An AuNPs-Based Fluorescent Sensor with Truncated Aptamer for Detection of Sulfaquinoxaline in Water
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Molecular Docking
2.3. Molecular Dynamic (MD) Simulations
2.4. Fluorescence Polarization-Based Binding Affinity and Specificity Assays
2.5. Synthesis and Characterization of AuNPs
2.6. Interactions between the Components of the Aptasensor
2.7. Optimization of Detection Conditions
2.8. Fluorescence Assay for SQX Detection
2.9. Analysis of Real Samples
3. Results
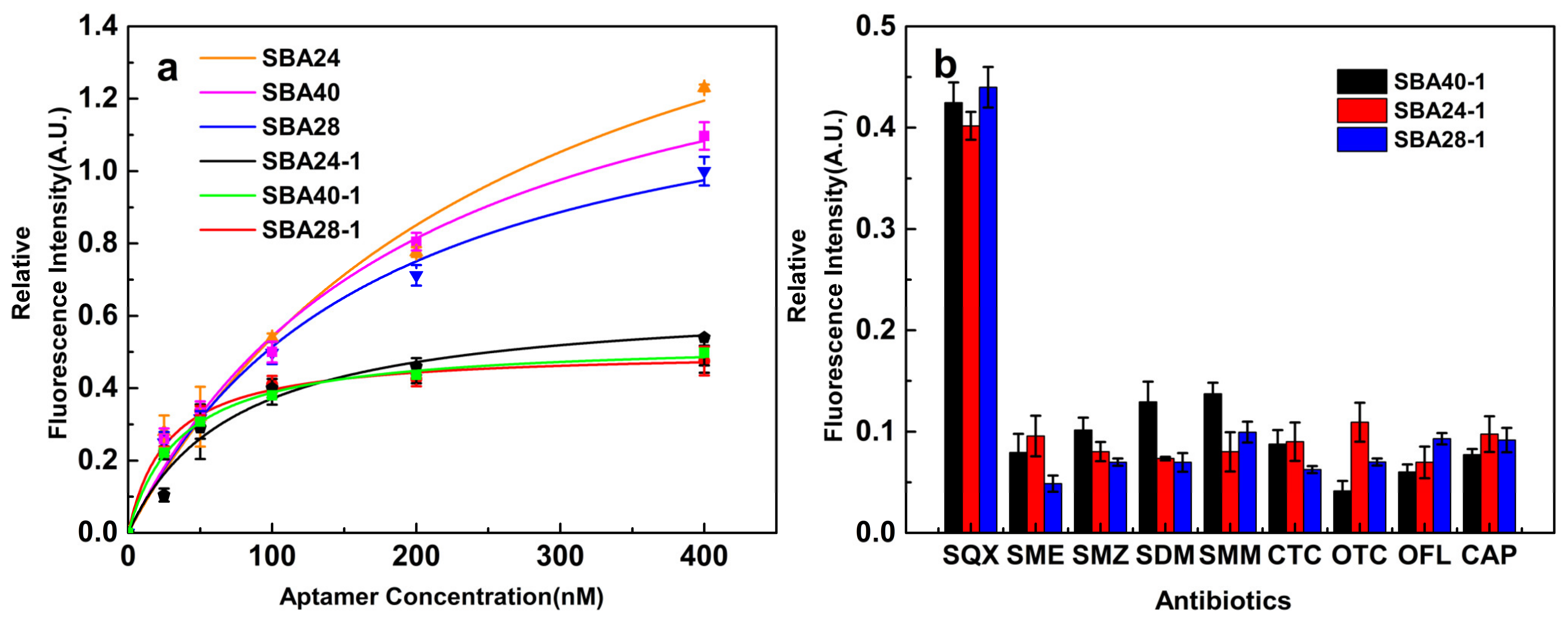
3.1. Truncation Strategy
3.2. Molecular Docking between Aptamers and SQX
3.3. MD Simulations of Aptamer-Target Interactions
3.4. Principle of the Aptasensor
3.5. Interactions between the Components of the Aptasensor
3.6. Optimization of Detection Conditions
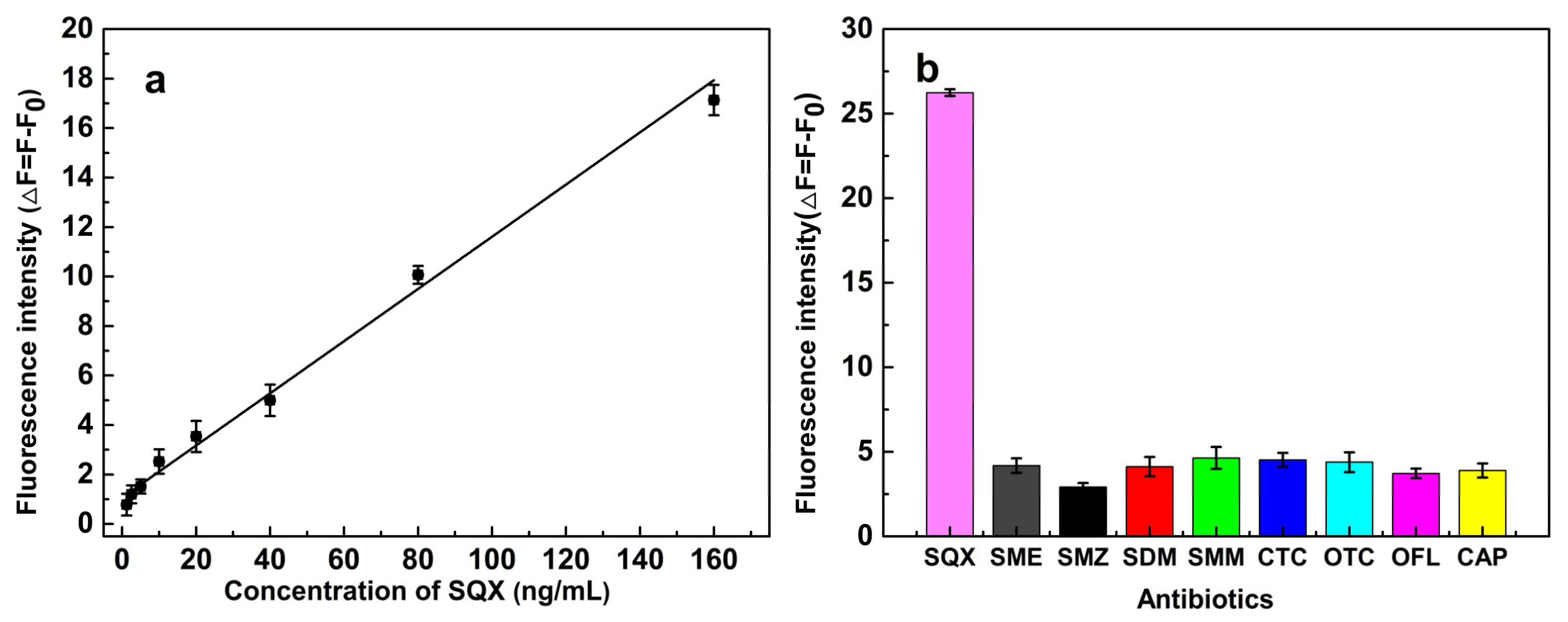
3.7. Properties of the Aptasensor
3.8. Validation of the Aptasensor
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aminov, R. History of antimicrobial drug discovery: Major classes and health impact. Biochem. Pharmacol. 2017, 133, 4–19. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Zhao, Y.; Wang, C.; Zhang, H.; Chen, Q.; Zhang, X.; Zhang, L.; Wu, J.; Wu, Z.; Zhou, Q. Removal performance of antibiotics and antibiotic resistance genes in swine wastewater by integrated vertical-flow constructed wetlands with zeolite substrate. Sci. Total Environ. 2020, 721, 137765. [Google Scholar] [CrossRef]
- Qiu, W.; Zheng, M.; Sun, J.; Tian, Y.; Fang, M.; Zheng, Y.; Zhang, T.; Zheng, C. Photolysis of enrofloxacin, pefloxacin and sulfaquinoxaline in aqueous solution by UV/H2O2, UV/Fe(II), and UV/H2O2/Fe(II) and the toxicity of the final reaction solutions on zebrafish embryos. Sci. Total Environ. 2019, 651 Pt 1, 1457–1468. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Kou, Q.; Wu, P.; Sun, Q.; Wu, J.; Le, T. Selection and Application of DNA Aptamers Against Sulfaquinoxaline Assisted by Graphene Oxide–Based SELEX. Food Anal. Methods 2020, 14, 250–259. [Google Scholar] [CrossRef]
