Colorimetric Systems for the Detection of Bacterial Contamination: Strategy and Applications
Abstract
:1. Introduction
2. Colorimetric Detection and Monitoring Strategies of Bacterial Contamination
3. Current Colorimetric Sensing Systems for the Detection of Bacteria and Toxins
3.1. Color Change of pH Indicators via External pH Change
3.2. Color Change of Chemicals via Metabolic Activity of Intracellular Enzymes
3.3. Color Change of Substrates via Enzyme-like Catalytic Reactions of NPs
3.4. Color Change via NP Aggregation-Based Reaction
3.5. Color Change via NP Accumulation-Based Reaction
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, J.; Andler, S.M.; Goddard, J.M.; Nugen, S.R.; Rotello, V.M. Integrating recognition elements with nanomaterials for bacteria sensing. Chem. Soc. Rev. 2016, 46, 1272–1283. [Google Scholar] [CrossRef] [PubMed]
- Florentin, A.; Lizon, J.; Asensio, E.; Forin, J.; Rivier, A. Water and surface microbiologic quality of point-of-use water filters: A comparative study. Am. J. Infect. Control 2016, 44, 1061–1062. [Google Scholar] [CrossRef]
- Lemarchand, K.; Lebaron, P. Occurrence of Salmonella spp. and Cryptosporidium spp. in a French coastal watershed: Relationship with fecal indicators. FEMS Microbiol. Lett. 2003, 218, 203–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farrell, M.L.; Joyce, A.; Duane, S.; Fitzhenry, K.; Hooban, B.; Burke, L.P.; Morris, D. Evaluating the potential for exposure to organisms of public health concern in naturally occurring bathing waters in Europe: A scoping review. Water Res. 2021, 206, 117711. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control. Outbreak of E. coli Infections Linked to Romaine Lettuce, E. coli Infections, E. coli, CDC. November 2019. Available online: https://www.cdc.gov/ecoli/2019/o157h7–11-19/ (accessed on 1 June 2022).
- Nguendo-Yongsi, H.B. Microbiological evaluation of drinking water in a sub-saharan urban community (Yaunde). Am. J. Biochem. Mol. Biol. 2011, 1, 61–81. [Google Scholar]
- Delaire, C.; Peletz, R.; Kumpel, E.; Kisiangani, J.; Bain, R.; Khus, R. How much will it cost to monitor microbial drinking water quality in sub-Saharan Africa? Environ. Sci. Technol. 2017, 51, 5869–5878. [Google Scholar] [CrossRef] [Green Version]
- Schirone, M.; Visciano, P.; Tofalo, R.; Suzzi, G. Editorial: Foodborne pathogens: Hygiene and safety. Front. Microbiol. 2019, 10, 1974. [Google Scholar] [CrossRef] [Green Version]
- Harris, M.; Alzua, M.L.; Osbert, N.; Pickering, A. Community-level sanitation coverage more strongly associated with child growth and household drinking water quality than access to a private toilet in rural Mali. Environ. Sci. Technol. 2017, 51, 7219–7227. [Google Scholar] [CrossRef] [Green Version]
- Du, H.; Wang, X.; Yang, Q.; Wu, W. Quantum dot: Lightning invisible foodborne pathogens. Trends Food Sci. Technol. 2021, 110, 1–12. [Google Scholar] [CrossRef]
- Li, T.; Ou, G.; Chen, X.; Li, Z.; Hu, R.; Li, Y.; Yang, Y.; Liu, M. Naked-eye based point-of-care detection of E. coli O157: H7 by a signal-amplified microfluidic aptasensor. Anal. Chim. Acta. 2020, 1130, 20–28. [Google Scholar] [CrossRef]
- Castle, L.M.; Schuh, D.A.; Reynolds, E.E.; Furst, A.L. Electrochemical sensors to detect bacterial foodborne pathogens. ACS Sens. 2021, 6, 1717–1730. [Google Scholar] [CrossRef] [PubMed]
- Luo, K.; Kim, H.-Y.; Oh, M.-H.; Kim, Y.-R. Paper-based lateral flow strip assay for the detection of foodborne pathogens: Principles, applications, technological challenges and opportunities. Crit. Rev. Food Sci. Nutr. 2020, 60, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Verma, M.S.; Rogowski, J.L.; Jones, L.; Gu, F.X. Colorimetric biosensing of pathogens using gold nanoparticles. Biotechnol. Adv. 2015, 33 Pt 1, 666–680. [Google Scholar] [CrossRef]
- Choi, Y.; Hwang, J.H.; Lee, S.Y. Recent trends in nanomaterials-based colorimetric detection of pathogenic bacteria and viruses. Small Methods 2018, 2, 1700351. [Google Scholar] [CrossRef]
- Filik, H.; Avan, A.A. Nanotechnology-based colorimetric approaches for pathogenic virus sensing: A review. Curr. Med. Chem. 2022, 29, 2691–2718. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Q.H.; Kim, M.I. Nanomaterial-mediated paper-based biosensors for colorimetric pathogen detection. Trends Analyt. Chem. 2020, 132, 116038. [Google Scholar] [CrossRef]
