patel, S.K.; Singh, P. Gold nanoparticle based immunochromatographic biosensor for rapid diagnosis of Mycobacterium avium subspecies paratuberculosis infection using recombinant protein. J. Microbiol. Methods 2020, 177, 106024. [Google Scholar] [CrossRef]
- Kim, S.H.; Lee, J.; Lee, B.H.; Song, C.-S.; Gu, M.B. Specific detection of avian influenza H5N2 whole virus particles on lateral flow strips using a pair of sandwich-type aptamers. Biosens. Bioelectron. 2019, 134, 123–129. [Google Scholar] [CrossRef]
- Li, X.; Lu, D.; Sheng, Z.; Chen, K.; Guo, X.; Jin, M.; Han, H.-Y. A fast and sensitive immunoassay of avian influenza virus based on label-free quantum dot probe and lateral flow test strip. Talanta 2012, 100, 1–6. [Google Scholar] [CrossRef]
- Wu, F.; Yuan, H.; Zhou, C.; Mao, M.; Liu, Q.; Shen, H.; Cen, Y.; Qin, Z.; Ma, L.; Li, L.S. Multiplexed detection of influenza A virus subtype H5 and H9 via quantum dot-based immunoassay. Biosens. Bioelectron. 2015, 77, 464–470. [Google Scholar] [CrossRef]
- Hanon, J.-B.; Vandenberge, V.; Deruelle, M.; De Leeuw, I.; De Clercq, K.; Van Borm, S.; Koenen, F.; Liu, L.; Hoffmann, B.; Batten, C.A.; et al. Inter-laboratory evaluation of the performance parameters of a Lateral Flow Test device for the detection of Bluetongue virus-specific antibodies. J. Virol. Methods 2016, 228, 140–150. [Google Scholar] [CrossRef]
- Huang, L.; Xiao, W.; Xu, T.; Chen, H.; Jin, Z.; Zhang, Z.; Song, Q.; Tang, Y. Miniaturized Paper-Based Smartphone Biosensor for Differential Diagnosis of Wild-type Pseudorabies Virus Infection versus Vaccination Immunization. Sens. Actuators B Chem. 2020, 327, 128893. [Google Scholar] [CrossRef]
- James, H.E.; Ebert, K.; McGonigle, R.; Reid, S.M.; Boonham, N.; Tomlinson, J.; Hutchings, G.H.; Denyer, M.; Oura, C.A.; Dukes, J.P.; et al. Detection of African swine fever virus by loop-mediated isothermal amplification. J. Virol. Methods 2010, 164, 68–74. [Google Scholar] [CrossRef]
- Choi, J.R.; Hu, J.; Tang, R.; Gong, Y.; Feng, S.; Ren, H.; Wen, T.; Li, X.; Abas, W.A.B.W.; Pingguan-Murphy, B.; et al. An integrated paper-based sample-to-answer biosensor for nucleic acid testing at the point of care. Lab Chip 2015, 16, 611–621. [Google Scholar] [CrossRef]
- Fu, Q.; Yuan, L.; Cao, F.; Zang, L.; Ji, D. Lateral flow strip biosensor based on streptavidin-coated gold nanoparticles with recombinase polymerase amplification for the quantitative point-of-care testing of Salmonella. Microchem. J. 2021, 171, 106859. [Google Scholar] [CrossRef]
- Eltzov, E.; Marks, R.S. Miniaturized Flow Stacked Immunoassay for Detecting Escherichia coli in a Single Step. Anal. Chem. 2016, 88, 6441–6449. [Google Scholar] [CrossRef]
- Eltzov, E.; Marks, R.S. Colorimetric stack pad immunoassay for bacterial identification. Biosens. Bioelectron. 2017, 87, 572–578. [Google Scholar] [CrossRef]
- Shen, H.; Chen, H.; Cheng, Z.; Ma, L.; Huang, L.; Xiao, M.; Xiao, W.; Xie, K.; Tang, Y. A novel fluorescent immunochromatographic strip combined with pocket fluorescence observation instrument for rapid detection of PRV. Anal. Bioanal. Chem. 2018, 410, 7655–7661. [Google Scholar] [CrossRef]
- Xiao, W.; Huang, C.; Xu, F.; Yan, J.; Bian, H.; Fu, Q.; Xie, K.; Wang, L.; Tang, Y. A simple and compact smartphone-based device for the quantitative readout of colloidal gold lateral flow immunoassay strips. Sens. Actuators B Chem. 2018, 266, 63–70. [Google Scholar] [CrossRef]
- Hou, P.; Zhao, G.; Wang, H.; He, C.; Huan, Y.; He, H. Development of a recombinase polymerase amplification combined with lateral-flow dipstick assay for detection of bovine ephemeral fever virus. Mol. Cell. Probes 2017, 38, 31–37. [Google Scholar] [CrossRef]
- Kameyama, K.; Sakoda, Y.; Tamai, K.; Igarashi, H.; Tajima, M.; Mochizuki, T.; Namba, Y.; Kida, H. Development of an immunochromatographic test kit for rapid detection of bovine viral diarrhea virus antigen. J. Virol. Methods 2006, 138, 140–146. [Google Scholar] [CrossRef]