- Li, S.; He, B.; Liang, Y.; Wang, J.; Jiao, Q.; Liu, Y.; Guo, R.; Wei, M.; Jin, H. Sensitive electrochemical aptasensor for determination of sulfaquinoxaline based on AuPd NPs@UiO-66-NH/CoSe and RecJf exonuclease-assisted signal amplification. Anal. Chim. Acta 2021, 1182, 338948. [Google Scholar] [CrossRef] [PubMed]
- Soleymanpour, A.; Rezvani, S. Development of a novel carbon paste sensor for determination of micromolar amounts of sulfaquinoxaline in pharmaceutical and biological samples. Mater. Sci. Eng. C Mater. Biol. Appl. 2016, 58, 504–509. [Google Scholar] [CrossRef] [PubMed]
- Hoff, R. Analysis of sulfonamide residues in bovine liver by liquid chromatography- tandem mass spectrometry without chemical extraction or clean-up procedures. Anal. Biochem. 2020, 611, 114011. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xiao, X.; Li, G. Porous molecularly imprinted monolithic capillary column for on-line extraction coupled to high-performance liquid chromatography for trace analysis of antimicrobials in food samples. Talanta 2014, 123, 63–70. [Google Scholar] [CrossRef]
- Mamani, M.; Amaya-Farfan, J.; Reyes, F.; Silva, J.; Rath, S. Use of experimental design and effective mobility calculations to develop a method for the determination of antimicrobials by capillary electrophoresis. Talanta 2008, 76, 1006–1014. [Google Scholar] [CrossRef]
- Le, T.; Yan, P.; Liu, J.; Wei, S. Simultaneous detection of sulfamethazine and sulfaquinoxaline using a dual-label time-resolved fluorescence immunoassay. Food Addit. Contam. Part A 2013, 30, 1264–1269. [Google Scholar] [CrossRef]
- Ye, H.; Duan, N.; Gu, H.; Wang, H.; Wang, Z. Fluorometric determination of lipopolysaccharides via changes of the graphene oxide-enhanced fluorescence polarization caused by truncated aptamers. Mikrochim. Acta 2019, 186, 173. [Google Scholar] [CrossRef]
- Sabrowski, W.; Dreymann, N.; Möller, A.; Czepluch, D.; Albani, P.; Theodoridis, D.; Menger, M. The use of high-affinity polyhistidine binders as masking probes for the selection of an NDM-1 specific aptamer. Sci. Rep. 2022, 12, 7936. [Google Scholar] [CrossRef]
- Gu, C.; Xiang, Y.; Guo, H.; Shi, H. Label-free fluorescence detection of melamine with a truncated aptamer. Analyst 2016, 141, 4511–4517. [Google Scholar] [CrossRef] [Green Version]