- Liu, B.; Zhuang, J.; Wei, G. Recent advances in the design of colorimetric sensors for environmental monitoring. Environ. Sci. Nano 2020, 7, 2195–2213. [Google Scholar] [CrossRef]
- Chen, Q.; Huang, F.; Cai, G.; Wang, M.; Lin, J. An optical biosensor using immunomagnetic separation, urease catalysis and pH indication for rapid and sensitive detection of Listeria monocytogenes. Sens. Actuators B-Chem. 2018, 258, 447–453. [Google Scholar] [CrossRef]
- Singh, P.; Kakkar, S.; Bharti; Kumar, R.; Bhalla, V. Rapid and sensitive colorimetric detection of pathogens based on silver-urease interactions. Chem. Commun. 2019, 55, 4765–4768. [Google Scholar] [CrossRef]
- Kumar, V.; Chopra, A.; Bisht, B.; Bhalla, V. Colorimetric and electrochemical detection of pathogens in water using silver ions as a unique probe. Sci. Rep. 2020, 10, 11986. [Google Scholar] [CrossRef]
- Yan, C.; Sun, Y.; Yao, M.; Jin, X.; Yang, Q.; Wu, W. pH-responsive nanoparticles and automated detection apparatus for dual detection of pathogenic bacteria. Sens. Acturator B-Chem. 2022, 354, 131117. [Google Scholar] [CrossRef]
- Abo Dena, A.S.; Khalid, S.A.; Ghanem, A.F.; Shehata, A.I.; El-Sherbiny, I.M. User-friendly lab-on-paper optical sensor for the rapid detection of bacterial spoilage in packaged meat products. RSC Adv. 2021, 11, 35165–35173. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Ran, X.; Lin, Y.; Ren, J.; Qu, X. Enzyme-regulated the changes of pH values for assembling a colorimetric and multistage interconnection logic network with multiple readouts. Anal. Chim. Acta 2015, 870, 92–98. [Google Scholar] [CrossRef]
- Jankowska, D.A.; Bannwarth, M.B.; Schulenburg, C.; Faccio, G.; Maniura-Weber, K.; Rossi, R.M.; Scherer, L.; Richter, M.; Boesel, L.F. Simultaneous detection of pH value and glucose concentrations for wound monitoring applications. Biosens. Bioelectron. 2017, 87, 312–319. [Google Scholar] [CrossRef]
- Song, Y.; Qu, K.; Zhao, C.; Ren, J.; Qu, X. Graphene oxide: Intrinsic peroxidase catalytic activity and its application to glucose detection. Adv. Mater. 2010, 22, 2206–2210. [Google Scholar] [CrossRef]
- Contin, A.; Frasca, S.; Vivekananthan, J.; Leimkühler, S.; Wollenberger, U.; Plumeré, N.; Schuhmann, W. A pH responsive redox hydrogel for electrochemical detection of redox silent biocatalytic processes. Control of Hydrogel Solvation. Electroanalysis 2015, 27, 938–944. [Google Scholar] [CrossRef]
- Bidmanova, S.; Hlavacek, A.; Damborsky, J.; Prokop, Z. Conjugation of 5(6)-carboxyfluorescein and 5(6)-carboxynaphthofluorescein with bovine serum albumin and their immobilization for optical pH sensing. Sens. Actuators B-Chem. 2012, 161, 93–99. [Google Scholar] [CrossRef]
- Chen, C.H.; Yang, K.L. A liquid crystal biosensor for detecting organophosphates through the localized pH changes induced by their hydrolytic products. Sens. Actuators B-Chem. 2013, 181, 368–374. [Google Scholar] [CrossRef]
- Lei, C.; Dai, H.; Fu, Y.; Ying, Y.; Li, Y. Colorimetric sensor array for thiols discrimination based on urease-metal ion pairs. Anal. Chem. 2016, 88, 8542–8547. [Google Scholar] [CrossRef]
- Kim, H.J.; Kwon, C.; Lee, B.S.; Noh, H. One-step sensing of foodborne pathogenic bacteria using a 3D paper-based device. Analyst 2019, 144, 2248–2255. [Google Scholar] [CrossRef]
- Kim, H.J.; Kwon, C.; Noh, H. Paper-based diagnostic system facilitating Escherichia coli assessments by duplex coloration. ACS Sens. 2019, 4, 2435–2441. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhong, Y.; Wang, D.; Lu, Z. A simple colorimetric method for viable bacteria detection based on cell counting Kit-8. Anal. Methods 2021, 13, 5211–5215. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Warden, A.R.; Huang, J.; Wang, W.; Ding, X. Colorimetric and electrochemical detection of Escherichia coli and antibiotic resistance based on a p-benzoquinone-mediated bioassay. Anal. Chem. 2019, 91, 7524–7530. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Huang, J.; Li, Y.; Lv, J.; Ding, X. A simple and rapid colorimetric bacteria detection method based on bacterial inhibition of glucose oxidase-catalyzed reaction. Talanta 2019, 197, 304–309. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Gao, X.; Wang, S.; Liu, Y. A smartphone-integrated paper sensing system for fluorescent and colorimetric dual-channel detection of foodborne pathogenic bacteria. Anal. Bioanal. Chem. 2020, 412, 611–620. [Google Scholar] [CrossRef]