- Ferris, N.P.; Nordengrahn, A.; Hutchings, G.H.; Reid, S.M.; King, D.; Ebert, K.; Paton, D.J.; Kristersson, T.; Brocchi, E.; Grazioli, S.; et al. Development and laboratory validation of a lateral flow device for the detection of foot-and-mouth disease virus in clinical samples. J. Virol. Methods 2009, 155, 10–17. [Google Scholar] [CrossRef]
- Ferris, N.P.; Nordengrahn, A.; Hutchings, G.H.; Paton, D.J.; Kristersson, T.; Brocchi, E.; Grazioli, S.; Merza, M. Development and laboratory validation of a lateral flow device for the detection of serotype SAT 2 foot-and-mouth disease viruses in clinical samples. J. Virol. Methods 2010, 163, 474–476. [Google Scholar] [CrossRef]
- Waters, R.A.; Fowler, V.L.; Armson, B.; Nelson, N.; Gloster, J.; Paton, D.J.; King, D. Preliminary Validation of Direct Detection of Foot-And-Mouth Disease Virus within Clinical Samples Using Reverse Transcription Loop-Mediated Isothermal Amplification Coupled with a Simple Lateral Flow Device for Detection. PLoS ONE 2014, 9, e105630. [Google Scholar] [CrossRef]
- Wang, H.-M.; Zhao, G.-M.; Hou, P.-L.; Yu, L.; He, C.-Q.; He, H.-B. Rapid detection of foot-and-mouth disease virus using reverse transcription recombinase polymerase amplification combined with a lateral flow dipstick. J. Virol. Methods 2018, 261, 46–50. [Google Scholar] [CrossRef]
- Miao, F.; Zhang, J.; Li, N.; Chen, T.; Wang, L.; Zhang, F.; Mi, L.; Zhang, J.; Wang, S.; Wang, Y.; et al. Rapid and Sensitive Recombinase Polymerase Amplification Combined With Lateral Flow Strip for Detecting African Swine Fever Virus. Front. Microbiol. 2019, 10, 1004. [Google Scholar] [CrossRef]
- Xie, K.; Chen, H.; Peng, B.; Jin, Z.; Xiao, W.; Zhang, Z.; Huang, B.; Song, Q.; Tang, Y. On-Site Determination of Classical Swine Fever Virus (CSFV) by a Fluorescent Microsphere-Based Lateral Flow Immunoassay Strip (FM-LFIAs) Based on Monoclonal Antibodies. Anal. Lett. 2020, 54, 2347–2362. [Google Scholar] [CrossRef]
- Chowdry, V.K.; Luo, Y.; Widén, F.; Qiu, H.-J.; Shan, H.; Belák, S.; Liu, L. Development of a loop-mediated isothermal amplification assay combined with a lateral flow dipstick for rapid and simple detection of classical swine fever virus in the field. J. Virol. Methods 2014, 197, 14–18. [Google Scholar] [CrossRef]
- Jin, Q.; Yang, J.; Lu, Q.; Guo, J.; Deng, R.; Wang, Y.; Wang, S.; Wang, S.; Chen, W.; Zhi, Y.; et al. Development of an immunochromatographic strip for the detection of antibodies against Porcine circovirus-2. J. Veter. Diagn. Investig. 2012, 24, 1151–1157. [Google Scholar] [CrossRef] [Green Version]
- Cui, S.; Zhou, S.; Chen, C.; Qi, T.; Zhang, C.; Oh, J. A simple and rapid immunochromatographic strip test for detecting antibody to porcine reproductive and respiratory syndrome virus. J. Virol. Methods 2008, 152, 38–42. [Google Scholar] [CrossRef]
- Mei, X.; Zhai, X.; Lei, C.; Ye, X.; Kang, Z.; Wu, X.; Xiang, R.; Wang, Y.; Wang, H. Development and application of a visual loop-mediated isothermal amplification combined with lateral flow dipstick (LAMP-LFD) method for rapid detection of Salmonella strains in food samples. Food Control 2019, 104, 9–19. [Google Scholar] [CrossRef]
- Hu, J.; Huang, R.; Sun, Y.; Wei, X.; Wang, Y.; Jiang, C.; Geng, Y.; Sun, X.; Jing, J.; Gao, H.; et al. Sensitive and rapid visual detection of Salmonella Typhimurium in milk based on recombinase polymerase amplification with lateral flow dipsticks. J. Microbiol. Methods 2019, 158, 25–32. [Google Scholar] [CrossRef]
- Li, S.; Liu, Y.; Wang, Y.; Wang, M.; Liu, C.; Wang, Y. Rapid Detection of Brucella spp. and Elimination of Carryover Using Multiple Cross Displacement Amplification Coupled With Nanoparticles-Based Lateral Flow Biosensor. Front. Cell. Infect. Microbiol. 2019, 9, 78. [Google Scholar] [CrossRef] [PubMed]