- Jia, M.; Sha, J.; Li, Z.; Wang, W.; Zhang, H. High affinity truncated aptamers for ultra-sensitive colorimetric detection of bisphenol A with label-free aptasensor. Food Chem. 2020, 317, 126459. [Google Scholar] [CrossRef]
- Kwon, Y.; Ahmad Raston, N.; Gu, M. An ultra-sensitive colorimetric detection of tetracyclines using the shortest aptamer with highly enhanced affinity. Chem. Commun. 2014, 50, 40–42. [Google Scholar] [CrossRef]
- Gao, S.; Zheng, X.; Jiao, B.; Wang, L. Post-SELEX optimization of aptamers. Anal. Bioanal. Chem. 2016, 408, 4567–4573. [Google Scholar] [CrossRef]
- Chinnappan, R.; AlZabn, R.; Mir, T.; Bader, M.; Zourob, M. Fluorometric determination of okadaic acid using a truncated aptamer. Mikrochim. Acta 2019, 186, 406. [Google Scholar] [CrossRef]
- Lang, P.; Brozell, S.; Mukherjee, S.; Pettersen, E.; Meng, E.; Thomas, V.; Rizzo, R.; Case, D.; James, T.; Kuntz, I. DOCK 6: Combining techniques to model RNA-small molecule complexes. RNA 2009, 15, 1219–1230. [Google Scholar] [CrossRef] [Green Version]
- Ma, P.; Guo, H.; Duan, N.; Ma, X.; Yue, L.; Gu, Q.; Wang, Z. Label free structure-switching fluorescence polarization detection of chloramphenicol with truncated aptamer. Talanta 2021, 230, 122349. [Google Scholar] [CrossRef]
- Oluyemi, W.M.; Samuel, B.B.; Adewumi, A.T.; Adekunle, Y.A.; Soliman, M.E.S.; Krenn, L. An Allosteric Inhibitory Potential of Triterpenes from Combretum racemosum on the Structural and Functional Dynamics of Plasmodium falciparum Lactate Dehydrogenase Binding Landscape. Chem. Biodivers. 2022, 19, e202100646. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R.; Open Source Drug Discovery, C.; Lynn, A. g_mmpbsa—A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yao, S.; Song, X.; Xu, K.; Wang, J.; Li, J.; Zhao, C.; Jin, M. One-step colorimetric detection of Staphylococcus aureus based on target-induced shielding against the peroxidase mimicking activity of aptamer-functionalized gold-coated iron oxide nanocomposites. Talanta 2021, 232, 122448. [Google Scholar] [CrossRef] [PubMed]