- Wu, S.; Duan, N.; Qiu, Y.; Li, J.; Wang, Z. Colorimetric aptasensor for the detection of Salmonella enterica serovar typhimurium using ZnFe2O4-reduced graphene oxide nanostructures as an effective peroxidase mimetics. Int. J. Food Microbiol. 2017, 261, 42–48. [Google Scholar] [CrossRef]
- Wang, L.; Liao, T.; Zhou, H.; Huang, Y.V.; Chen, P.; Yang, X.; Chen, X. Colorimetric method for Salmonella spp. detection based on peroxidase-like activity of Cu(II)-rGO nanoparticles and PCR. Anal. Biochem. 2021, 615, 114068. [Google Scholar] [CrossRef]
- Tarokh, A.; Pebdeni, A.B.; Othman, H.O.; Salehnia, F.; Hosseini, M. Sensitive colorimetric aptasensor based on g-C3N4@Cu2O composites for detection of Salmonella typhimurium in food and water. Mikrochim. Acta. 2021, 188, 87. [Google Scholar] [CrossRef]
- Yao, S.; Li, J.; Pang, B.; Wang, X.; Shi, Y.; Song, X.; Xu, K.; Wang, J.; Zhao, C. Colorimetric immunoassay for rapid detection of Staphylococcus aureus based on etching-enhanced peroxidase-like catalytic activity of gold nanoparticles. Mikrochim. Acta. 2020, 187, 504. [Google Scholar] [CrossRef]
- Lai, W.; Wei, Q.; Zhuang, J.; Lu, M.; Tang, D. Fenton reaction-based colorimetric immunoassay for sensitive detection of brevetoxin B. Biosens. Bioelectron. 2016, 80, 249–256. [Google Scholar] [CrossRef] [Green Version]
- Lai, W.; Guo, J.; Wu, Q.; Chen, Y.; Cai, Q.; Wu, L.; Wang, S.; Song, J.; Tang, D. A novel colorimetric immunoassay based on enzyme-regulated instant generation of Turnbull’s blue for the sensitive determination of ochratoxin A. Analyst 2020, 145, 2420–2424. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Gao, R.; Jia, L. Enhancement of the peroxidase-like activity of aptamers modified gold nanoclusters by bacteria for colorimetric detection of Salmonella typhimurium. Talanta 2021, 221, 121476. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhao, C.; Zhao, W.; Zhang, H.; Yao, S.; Shi, Y.; Li, J.; Wang, J. Multi-functional MnO2-doped Fe3O4 nanoparticles as an artificial enzyme for the colorimetric detection of bacteria. Anal. Bioanal. Chem. 2020, 412, 3135–3140. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, X.; Wang, Q.; Lou, Z.; Li, S.; Zhu, Y.; Qin, L.; Wei, H. Nanomaterials with enzyme-like characteristics (nanozymes): Next-generation artificial enzymes (II). Chem. Soc. Rev. 2019, 48, 1004–1076. [Google Scholar] [CrossRef]
- Bhardwaj, N.; Bhardwaj, S.K.; Bhatt, D.; Lim, D.K.; Kim, K.H.; Deep, A. Optical detection of waterborne pathogens using nanomaterials. TrAC Trends Anal. Chem. 2019, 113, 280–300. [Google Scholar] [CrossRef]
- Wei, H.; Wang, E. Nanomaterials with enzyme-like characteristics (nanozymes): Next-generation artificial enzymes. Chem. Soc. Rev. 2013, 42, 6060–6093. [Google Scholar] [CrossRef]
- Huang, J.; Sun, J.; Warden, A.R.; Ding, X. Colorimetric and photographic detection of bacteria in drinking water by using 4-mercaptophenylboronic acid functionalized AuNPs. Food Control 2020, 108, 106885. [Google Scholar] [CrossRef]
- Feng, J.; Shen, Q.; Wu, J.; Dai, Z.; Wang, Y. Naked-eyes detection of Shigella flexneri in food samples based on a novel gold nanoparticle-based colorimetric aptasensor. Food Control 2019, 98, 333–341. [Google Scholar] [CrossRef]
- Jalalian, S.H.; Lavaee, P.; Ramezani, M.; Danesh, N.M.; Alibolandi, M.; Abnous, K.; Taghdisi, S.M. An optical aptasensor for aflatoxin M1 detection based on target-induced protection of gold nanoparticles against salt-induced aggregation and silica nanoparticles. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 246, 119062. [Google Scholar] [CrossRef]
- Fu, K.; Zheng, Y.; Li, J.; Liu, Y.; Pang, B.; Song, X.; Xu, K.; Wang, J.; Zhao, C. Colorimetric immunoassay for rapid detection of Vibrio parahemolyticus based on Mn2+ mediates the assembly of gold nanoparticles. J. Agric. Food Chem. 2018, 66, 9516–9521. [Google Scholar] [CrossRef]
- Peng, H.; Chen, I.A. Rapid colorimetric detection of bacterial species through the capture of gold nanoparticles by chimeric phages. ACS Nano 2019, 13, 1244–1252. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Borg, R.E.; Nguyen, A.B.N.; Chen, I.A. Chimeric phage nanoparticles for rapid characterization of bacterial pathogens, detection in complex biological samples and determination of antibiotic sensitivity. ACS Sens. 2020, 5, 1491–1499. [Google Scholar] [CrossRef]