- Wadl, M.; Pölzler, T.; Flekna, G.; Thompson, L.; Slaghuis, J.; Köfer, J.; Hein, I.; Wagner, M. Easy-to-Use Rapid Test for Direct Detection of Campylobacter spp. in Chicken Feces. J. Food Prot. 2009, 72, 2483–2488. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Wang, H.; Hou, P.; He, C.; He, H. Rapid Visual Detection of Mycobacterium Avium Subsp. Paratuberculosis by Recombinase Polymerase Amplification Combined with a Lateral Flow Dipstick. J. Vet. Sci. 2018, 19, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Cao, J.; Zhu, M.; Xu, M.; Shi, F. Loop-mediated isothermal amplification-lateral-flow dipstick (LAMP-LFD) to detect Mycoplasma ovipneumoniae. World J. Microbiol. Biotechnol. 2019, 35, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Cordero, J.L.; Barrett, L.M.; O’Kennedy, R.; Ricco, A.J. Microfluidic sedimentation cytometer for milk quality and bovine mastitis monitoring. Biomed. Microdevices 2010, 12, 1051–1059. [Google Scholar] [CrossRef]
- Kim, B.; Lee, Y.J.; Park, J.G.; Yoo, D.; Hahn, Y.K.; Choi, S. A portable somatic cell counter based on a multi-functional counting chamber and a miniaturized fluorescence microscope. Talanta 2017, 170, 238–243. [Google Scholar] [CrossRef]
- Moon, J.; Koo, H.; Joo, Y.; Jeon, S.; Hur, D.; Chung, C.; Jo, H.; Park, Y. Application of a New Portable Microscopic Somatic Cell Counter with Disposable Plastic Chip for Milk Analysis. J. Dairy Sci. 2007, 90, 2253–2259. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, A.C.; Duarte, C.M.; Cardoso, F.A.; Bexiga, R.; Cardoso, S.; Freitas, P.P. Lab-on-Chip Cytometry Based on Magnetoresistive Sensors for Bacteria Detection in Milk. Sensors 2014, 14, 15496–15524. [Google Scholar] [CrossRef] [Green Version]
- Duarte, C.; Costa, T.; Carneiro, C.; Soares, R.; Jitariu, A.; Cardoso, S.; Piedade, M.; Bexiga, R.; Freitas, P. Semi-Quantitative Method for Streptococci Magnetic Detection in Raw Milk. Biosensors 2016, 6, 19. [Google Scholar] [CrossRef] [Green Version]
- Takekawa, J.Y.; Iverson, S.A.; Schultz, A.K.; Hill, N.J.; Cardona, C.J.; Boyce, W.M.; Dudley, J.P. Field detection of avian influenza virus in wild birds: Evaluation of a portable rRT-PCR system and freeze-dried reagents. J. Virol. Methods 2010, 166, 92–97. [Google Scholar] [CrossRef]
- Jung, J.H.; Park, B.H.; Oh, S.J.; Choi, G.; Seo, T.S. Integrated centrifugal reverse transcriptase loop-mediated isothermal amplification microdevice for influenza A virus detection. Biosens. Bioelectron. 2014, 68, 218–224. [Google Scholar] [CrossRef]
- Fang, X.; Liu, Y.; Kong, J.; Jiang, X. Loop-Mediated Isothermal Amplification Integrated on Microfluidic Chips for Point-of-Care Quantitative Detection of Pathogens. Anal. Chem. 2010, 82, 3002–3006. [Google Scholar] [CrossRef]
- El Wahed, A.A.; Weidmann, M.; Hufert, F.T. Diagnostics-in-a-Suitcase: Development of a portable and rapid assay for the detection of the emerging avian influenza A (H7N9) virus. J. Clin. Virol. 2015, 69, 16–21. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.-H.; Park, J.; Kim, C.-J.; Cho, Y.-K. Fully Integrated Lab-on-a-Disc for Nucleic Acid Analysis of Food-Borne Pathogens. Anal. Chem. 2014, 86, 3841–3848. [Google Scholar] [CrossRef]
- Ye, X.; Li, L.; Li, J.; Wu, X.; Fang, X.; Kong, J. Microfluidic-CFPA Chip for the Point-of-Care Detection of African Swine Fever Virus with a Median Time to Threshold in about 10 min. ACS Sens. 2019, 4, 3066–3071. [Google Scholar] [CrossRef]
- He, Q.; Yu, D.; Bao, M.; Korensky, G.; Chen, J.; Shin, M.; Kim, J.; Park, M.; Qin, P.; Du, K. High-throughput and all-solution phase African Swine Fever Virus (ASFV) detection using CRISPR-Cas12a and fluorescence based point-of-care system. Biosens. Bioelectron. 2020, 154, 112068. [Google Scholar] [CrossRef]
- Cui, X.; Hu, J.; Choi, J.R.; Huang, Y.; Wang, X.; Lu, T.J.; Xu, F. A volumetric meter chip for point-of-care quantitative detection of bovine catalase for food safety control. Anal. Chim. Acta 2016, 935, 207–212. [Google Scholar] [CrossRef]
- Jeong-Woo Choi; Kim Young-Kee; Kim Hee-Joo; Lee Woochang; Seong Gi-Hun Lab-on-a-Chip for Monitoring the Quality of Raw Milk. J. Microbiol. Biotechnol. 2006, 16, 1229–1235.