- Sokullu, E.; Pinsard, M.; Zhang, J.; Plathier, J.; Kolhatkar, G.; Blum, A.; Légaré, F.; Ruediger, A.; Ozaki, T.; Gauthier, M. Plasmonic Enhancement of Two-Photon Excitation Fluorescence by Colloidal Assemblies of Very Small AuNPs Templated on M13 Phage. Biomacromolecules 2020, 21, 2705–2713. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Ni, P.; Dai, H.; Sun, Y.; Wang, Y.; Jiang, S.; Li, Z. Aptamer-based colorimetric biosensing of abrin using catalytic gold nanoparticles. Analyst 2015, 140, 3581–3586. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, Y.; Li, H.; Wen, Y.; Fan, X.; Lin, F.; Tan, L.; Yao, S. Ultrasensitive electrochemical aptasensor for thrombin based on the amplification of aptamer-AuNPs-HRP conjugates. Biosens. Bioelectron. 2011, 26, 2297–2303. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, L.; Jiang, W. Label free DNA detection based on gold nanoparticles quenching fluorescence of Rhodamine B. Talanta 2011, 85, 725–729. [Google Scholar] [CrossRef]
- Ma, P.; Ye, H.; Guo, H.; Ma, X.; Yue, L.; Wang, Z. Aptamer truncation strategy assisted by molecular docking and sensitive detection of T-2 toxin using SYBR Green I as a signal amplifier. Food Chem. 2022, 381, 132171. [Google Scholar] [CrossRef]
- Yan, X.; Wang, Y.; Kou, Q.; Sun, Q.; Tang, J.; Yang, L.; Chen, X.; Xu, W.; Le, T. A novel aptasensor based on Fe3O4/Au/g-C3N4 for sensitive detection of sulfameter in food matrices. Sens. Actuators B Chem. 2022, 353, 131148. [Google Scholar] [CrossRef]
- Grabar, K.; Brown, K.; Keating, C.; Stranick, S.; Tang, S.; Natan, M. Nanoscale characterization of gold colloid monolayers:A comparison of four techniques. Anal. Chem. 1997, 69, 471–477. [Google Scholar] [CrossRef]
- Zhao, W.; Brook, M.; Li, Y. Design of gold nanoparticle-based colorimetric biosensing assays. Chembiochem 2008, 9, 2363–2371. [Google Scholar] [CrossRef]
- Wang, Y.; Yan, X.; Kou, Q.; Sun, Q.; Wang, Y.; Wu, P.; Yang, L.; Tang, J.; Le, T. An Ultrasensitive Label-Free Fluorescent Aptasensor Platform for Detection of Sulfamethazine. Int. J. Nanomed. 2021, 16, 2751–2759. [Google Scholar] [CrossRef]
- Su, L.; Wang, S.; Wang, L.; Yan, Z.; Yi, H.; Zhang, D.; Shen, G.; Ma, Y. Fluorescent aptasensor for carbendazim detection in aqueous samples based on gold nanoparticles quenching Rhodamine B. Spectrochim Acta A Mol. Biomol Spectrosc. 2020, 225, 117511. [Google Scholar] [CrossRef]