- Xu, X.; Yuan, Y.; Hu, G.; Wang, X.; Qi, P.; Wang, Z.; Wang, Q.; Wang, X.; Fu, Y.; Li, Y.; et al. Exploiting pH-regulated dimer-tetramer transformation of concanavalin A to develop colorimetric biosensing of bacteria. Sci. Rep. 2017, 7, 1452. [Google Scholar] [CrossRef] [PubMed]
- Chotchuang, T.; Cheewasedtham, W.; Jayeoye, T.J.; Rujiralai, T. Colorimetric determination of fumonisin B1 based on the aggregation of cysteamine-functionalized gold nanoparticles induced by a product of its hydrolysis. Mikrochim. Acta 2019, 186, 655. [Google Scholar] [CrossRef]
- Yang, H.; Xiao, M.; Lai, W.; Wan, Y.; Li, L.; Pei, H. Stochastic DNA dual-walkers for ultrafast colorimetric bacteria detection. Anal. Chem. 2020, 92, 4990–4995. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zong, X.; Zheng, J.; Zhang, J.; Zhou, M.; Chen, Q.; Man, C.; Jiang, Y. A colorimetric strategy based on aptamer-catalyzed hairpin assembly for the on-site detection of Salmonella typhimurium in milk. Foods 2021, 10, 2539. [Google Scholar] [CrossRef] [PubMed]
- Albanese, A.; Chan, W.C. Effect of gold nanoparticle aggregation on cell uptake and toxicity. ACS Nano 2011, 5, 5478–5489. [Google Scholar] [CrossRef] [PubMed]
- Thuy Nguyen, T.T.; Han, O.A.; Lim, E.B.; Haam, S.; Park, J.S.; Lee, S.W. The effect of pH and transition metal ions on cysteine-assisted gold aggregation for a distinct colorimetric response. RSC Adv. 2021, 11, 9664–9674. [Google Scholar] [CrossRef] [PubMed]
- Dodero, G.; De Michieli, L.; Cavalleri, O.; Rolandi, R.; Oliveri, L.; Daccà, A.; Parodi, R. L-Cysteine chemisorption on gold: An XPS and STM study. Colloids Surf. A 2000, 175, 121–128. [Google Scholar] [CrossRef]
- Aryal, S.; Remant, B.K.C.; Dharmaraj, N.; Bhattarai, N.; Kim, C.H.; Kim, H.Y. Spectroscopic identification of S-Au interaction in cysteine capped gold nanoparticles. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2006, 63, 160–163. [Google Scholar] [CrossRef]
- Kreibig, U.; Genzel, L. Optical absorption of small metallic particles. Surf. Sci. 1985, 156, 678–700. [Google Scholar] [CrossRef]
- Brovko, L.Y.; Anany, H.; Griffiths, M.W. Bacteriophages for detection and control of bacterial pathogens in food and food-processing environment. Adv. Food Nutr. Res. 2012, 67, 241–288. [Google Scholar] [PubMed]
- Yang, X.; Wisuthiphaet, N.; Young, G.M.; Nitin, N. Rapid detection of Escherichia coli using bacteriophage-induced lysis and image analysis. PLoS ONE 2020, 15, e0233853. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.H.; Huang, W.C.; Shyu, R.H.; Chang, S.C. Silver nanoparticle-base lateral flow immunoassay for rapid detection of Staphylococcal enterotoxin B in milk and honey. J. Inorg. Biochem. 2020, 210, 111163. [Google Scholar] [CrossRef]
- Bu, T.; Huang, Q.; Yan, L.; Huang, L.; Zhang, M.; Yang, Q.; Yang, B.; Wang, J.; Zhang, D. Ultra technically-simple and sensitive detection for Salmonella enteritidis by immunochromatographic assay based on gold growth. Food Control 2018, 84, 536–543. [Google Scholar] [CrossRef]
- Rodriguez-Quijada, C.; Lyons, C.; Santamaria, C.; Quinn, S.; Tlusty, M.F.; Shiaris, M.; Hamad-Schifferli, K. Optimization of paper-based nanoparticle immunoassays for direct detection of the bacterial pathogen V. parahaemolyticus in oyster hemolymph. Anal. Methods 2020, 12, 3056–3063. [Google Scholar] [CrossRef]
- Lu, C.; Gao, X.; Chen, Y.; Ren, J.; Liu, C. Aptamer-based lateral flow test strip for the simultaneous detection of Salmonella typhimurium, Escherichia coli O157:H7 and Staphylococcus aureus. Anal. Lett. 2020, 53, 646–659. [Google Scholar] [CrossRef]
- Ying, N.; Wang, Y.; Song, X.; Yang, L.; Qin, B.; Wu, Y.; Fang, W. Lateral flow colorimetric biosensor for detection of Vibrio parahaemolyticus based on hybridization chain reaction and aptamer. Mikrochim. Acta 2021, 188, 381. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Zhou, Y.; Huang, X.; Zhang, W.; Wu, Y.; Fang, H.; Zhang, C.; Xiong, Y. Dramatically enhanced immunochromatographic assay using cascade signal amplification for ultrasensitive detection of Escherichia coli O157:H7 in milk. J. Agric. Food Chem. 2020, 68, 1118–1125. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Zhang, L.; Hu, L.; Xing, K.; Lu, X.; Huang, Y.; Zhang, J.; Lai, W.; Chen, T. Nanozyme-based lateral flow assay for the sensitive detection of Escherichia coli O157:H7 in milk. J. Dairy Sci. 2018, 101, 5770–5779. [Google Scholar] [CrossRef] [Green Version]