- Bhatta, D.; Villalba, M.M.; Johnson, C.; Emmerson, G.; Ferris, N.; King, D.; Lowe, C. Rapid Detection of Foot-and-Mouth Disease Virus with Optical Microchip Sensors. Procedia Chem. 2012, 6, 2–10. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Li, W.; Wang, T.; Lin, Z.; Jiang, M.; Hu, F. Development of a label-free and innovative approach based on surface plasmon resonance biosensor for on-site detection of infectious bursal disease virus (IBDV). Biosens. Bioelectron. 2012, 31, 475–479. [Google Scholar] [CrossRef]
- Hu, J.; Wang, T.; Wang, S.; Chen, M.; Wang, M.; Mu, L.; Chen, H.; Hu, X.; Liang, H.; Zhu, J.; et al. Development of a Surface Plasmon Resonance Biosensing Approach for the Rapid Detection of Porcine Circovirus Type2 in Sample Solutions. PLoS ONE 2014, 9, e111292. [Google Scholar] [CrossRef]
- Manessis, G.; Mourouzis, C.; Griol, A.; Zurita-Herranz, D.; Peransi, S.; Sanchez, C.; Giusti, A.; Gelasakis, A.I.; Bossis, I. Integration of Microfluidics, Photonic Integrated Circuits and Data Acquisition and Analysis Methods in a Single Platform for the Detection of Swine Viral Diseases. Animals 2021, 11, 3193. [Google Scholar] [CrossRef]
- Manessis, G.; Frant, M.; Wozniakowski, G.; Nannucci, L.; Benedetti, M.; Denes, L.; Gyula, B.; Gelasakis, A.I.; Squires, C.; Recuero, S.; et al. Point-of-Care and Label-Free Detection of Porcine Reproductive and Respiratory Syndrome and Swine Influenza Viruses Using a Microfluidic Device with Photonic Integrated Circuits. Viruses 2022, 14, 988. [Google Scholar] [CrossRef]
- Potockova, H.; Dohnal, J.; Thome-Kromer, B. Regulation of veterinary point-of-care testing in the European Union, the United States of America and Japan. Rev. Sci. Tech. 2020, 39, 699–709. [Google Scholar] [CrossRef]
- Morgan, A.P. Regulatory Control of Veterinary Diagnostic Test Kits. Rev. Sci. Tech. 1998, 17, 562–567. [Google Scholar] [CrossRef]
- Teles, F.; Fonseca, L. Nucleic-Acid Testing, New Platforms and Nanotechnology for Point-of-Decision Diagnosis of Animal Pathogens. In Veterinary Infection Biology: Molecular Diagnostics and High-Throughput Strategies; Springer: New York, NY, USA, 2014; pp. 253–283. ISBN 9781493920044. [Google Scholar]
- Chin, C.D.; Linder, V.; Sia, S.K. Commercialization of microfluidic point-of-care diagnostic devices. Lab Chip 2012, 12, 2118–2134. [Google Scholar] [CrossRef]
- Srinivasan, B.; Tung, S. Development and Applications of Portable Biosensors. J. Lab. Autom. 2015, 20, 365–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hobbs, E.C.; Colling, A.; Gurung, R.B.; Allen, J. The potential of diagnostic point-of-care tests (POCTs) for infectious and zoonotic animal diseases in developing countries: Technical, regulatory and sociocultural considerations. Transbound. Emerg. Dis. 2020, 68, 1835–1849. [Google Scholar] [CrossRef] [PubMed]
- Glas, A.S.; Lijmer, J.G.; Prins, M.H.; Bonsel, G.J.; Bossuyt, P.M.M. The diagnostic odds ratio: A single indicator of test performance. J. Clin. Epidemiol. 2003, 56, 1129–1135. [Google Scholar] [CrossRef]
- Neethirajan, S. Transforming the Adaptation Physiology of Farm Animals through Sensors. Animals 2020, 10, 1512. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, J.; Dunn, S.; Best, A.; Mirza, J.; Percival, B.; Mayhew, M.; Megram, O.; Ashford, F.; White, T.; Moles-Garcia, E.; et al. Validation testing to determine the sensitivity of lateral flow testing for asymptomatic SARS-CoV-2 detection in low prevalence settings: Testing frequency and public health messaging is key. PLoS Biol. 2021, 19, e3001216. [Google Scholar] [CrossRef]
- Guo, J.; Chen, S.; Guo, J.; Ma, X. Nanomaterial Labels in Lateral Flow Immunoassays for Point-of-Care-Testing. J. Mater. Sci. Technol. 2020, 60, 90–104. [Google Scholar] [CrossRef]
- Nguyen, Q.H.; Kim, M.I. Nanomaterial-Mediated Paper-Based Biosensors for Colorimetric Pathogen Detection. TrAC Trends Anal. Chem. 2020, 132, 116038. [Google Scholar] [CrossRef]
- Lou, D.; Fan, L.; Jiang, T.; Zhang, Y. Advances in nanoparticle-based lateral flow immunoassay for point-of-care testing. View 2022, 3, 20200125. [Google Scholar] [CrossRef]
- Ye, H.; Liu, Y.; Zhan, L.; Liu, Y.; Qin, Z. Signal amplification and quantification on lateral flow assays by laser excitation of plasmonic nanomaterials. Theranostics 2020, 10, 4359–4373. [Google Scholar] [CrossRef]