- Koo, K.M.; Sina, A.A.I.; Carrascosa, L.G.; Shiddiky, M.J.A.; Trau, M. DNA–bare gold affinity interactions: Mechanism and applications in biosensing. Anal. Methods 2015, 7, 7042–7054. [Google Scholar] [CrossRef]




| Aptamer | Binding Energy (kcal/mol) | The Base that Binds to the Ligand | The Lengths of the Hydrogen Bond (Å) | The Lengths of the π-Sulfur/π-π T-Shaped/π-Anion (Å) |
|---|---|---|---|---|
| SBA 40-1 | −3.76 | C-7 | 2.90 | - |
| T-8 | 1.99 | - | ||
| G-9 | 2.05/2.45 | 4.51/5.10 | ||
| C-10 | - | 4.59 | ||
| SBA 24-1 | −3.56 | G-1 | - | 4.79 |
| C-2 | - | 4.83 | ||
| G-3 | 2.30 | - | ||
| G-5 | 2.84 | - | ||
| SBA 28-1 | −4.01 | A-5 | 1.81/2.05 | 5.92/5.32 |
| G-6 | 1.79 | 4.16/4.88/4.84 | ||
| G-7 | 2.68 | - |
| Sample | Spiked Concentration (ng/mL) | Mean Recovery (%) ± SD | CV(%) |
|---|---|---|---|
| Lake Water | 50 | 96.1 ± 9.8 | 10.2 |
| 100 | 96.2 ± 6.2 | 6.4 | |
| 150 | 98.4 ± 2.1 | 2.2 | |
| Tap Water | 50 | 93.1 ± 4.5 | 4.9 |
| 100 | 98.8 ± 6.1 | 6.2 | |
| 150 | 100.1 ± 3.3 | 3.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Yang, L.; Tang, J.; Wen, X.; Zheng, X.; Chen, L.; Li, J.; Xie, Y.; Le, T. An AuNPs-Based Fluorescent Sensor with Truncated Aptamer for Detection of Sulfaquinoxaline in Water. Biosensors 2022, 12, 513. https://doi.org/10.3390/bios12070513
Chen X, Yang L, Tang J, Wen X, Zheng X, Chen L, Li J, Xie Y, Le T. An AuNPs-Based Fluorescent Sensor with Truncated Aptamer for Detection of Sulfaquinoxaline in Water. Biosensors. 2022; 12(7):513. https://doi.org/10.3390/bios12070513
Chicago/Turabian StyleChen, Xingyue, Lulan Yang, Jiaming Tang, Xu Wen, Xiaoling Zheng, Lingling Chen, Jiaqi Li, Yong Xie, and Tao Le. 2022. "An AuNPs-Based Fluorescent Sensor with Truncated Aptamer for Detection of Sulfaquinoxaline in Water" Biosensors 12, no. 7: 513. https://doi.org/10.3390/bios12070513
APA StyleChen, X., Yang, L., Tang, J., Wen, X., Zheng, X., Chen, L., Li, J., Xie, Y., & Le, T. (2022). An AuNPs-Based Fluorescent Sensor with Truncated Aptamer for Detection of Sulfaquinoxaline in Water. Biosensors, 12(7), 513. https://doi.org/10.3390/bios12070513