- Jiang, T.; Song, Y.; Wei, T.; Li, H.; Du, D.; Zhu, M.J.; Lin, Y. Sensitive detection of Escherichia coli O157: H7 using Pt-Au bimetal nanoparticles with peroxidase-like amplification. Biosens. Bioelectron. 2016, 77, 687–694. [Google Scholar] [CrossRef]
- Park, J.; Shin, J.H.; Park, J.-K. Pressed paper-based dipstick for detection of foodborne pathogens with multistep reactions. Anal. Chem. 2016, 88, 3781–3788. [Google Scholar] [CrossRef] [PubMed]
- Prakash, C.; Kumar, B.; Singh, R.P.; Singh, P.; Shrinet, G.; Das, A.; Ashmi, M.; Abhishek Singh, K.P.; Singh, M.K.; Gupta, V.K. Development and evaluation of a gold nanoparticle based lateral flow assay (LFA) strip test for detection of Brucella spp. J. Microbiol. Methods 2021, 184, 106185. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q. Simultaneous quantitative analysis of Listeria monocytogenes and Staphylococcus aureus based on antibiotic-introduced lateral flow immunoassay. Anal. Methods 2021, 13, 5866–5874. [Google Scholar] [CrossRef]
- Porras, J.C.; Bernuz, M.; Marfa, J.; Pallares-Rusiñol, A.; Martí, M.; Pividori, M.I. Comparative study of gold and carbon nanoparticles in nucleic acid lateral flow assay. Nanomaterials 2021, 11, 741. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.H.; Huang, W.C.; Chang, S.C.; Shyu, R.H. Colloidal silver-based lateral flow immunoassay for detection of profenofos pesticide residue in vegetables. RSC Adv. 2022, 12, 13035–13044. [Google Scholar] [CrossRef]
- Liang, P.; Guo, Q.; Zhao, T.; Wen, C.Y.; Tian, Z.; Shang, Y.; Xing, J.; Jiang, Y.; Zeng, J. Ag nanoparticles with ultrathin Au shell-based lateral flow immunoassay for colorimetric and SERS dual-mode detection of SARS-CoV-2 IgG. Anal. Chem. 2022, 94, 8466–8473. [Google Scholar] [CrossRef]
- Shi, F.; Zhao, Y.; Sun, Y.; Chen, C. Development and application of a colloidal carbon test strip for the detection of antibodies against Mycoplasma bovis. World J. Microbiol. Biotechnol. 2020, 36, 157. [Google Scholar] [CrossRef]
- Li, C.M.; Li, Y.F.; Wang, J.; Huang, C.Z. Optical investigations on ATP-induced aggregation of positive-charged gold nanoparticles. Talanta 2010, 81, 1339–1345. [Google Scholar] [CrossRef]
- Du, X.J.; Zhou, T.J.; Li, P.; Wang, S. A rapid Salmonella detection method involving thermophilic helicase-dependent amplification and a lateral flow assay. Mol. Cell. Probes 2017, 34, 37–44. [Google Scholar] [CrossRef]
- Özay, B.; McCalla, E.S. A review of reaction enhancement strategies for isothermal nucleic acid amplification reactions. Sens. Actuator Rep. 2021, 3, 100033. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, Y.; Xie, H.; Zhao, L.; Zheng, L.; Ye, H. Applications of catalytic hairpin assembly reaction in biosensing. Small 2019, 15, 1902989. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Fu, C.; Shi, W.; Chen, J. Recent advances in catalytic hairpin assembly signal amplification-based sensing strategies for microRNA detection. Talanta 2021, 235, 122735. [Google Scholar] [CrossRef]
- Chen, Q.; Lin, J.; Gan, C.; Wang, Y.; Wang, D.; Xiong, Y.; Lai, W.; Li, Y.; Wang, M. A sensitive impedance biosensor based on immunomagnetic separation and urease catalysis for rapid detection of Listeria monocytogenes using an immobilization-free interdigitated array microelectrode. Biosens. Bioelectron. 2015, 74, 504–511. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, D.; Cai, G.; Xiong, Y.; Li, Y.; Wang, M.; Huo, H.; Lin, J. Fast and sensitive detection of foodborne pathogen using electrochemical impedance analysis, urease catalysis and microfluidics. Biosens. Bioelectron. 2016, 86, 770–776. [Google Scholar] [CrossRef] [PubMed]
- Du, N.; Chen, M.; Liu, Z.; Sheng, L.; Xu, H.; Chen, S. Kinetics and mechanism of jack bean urease inhibition by Hg2+. Chem. Cent. J. 2012, 6, 154. [Google Scholar] [CrossRef] [Green Version]
- Ren, R.; Cai, G.; Yu, Z.; Zeng, Y.; Tang, D. Metal-polydopamine framework: An innovative signal-generation tag for colorimetric immunoassay. Anal. Chem. 2018, 90, 11099–11105. [Google Scholar] [CrossRef]
- Jiao, L.; Yan, H.; Xu, W.; Wu, Y.; Gu, W.; Li, H.; Du, D.; Lin, Y.; Zhu, C. Self-assembly of all-inclusive allochroic nanoparticles for the improved ELISA. Anal. Chem. 2019, 91, 8461–8465. [Google Scholar] [CrossRef]