- Denmark, D.J.; Mohapatra, S.; Mohapatra, S.S. Point-of-Care Diagnostics: Molecularly Imprinted Polymers and Nanomaterials for Enhanced Biosensor Selectivity and Transduction. EuroBiotech J. 2020, 4, 184–206. [Google Scholar] [CrossRef]
- Pirzada, M.; Altintas, Z. Nanomaterials for Healthcare Biosensing Applications. Sensors 2019, 19, 5311. [Google Scholar] [CrossRef] [Green Version]
- Mejía-Salazar, J.R.; Rodrigues Cruz, K.; Materon Vasques, E.M. Microfluidic Point-of-Care Devices: New Trends and Future Prospects for eHealth Diagnostics. Sensors 2020, 20, 1951. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.-M.; Lv, S.; Zhang, W.; Cui, Y. Microfluidic Point-of-Care (POC) Devices in Early Diagnosis: A Review of Opportunities and Challenges. Sensors 2022, 22, 1620. [Google Scholar] [CrossRef]
- Liu, D.; Wang, J.; Wu, L.; Huang, Y.; Zhang, Y.; Zhu, M.; Wang, Y.; Zhu, Z.; Yang, C. Trends in miniaturized biosensors for point-of-care testing. TrAC Trends Anal. Chem. 2019, 122, 115701. [Google Scholar] [CrossRef]
- Tran, V.; Walkenfort, B.; König, M.; Salehi, M.; Schlücker, S. Schnelle, quantitative und hochempfindliche patientennahe Labordiagnostik: Ein tragbares Raman-Lesegerät für seitliche Flusstests in der klinischen Chemie. Angew. Chem. 2018, 131, 450–455. [Google Scholar] [CrossRef]
- Ong, D.; Poljak, M. Smartphones as mobile microbiological laboratories. Clin. Microbiol. Infect. 2020, 26, 421–424. [Google Scholar] [CrossRef]
- Xu, X.; Akay, A.; Wei, H.; Wang, S.; Pingguan-Murphy, B.; Erlandsson, B.-E.; Li, X.; Lee, W.; Hu, J.; Wang, L.; et al. Advances in Smartphone-Based Point-of-Care Diagnostics. In Proceedings of the IEEE, Santiago, Chile, 7–13 December 2015; Volume 103, pp. 236–247. [Google Scholar] [CrossRef]
- Gil Rosa, B.; Akingbade, O.E.; Guo, X.; Gonzalez-Macia, L.; Crone, M.A.; Cameron, L.P.; Freemont, P.; Choy, K.-L.; Güder, F.; Yeatman, E.; et al. Multiplexed immunosensors for point-of-care diagnostic applications. Biosens. Bioelectron. 2022, 203, 114050. [Google Scholar] [CrossRef]
- Vashist, S.K. Trends in Multiplex Immunoassays for In Vitro Diagnostics and Point-of-Care Testing. Diagnostics 2021, 11, 1630. [Google Scholar] [CrossRef] [PubMed]


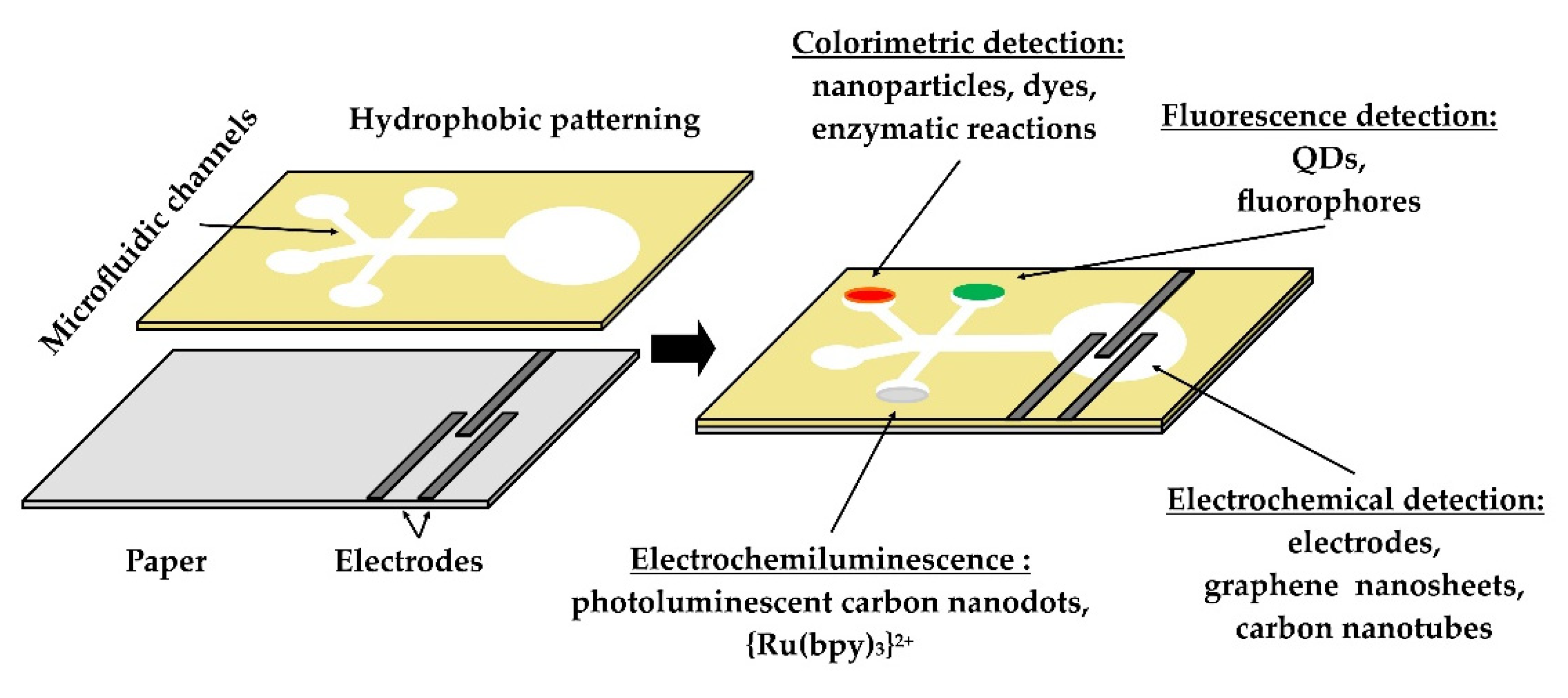

| Targeted Analyte | Recognition Element | Materials | Detection Technique | Detection Matrix | Performance | Reference |
|---|---|---|---|---|---|---|
| Electrochemical biosensors | ||||||
| Haptoglobin | Goat anti-bovine Hp polyclonal antibody (Abcam) | Functionalized gold electrode | Amperometric detection | Skimmed milk | LOD 1 of 0.63 ng/mL. Linear response range: 15–100 mg/L. Detection in 5 min | [104] |
| Haptoglobin | Anti-Hp antibody | Functionalized liquid-exfoliated two-dimensional phosphorene (Ph) nanosheets electrodeposited on screen-printed electrode | Differential pulse voltammetry | Spiked serum samples | LOD of 11 ng/mL. Linear response range: 10–10 × 103 ng/mL. Detection in 60 s | [105] |
| Mouse polyclonal anti-M1 antibodies | Functionalized gold electrodes | Electrochemical impedance spectroscopy | PBS 2 buffer | LOD of 2 × 10−2 ng/mL. Detection in 30 min | [106] | |
| Avian influenza A H5N1 virus | Monoclonal antibodies (produced in mouse myeloma cells) | Protein-A-modified interdigitated array microelectrode | Electrochemical impedance spectroscopy | Tracheal and cloacal swabs | LOD of 2−1 HAU 3/50 μL. Linear response range: 2−1–24 HAU/50 μL. Detection in 1 h | [107] |
| Avian influenza A H5N1 virus | H5N1-specific aptamer | Aptamer-modified magnetic beads, concanavalin A-glucose oxidase-Au nanoparticle complexes, glucose solution, screen-printed interdigitated array electrode | Electrochemical impedance spectroscopy | PBS buffer | LOD of 8 × 10–4 HAU in 200 μL | [108] |
| Avian influenza A H7N1 virus | Rabbit anti-H7N1 polyclonal antibodies (affinity-chromatography purified) | Functionalized gold electrodes | Electrochemical impedance spectroscopy | Antigen extracted from vaccine diluted in buffer | LOD of 5 × 103 ng/mL | [109] |
| Avian influenza A H7N9 single-stranded (ss)DNA | DNA tetrahedral probe | Biotinylated-ssDNA oligonucleotide (detection probe), avidin–horseradish peroxidase (HRP) | Amperometric detection | ssDNA (PCR product in buffer) | Sensitivity of 10−7 μM. Detection in under 80 min | [110] |