- Courbat, J.; Briand, D.; Damon-Lacoste, J.; Wöllenstein, J.; de Rooij, N.F. Evaluation of pH indicator-based colorimetric films for ammonia detection using optical waveguides. Sens. Actuators B Chem. 2009, 143, 62–70. [Google Scholar] [CrossRef]
- Xu, W.; Jiao, L.; Ye, H.; Guo, Z.; Wu, Y.; Yan, H.; Gu, W.; Du, D.; Lin, Y.; Zhu, C. pH-responsive allochroic nanoparticles for the multicolor detection of breast cancer biomarkers. Biosens. Bioelectron. 2020, 148, 111780. [Google Scholar] [CrossRef]
- Hodgkinson, V.; Petris, M.J. Copper homeostasis at the host-pathogen interface. J. Biol. Chem. 2012, 287, 13549–13555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rensing, C.; Grass, G. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 2003, 27, 197–213. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Li, M.; Qin, Y.; Chu, Z.; Zhao, S. A Convenient label free colorimetric assay for pyrophosphatase activity based on a pyrophosphate-inhibited Cu2+-ABTS-H2O2 reaction. Analyst 2014, 139, 6298–6303. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Huang, Y.; Liu, W.; Zuo, W.; Ye, F.; Zhao, S. Colorimetric detection of salicylic acid in aspirin using MIL-53(Fe) nanozyme. Front. Chem. 2020, 8, 671. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hou, Y.; Guo, X.; Liu, W.; Lv, C.; Zhang, C.; Jin, Y.; Li, B. Fe(III) bipyridyl or phenanthroline complexes with oxidase-like activity for sensitive colorimetric detection of glutathione. Luminescence 2020, 35, 1350–1359. [Google Scholar] [CrossRef]
- Gao, L.; Zhuang, J.; Nie, L.; Zhang, J.; Zhang, Y.; Gu, N.; Wang, T.; Feng, J.; Yang, D.; Perrett, S.; et al. Intrinsic peroxidase-like activity of ferromagnetic nanoparticles. Nat. Nanotechnol. 2007, 2, 577–583. [Google Scholar] [CrossRef]
- Jia, H.; Yang, D.; Han, X.; Cai, J.; Liu, H.; He, W. Peroxidase-like activity of the Co3O4 nanoparticles used for biodetection and evaluation of antioxidant behavior. Nanoscale 2016, 8, 5938–5945. [Google Scholar] [CrossRef]
- Chen, W.; Fang, X.; Li, H.; Cao, H.; Kong, J. DNA-mediated inhibition of peroxidase-like activities on platinum nanoparticles for simple and rapid colorimetric detection of nucleic acids. Biosens. Bioelectron. 2017, 94, 169–175. [Google Scholar] [CrossRef]
- Li, J.; Liu, W.; Wu, X.; Gao, X. Mechanism of pH-switchable peroxidase and catalase-like activities of gold, silver, platinum and palladium. Biomaterials 2015, 48, 37–44. [Google Scholar] [CrossRef]
- Wang, S.; Cazelles, R.; Liao, W.C.; Vázquez-González, M.; Zoabi, A.; Abu-Reziq, R.; Willner, I. Mimicking horseradish peroxidase and NADH peroxidase by heterogeneous Cu2+-modified graphene oxide nanoparticles. Nano Lett. 2017, 17, 2043–2048. [Google Scholar] [CrossRef]
- Lai, W.; Tang, D.; Zhuang, J.; Chen, G.; Yang, H. Magnetic bead-based enzyme-chromogenic substrate system for ultrasensitive colorimetric immunoassay accompanying cascade reaction for enzymatic formation of squaric acid-iron(III) chelate. Anal. Chem. 2014, 86, 5061–5068. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Du, J. Selective sensing of submicromolar iron(III) with 3,3′,5,5′-tetramethylbenzidine as a chromogenic probe. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2016, 158, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Zheng, A.; Zhang, X.; Gao, J.; Liu, X.; Liu, J. Peroxidase-like catalytic activity of copper ions and its application for highly sensitive detection of glypican-3. Anal. Chim. Acta 2016, 941, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Zou, Y.; Huang, M.; Dong, S.; Wu, X.; Liang, G.; Han, Z.; Zhang, Z. A sensitive, colorimetric immunosensor based on Cu-MOFs and HRP for detection of dibutyl phthalate in environmental and food samples. Talanta 2018, 186, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Pang, H.; Dong, Y.; Chi, Y.; Fu, F. Colorimetric determination of glutathione by using a nanohybrid composed of manganese dioxide and carbon dots. Mikrochim. Acta 2018, 185, 291. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; He, J.; Niu, Y.; Chen, J.; Zhao, Y.; Zhang, Y.; Yu, C. Sandwich-type biosensor for the detection of alpha2, 3-sialylated glycans based on fullerene-palladium-platinum alloy and 4-mercaptophenylboronic acid nanoparticle hybrids coupled with Au-methylene blue-MAL signal amplification. Biosens. Bioelectron. 2018, 102, 321–327. [Google Scholar] [CrossRef]
- Bryła, M.; Roszko, M.; Szymczyk, K.; Jędrzejczak, R.; Obiedziński, M.W. Fumonisins and their masked forms in maize products. Food Control 2016, 59, 619–627. [Google Scholar] [CrossRef]
- Voss, K.A.; Smith, G.W.; Haschek, W.M. Fumonisins: Toxicokinetics, mechanism of action and toxicity. Anim. Feed Sci. Technol. 2007, 137, 299–325. [Google Scholar] [CrossRef]
- Li, Y.S.; Zhou, Y.; Lu, S.Y.; Guo, D.J.; Ren, H.L.; Meng, X.M.; Zhi, B.H.; Lin, C.; Wang, Z.; Li, X.B.; et al. Development of a one-step test strip for rapid screening of fumonisins B1, B2 and B3 in maize. Food Control 2012, 24, 72–77. [Google Scholar] [CrossRef]
- Shi, F.; Sun, Y.; Wu, Y.; Zhu, M.; Feng, D.; Zhang, R.; Peng, L.; Chen, C. A novel, rapid and simple method for detecting brucellosis based on rapid vertical flow technology. J. Appl. Microbiol. 2020, 128, 794–802. [Google Scholar] [CrossRef]
- Morbioli, G.G.; Mazzu-Nascimento, T.; Stockton, A.M.; Carrilho, E. Technical aspects and challenges of colorimetric detection with microfluidic paper-based analytical devices (μPADs)—A review. Anal. Chim. Acta 2017, 970, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Kanchi, S.; Sabela, M.I.; Mdluli, P.S.; Inamuddin; Bisetty, K. Smartphone based bioanalytical and diagnosis applications: A review. Biosens. Bioelectron. 2018, 102, 136–149. [Google Scholar] [CrossRef] [PubMed]

| Material | Receptor | Target | Linear Range | LoD | Assay Time | Real Sample to Be Tested | Driving Force of Color Change | Feature | References |
|---|---|---|---|---|---|---|---|---|---|
| AuNPs, magnetic beads | Monoclonal antibody, polyclonal antibody | Listeria monocytogenes | 1.1 × 102 CFU/mL–1.1 × 106 CFU/mL | 100 CFU/mL | 30 min | Lettuce samples | Induction of pH change | Use of magnetic nanobeads modified with urease and monoclonal antibodies. | [19] |
| Use of AuNPs modified with urease and polyclonal antibodies. | |||||||||
| Use of BCP. | |||||||||
| AgNPs | Monoclonal antibody | Salmonella typhimurium | 1 × 108 CFU/mL–1 × 101 CFU/mL | 100 CFU/mL | Apple juice, lake water sample | Induction of pH change | Based on the competitive binding ability of urease and bacterial cells to PEI-functionalized AgNPs. | [20] | |
| Ag ion | None | S. typhimurium | 1 × 107 CFU/mL–1 × 101 CFU/mL | 100 CFU/mL | Tap water | Induction of pH change | Based on the Ag-induced inhibition of urease activity and Ag ion utilization. | [21] | |
| Combined with electrochemical sensing. | |||||||||
| NPs | Aptamer | Escherichia coli., S. typhimurium | 1 × 105 CFU/mL–1 × 101 CFU/mL | 1 CFU/mL | <1 h | Milk | Induction of pH change | Use of pH-responsive NPs made of phenolphthalein (PP) and thymolphthalein (TP) indicators. | [22] |
| Combined with automated equipment. | |||||||||
| Allows multiplexing detection. | |||||||||
| Filter paper | None | Bacteria | 11.2 × 103–1.12 × 106 CFU/g (using BTB), 38.0 × 103–1.12 × 106 CFU/g (using BCP) | 11.64 × 103 CFU/g | Chicken and meat samples | Induction of pH change | Monitoring of bacterial contamination level using paper-based pH indicators, BTB, and BCP. | [23] | |
| Sensing of external pH change caused by volatile basic nitrogen generated from bacterial spoilage. | |||||||||
| Use of RGB analysis software on a smartphone. | |||||||||
| Filter paper | None | E. coli, E. coli O157:H7, L. monocytogenes, Vibrio vulnificus | 1 × 106–1 × 108 CFU/mL | 10 CFU/mL | 1 h | Milk | Chemical reaction between intracellular enzymes and their chromogenic substrates | One-step-based 3D paper sensor functionalized with lysing and oxidizing agents. | [31] |
| Filter paper | None | E. coli, E. coli O157:H7 | 1 × 106–1 × 109 CFU/mL | 10 CFU/mL | <4 h | Milk | Chemical reaction between intracellular enzymes and their chromogenic substrates | Use of a multi-layered paper structure. | [32] |
| Use of β-glucuronidase and β-galactosidase-based enzymatic reactions. | |||||||||
| None | None | Staphylococcus aureus, E. coli | 2.6 × 102–1.16 × 109 CFU/mL (for E. coli), 9.75 × 102–6 × 109 CFU/mL (for S. aureus) | ND | 2 h | Drinking water, milk | Redox reaction between the cell counting kit-8 (CCK-8) solution and dehydrogenase | Measurement of formazan generated from the reduction reaction between dehydrogenase and CCK-8 (containing WST-8 and 1-methoxy-5-methylphenazinium methyl sulfate). | [33] |
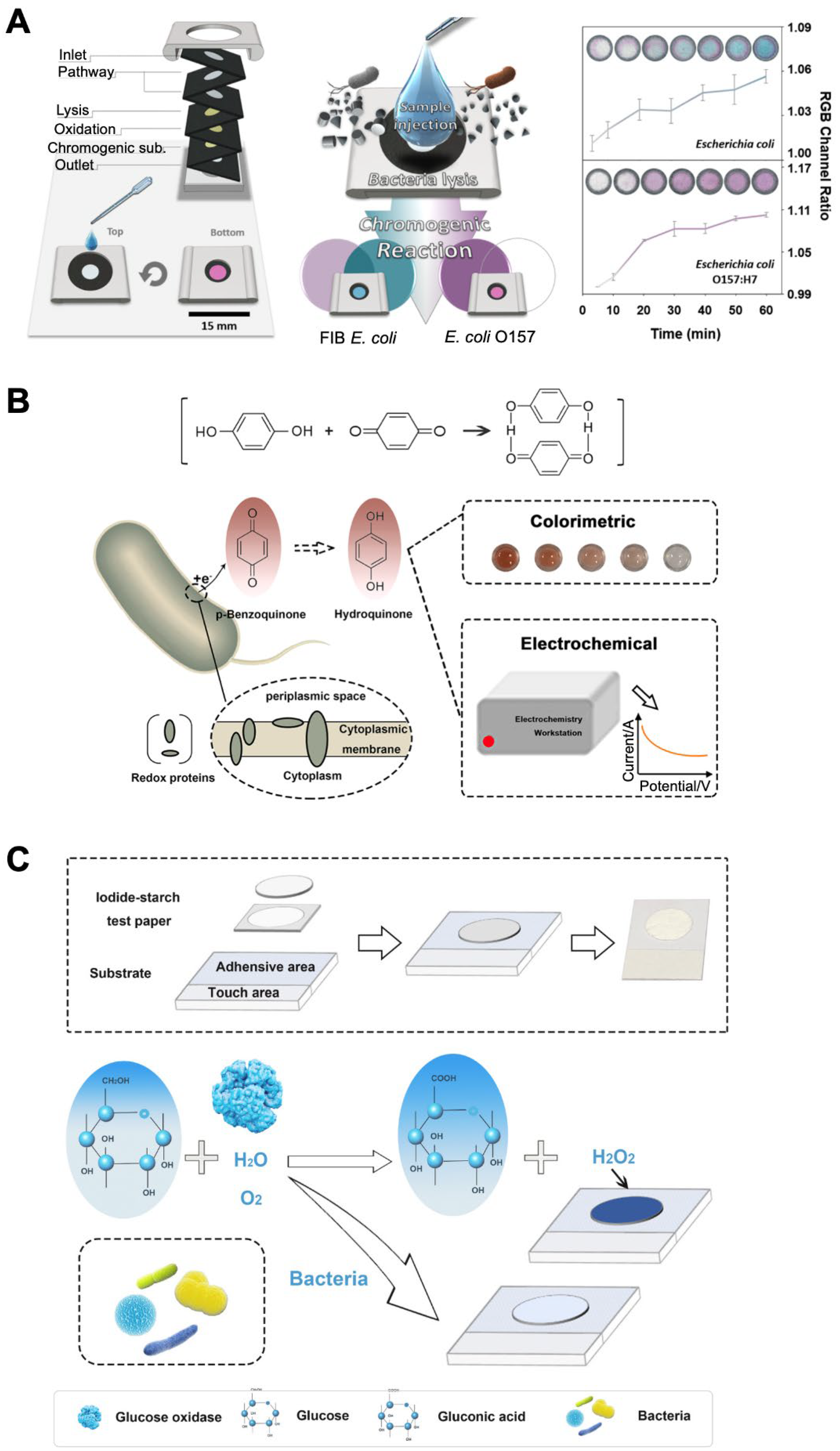
| None | None | E. coli | 1 × 104–1 × 109 CFU/mL | 1 × 104 CFU/mL | 1 h | Unfiltered tap water | Reduction reaction of p-benzoquinone by intracellular enzymes | Use of RGB analysis software on a smartphone for quantification. | [34] |