| Quantum-dot-modified influenza hemagglutinin | Biotinylated glycans | Streptavidin-modified magnetic particles, glassy carbon microelectrode, 3D microfluidic chip | Differential pulse voltammetry | Vaccine hemagglutinin in buffer | Accuracy 80%. Linear response range: 60–500 μM. Detection in 45 min | [111] |
| Bovine viral diarrhea (BVD) antibodies | BVD virus | Functionalized nanowire sensor integrated on chip | Electrochemical impedance spectroscopy, cyclic voltammetry | Serum | Detection of 103 ng/mL. Detection in 20 min | [112] |
| BVD virus, anti-BVD antibodies | BVDV-1 monoclonal antibody (RAE0823), recombinant purified BVDV-1 Erns protein (BVDR16-R-10) | Six gold nanoband electrodes, silicon-chip-based biosensor platform | Electrochemical impedance spectroscopy | Serum | Detection in 20 min | [113] |
| Fowl adenovirus-9 | Anti-adenovirus, group II polyclonal antibody | Functionalized graphene quantum dots, functionalized gold nanobundles, carbon electrodes, UV–visible light irradiation | Voltammetry, local electric signal enhancement by light–matter interaction (graphene-mediated) | Serum | LOD of 10 PFU 4/mL in buffer and 50 PFU/mL | [114] |
| Protective antigen (Anthrax biomarker) | Short-chain peptide | Functionalized gold electrodes | Square-wave voltammetry | Antigen diluted in PBS + BSA 5 | LOD of 5.2 × 10−6 μM. Detection in 60–100 min | [115] |
| Streptococcus suis serotype 2 | Antibodies (sandwich immunoassay) | Antibodies immobilized on gold nanoparticles electrodeposited on a glassy carbon electrode, l-cysteine/hollow PtPd nanochains/glucose oxidase/antibody bioconjugates (HRP-mimicking), d-glucose solution | Peroxydisulfate electrochemiluminescence | Antigen diluted in serum | LOD of 33 × 10−6 ng/mL. Linear response range: 0.0001–100 ng/mL. Detection in 40 min | [116] |
| Gram-negative bacteria | Anti-LPS antibodies (mouse monoclonal and goat polyclonal, Abcam) | Functionalized magnetic nanoparticles, interdigitated microelectrodes | Conductometry | 1% serum in PBS | Detection range: 10–103 CFU 6/mL | [117] |
| Salmonella spp. | Anti-Salmonella magnetic beads (prod. no. 710.02, Dynal Biotech). Anti-Salmonella-HRP (rabbit polyclonal, prod. no. ab20771, Abcam) | Antibody-functionalized magnetic particle, polyclonal anti-Salmonella-HRP antibody, graphite-epoxy composite magneto-sensor | Amperometric detection | Skimmed milk | LOD of 7.5 × 103 CFU/mL. Detection in 50 min | [118] |
| Brucella melitensis | Anti-brucella antibodies | Gold nanoparticle-modified screen-printed carbon electrodes | Cyclic voltammetry, electrochemical impedance spectroscopy | Milk | LOD of 4 × 105 CFU/mL. Linear response range: 4 × 104–4 × 106 CFU/mL. Detection in 90 min | [119] |
| Brucella abortus | Anti-lipopolysaccharide antibody (Abcam 3535) | Screen-printed gold-plated electrodes, copper-doped nickel and zirconium oxide nanoparticles. | Cyclic voltammetry, electrochemical impedance spectroscopy | Phosphate buffer | Detection range: 103 CFU/mL–2 × 106 CFU/mL | [120] |
| Babesia bovis circulating antibodies | Recombinant version of the C-terminal portion of RAP-1 (Portuguese B. bovis Santarém strain) | Functionalized gold electrodes | Electrochemical impedance spectroscopy | Serum | Detection range: 16.7–500 μM | [121] |
| Optical biosensors | ||||||
| Influenza A H1N1 virus | FAM-labeled aptamers | Aptamer-modified magnetic beads for magnetic separation, fully integrated microfluidic chip, optical detection unit | Fluorescent measurements | PBS | LOD of 0.032 HAU units. Detection in 30 min | [122] |
| Swine-origin influenza A H1N1 virus | Anti-H1 antibody (ProSci, Poway, CA, USA) | SPR chip (BK7 glass slide coated with a laminated Ag/Au 37/8 nm, metal layer), paired-surface plasma-wave biosensor | Surface plasmon resonance (SPR) | Mimic solution (human mucosa in PBS) | Theoretical LOD of 30 PFU/mL, 1.8 × 102 PFU/mL. Detection in 20 min | [123] |
| Avian influenza A H5N1 virus | Anti-H5N1 hemagglutinin antibody 2B7 (ab135382), anti-H5N1 neuraminidase polyclonal antibody (Cat. PA5-34949) | Anti-H5N1 hemagglutinin antibody functionalized chiral gold nanohybrids, anti-H5N1 neuraminidase functionalized quantum dots | Circular dichroism spectra | Serum | LOD of 10−3 ng/mL | [124] |
| Infectious bronchitis virus (IBV) | Anti-IBV antibodies | Alexa Fluor 488 labeled anti-IBV antibody, anti-IBV antibody conjugated with molybdenum disulfide (quencher) and immobilized on a cotton-thread-based microfluidic platform | Fluorescence-resonance energy transfer (FRET) | Serum | LOD of 4.6 × 102 EID50 7/mL. Linear response range: 102–106 EID50/mL | [125] |
| Muscovy duck parvovirus | ssDNA aptamer | Unmodified gold nanoparticles | Spectrophotometry or visual observation | Allantoic fluids | LOD of 1.5 EID50 for spectrophotometry or 3 EID50 for visual observation. Detection in 70 min | [126] |
| PRRSV 9 | Anti-PRRSV monoclonal antibody (SDOW17) | Fluorescent (Alexa Fluor 546) labeled antibody/Protein A/gold nanoparticles or quantum dots (catskill green) complexes | Fluorescence resonance energy transfer (FRET) | PBS | Detection limit of 3 viral particles/μl | [127] |
| PRRSV | Anti-PRRSV antibody (lgG2b isotype) | CdTe:Zn2+ quantum dots, antibody modified platinum nanotubes (quencher) | Fluorescence | Serum diluted in PBS | LOD of 2.4 ng/mL. Linear response range: 5.6 ng/mL–66.6 ng/mL | [128] |
| Bovine viral diarrhea (BVD) virus | Anti-BVD virus monoclonal antibodies (9021 Jeno Biotech or 244-FA National Veterinary Service Laboratories, USA | Functionalized highly carboxylated polystyrene microparticles, y-channel microfluidic chip with optical fibers | Static forward light scattering | Tissue culture media and fetal calf serum diluted in PBS | LOD of 10 TCID50 8/mL | [129] |
| Foot and mouth disease (FMD) antibodies | FMD antigen (O, A and Asia-1 serotypes from commercial vaccine) | Anti-bovine IgG functionalized gold nanoparticles, nitrocellulose or nylon membrane | Dot-blot assay, visual observation | Serum | 10−4 dilution of serum samples | [130] |
| Vesicular stomatitis virus (VSV) | Anti-VSV-G (monoclonal 8G5, monoclonal 1E9), anti-VSV-M (monoclonal 23H12), anti-VSV-N (monoclonal 10G4) | Interferometric reflectance imaging sensor (IRIS), thermally grown SiO2 on Si, CCD camera | Spectral reflectance imaging | Cell lysate | 3.5 × 105 PFU/mL | [131] |
| Brucella DNA | Nucleotide probe | Ionic self-assembled multilayer, long-period grating optical fiber | Optical spectrum analysis of the refractive index | Culture and tissue lysates | LOD of 100 cells/mL. Detection in 30 min | [132] |
| Salmonella typhimurium | Goat anti-Salmonella antibodies (Kirkegaard and Perry Laboratories) | Labeled (donor Alexa Fluor 546) anti-Salmonella antibodies, labeled (acceptor Alexa Fluor 594) protein G, fiber-optic biosensor | Fluorescence resonance energy transfer (FRET) | Ground pork | LOD of 105 CFU/g of ground pork. Detection in 5 min | [133] |
| Targeted Analyte | Materials and Methods | Equipment | Samples and Handling | Performance | Reference |
|---|---|---|---|---|---|
| PRV | Fluorescent immunochromatographic strip, anti-PRV gB monoclonal antibodies, 3D-printed customized pocket fluorescence observation instrument | None | Homogenized pig tissues | LOD of 0.13 ng/mL. Detection within 13 min | [147] |
| Porcine epidemic diarrhea virus (PEDV) | LFA test, antibody-functionalized gold nanoparticles, 3D-printed transmittance reader, image analysis | Smartphone | PEDV solution | LOD of 55 ng/mL. Linear detection range: 78–20 × 103 ng/mL | [148] |
| Bovine ephemeral fever virus (BEFV) | RPA 1, FAM 2, and biotin labeled amplicons, LFA | TwistAmp NFO kit for RPA amplification, heat block | RNA isolation from clinical samples and reverse transcription | LOD of eight copies per reaction. Coincidence rate with real-time PCR of 96.09%. Detection in 25 min | [149] |
| BVDV | Immunochromato-graphic test strip, anti-NS3 monoclonal antibody 46/1-conjugated gold nanoparticles | None | Leukocyte extracts | Sensitivity and specificity of 100% and 97.2%, respectively. Detection in 15 min | [150] |
| FMDV | LFA test, gold nanoparticles, monoclonal anti-FMDV antibody 1F10 or 2H6 | None | Homogenized epithelial suspensions | Sensitivity of 84% for 1F10 and 88% for 2H6. Specificity of 99% for both antibodies | [151,152] |
| FMDV viral RNA | RT-LAMP, FIP 3 and BIP 4 labeling at the 5′ terminus with fluorescein and biotin, LFA test | Water bath | RNA, epithelial suspensions spiked with FMD virus, epithelial samples, air samples, RNA isolation | LOD of 10 viral copies | [153] |
| FMDV viral RNA | RT-RPA-, FAM-, and biotin-labeled amplicons, LFA | TwistAmp NFO kit for RPA amplification, water bath | cDNA, reverse transcription, RNA isolation | LOD of 10 copies (plasmid DNA), 98.6% concordance with real-time PCR | [154] |
| ASFV DNA | RPA, FITC 5, and biotin labeled amplicons, LFA | TwistAmp NFO kit for RPA amplification, thermocycler | DNA isolated with a magnetic bead-based kit | Positive agreement of 100% with PCR. Detection in 15 min | [155] |
| Classical Swine Fever (CSFV) | Fluorescent microsphere (FM)-based LFA, monoclonal-antibody-functionalized FMs | Fluorescent immunochromatographic strip reader, fluorescent camera | Tissue extracts | LOD of 5.28 ng/mL, positive coincidence rate, negative coincidence rate, and total coincidence rate of 95.8%, 100%, and 98%, respectively. Detection within 15 min | [156] |
| CSFV RNA | RT-LAMP-, DIG 6-, and FITC-labeled amplicons, LFA | Thermocycler | Cell-culture supernatants, serum, RNA isolation | LOD of 100 copies per reaction. Detection in 70 min | [157] |
| PCV-2 antibodies | Immunochromatographic test strip, recombinant-Cap-protein-labeled colloidal gold | None | Serum samples | Agreement of 94% with commercial ELISA. Sensitivity and specificity of 93.14% and 98.70%, respectively. Detection in 5 min | [158] |
| PRRSV antibodies | Immunochromatographic test strip, PRRSV recombinant membrane and nucleocapsid proteins, Protein-G-conjugated gold nanoparticles | None | Serum samples | Sensitivity of 98.6%, specificity of 97.8%, accuracy of 98.3% | [159] |
| Salmonella hilA gene | LAMP-, FITC-, and biotin-labeled amplicons, LFA test | Heating block | DNA isolated with commercial kit | LOD values of 13.5 × 10−3 ng/mL of genomic DNA and 6.7 CFU/mL. Detection in 40 min | [160] |
| Salmonella Typhimurium DNA | RPA-, DIG-, and FAM-labeled amplicons, LFA | TwistAmp RPA reaction kit. Thermostatic water bath | DNA isolated with commercial kit | LOD of 10−6 ng (genomic DNA) and 1.95 CFU/mL in milk samples. Detection in less than 20 min | [161] |
| Brucella spp. | Multiple cross-displacement amplification, FITC- and biotin-labeling of amplicons, LFA utilizing dye streptavidin-coated polymer nanoparticles | Water bath or heat block | Human- and goat-serum samples, DNA extraction | LOD of 10−5 ng of templates (pure cultures). Detection in 70 min | [162] |
| Campylobacter jejuni and Campylobacter coli | LFA test, gold nanoparticles, monoclonal mouse anti–Campylobacter A and/or B | None | Chicken feces, dilution with saline, filtration, sedimentation for 10 min | LOD of 6.7 log CFU/g for Campylobacter jejuni or 7.1 log CFU/g for Campylobacter coli. Detection in 20 min | [163] |
| Mycobacterium avium subsp. paratuberculosis | RPA, labeled amplicons, LFA | TwistAmp RPA reaction kit, thermostatic water tank | DNA extracted with commercial kit | LOD of eight copies per reaction. Sensitivity and specificity of 100% and 97.63%, respectively. Detection in 35 min | [164] |
| Mycoplasma ovipneumoniae DNA | LAMP-, DIG-, and biotin-labeled amplicons, LFA | Water bath | Lung tissue sample, DNA extraction | LOD of 100 CFU/mL. Sensitivity of 86% in clinical samples, Detection in 70 min | [165] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manessis, G.; Gelasakis, A.I.; Bossis, I. Point-of-Care Diagnostics for Farm Animal Diseases: From Biosensors to Integrated Lab-on-Chip Devices. Biosensors 2022, 12, 455. https://doi.org/10.3390/bios12070455
Manessis G, Gelasakis AI, Bossis I. Point-of-Care Diagnostics for Farm Animal Diseases: From Biosensors to Integrated Lab-on-Chip Devices. Biosensors. 2022; 12(7):455. https://doi.org/10.3390/bios12070455
Chicago/Turabian StyleManessis, Georgios, Athanasios I. Gelasakis, and Ioannis Bossis. 2022. "Point-of-Care Diagnostics for Farm Animal Diseases: From Biosensors to Integrated Lab-on-Chip Devices" Biosensors 12, no. 7: 455. https://doi.org/10.3390/bios12070455
APA StyleManessis, G., Gelasakis, A. I., & Bossis, I. (2022). Point-of-Care Diagnostics for Farm Animal Diseases: From Biosensors to Integrated Lab-on-Chip Devices. Biosensors, 12(7), 455. https://doi.org/10.3390/bios12070455