| Filter paper | None | E. coli, S. aureus, Enterococcus faecalis, Streptococcus mutans, Salmonella pullorum | 1 × 104–1 × 108 CFU/mL | 7.48 × 103 CFU/mL (for E. coli) and 3.3 × 103 CFU/mL (for S. aureus) | 20 min | Inhibition of GOx activity by glucose uptake of bacterial cells | Use of starch–iodide doping paper as a substrate. | [35] | |
| Based on the conversion from iodide to iodine by H2O2 involving GOx-mediated glucose oxidation (causing color change of starch–iodine) and glucose uptake of bacterial cells (causing inhibition of color change of starch–iodine). | |||||||||
| Filter paper | None | E. coli | 1 × 102–1 × 106 CFU/mL | 44 CFU/mL | Tap water, degrease milk | Inhibition of color change of OPD via Cu2+ reduction by intracellular enzymes | Use of paper as a substrate. | [36] | |
| Based on the competitive reaction between the oxidation of OPD by Cu2+ (causing color change of OPD) and the reduction of Cu2+ by bacteria (causing inhibition of color change of OPD). | |||||||||
| Use of RGB analysis software on a smartphone for quantification. | |||||||||
| Allows dual-readout assay (colorimetry and fluorescence). | |||||||||
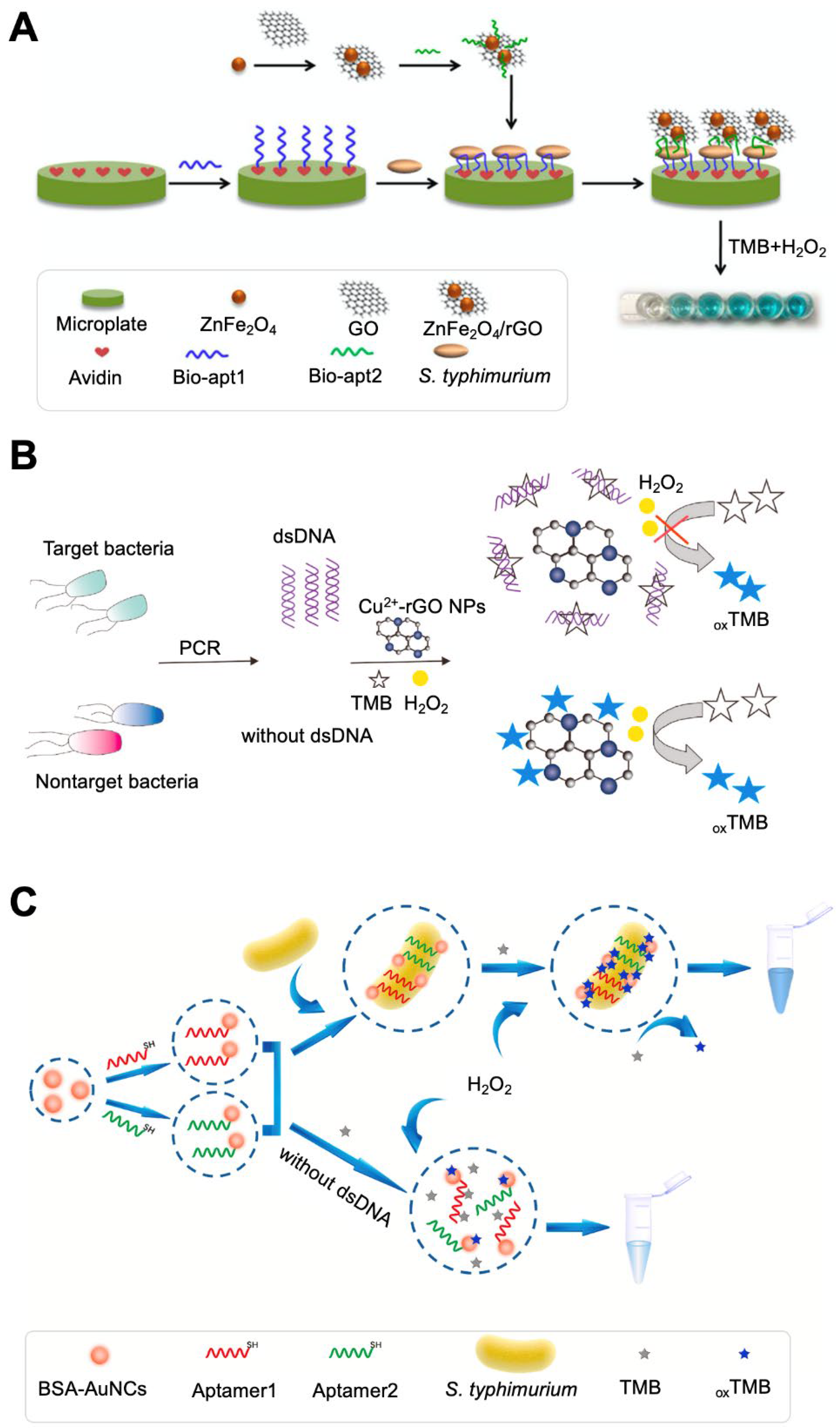
| ZnFe2O4/rGO | Aptamer | S. typhimurium | 11–1.10 × 105 CFU/mL | 11 CFU/mL | None | Peroxidase-like catalytic reaction of the ZnFe2O4/rGO nanostructure | Shows highly stable catalytic activity at low pH (over 5.5) and high temperature (over 50 °C). | [37] | |
| Cu2-rGO NPs | None | Salmonella spp. | 1.93 × 101–1.93 × 105 CFU/mL | 0.51 CFU/mL | Milk | Peroxidase-like catalytic reaction of GO | Use of dsDNA amplified via PCR from cells. Based on the competitive binding of bacterial dsDNA and Cu2-rGO NPs to TMB. | [38] | |
| Graphitic-C3N4@Cu2O | Aptamer | S. typhimurium | 1.5 × 101–1.5 × 105 CFU/mL | 15 CFU/mL | 6 min | Milk | Peroxidase-like catalytic reaction of the g-C3N4@Cu2O nanostructure | Based on the competitive binding of the aptamer and g-C3N4@Cu2O to TMB. | [39] |
| Fe3O4/Au magnetic nanocomposite | Antibody, aptamer | S. aureus | 1 × 101–1 × 106 CFU/mL | 10 CFU/mL | Pork, milk | Peroxidase-like catalytic reaction of AuNPs by H2O2 etching | Use of a magnetic nanocomposite consisting of a Fe3O4 core and an Au shell as a capture probe. | [40] | |
| Use of Apt–AuNPs as a signal amplifier. | |||||||||
| AuNPs, magnetic beads | Antibody | Brevotoxin B | 0.1–150 ng/kg | 0.076 ng/kg | Seafood sample | Peroxidase-based TMB oxidation reaction | Addition of Fe2+ for color signal amplification. | [41] | |
| Magnetic beads | Antibody | Ochratoxin A | 0.01–10 ng/mL | 8.3 pg/mL | 30 min (for color development) | Red wine sample | Enzyme-controlled Turnbull’s blue generation | Based on the formation or inhibition of Prussian blue from K3[Fe(CN)6] via GOx-catalyzed H2O2 production. | [42] |
| Aptamer@ BSA- AuNCs | Aptamer | S. typhimurium | 1 × 101–1 × 106 CFU/mL | 1 CFU/mL | Eggshell, Egg white | Peroxidase-like catalytic reaction of AuNCs | Based on the enhanced catalytic activity of a cell-bound nanostructure (cell-aptamer@BSA-AuNC composite). | [43] | |
| MnO2-doped Fe3O4 NPs | None | S. aureus, Vibrio parahaemolyticus | 1 × 101–1 × 106 CFU/mL | 1 × 102 CFU/mL | Lake water sample | Peroxidase-like catalytic reaction | Use of multifunctional NPs for recognition, absorption, and separation of the analyte. | [44] | |
| Exhibits the catalytic activity of TMB in the presence of oxygen in a solution without H2O2. | |||||||||
| AuNPs | 4-MPBA | E. coli | 1 × 104–1 × 107 CFU/mL | 1.02 × 103 CFU/mL | 20 min | Drinking water | Salt-induced aggregation | Use of AuNPs functionalised with 4-MPBA, which binds to LPS and peptidoglycan existing on the surface of gram-negative and gram-positive bacterial cells, respectively. | [48] |
| Use of RGB analysis software on a smartphone for quantification. | |||||||||
| AuNPs | Aptamer | Shigella flexneri | 1 × 102–1 × 106 CFU/mL | 80 CFU/mL | 20 min | Salmon | Salt-induced aggregation | Use of aptamers that can bind to bacterial cells rather than AuNPs. | [49] |
| AuNPs, silica nanoparticles (SNPs) | Aptamer | Aflatoxin M1 | 300–75,000 ng/L | 30 ng/L | Milk | Salt-induced AuNP aggregation | Salt-induced aggregation by releasing complementary strands from aptamer-modified SNPs in the presence of the target. | [50] | |
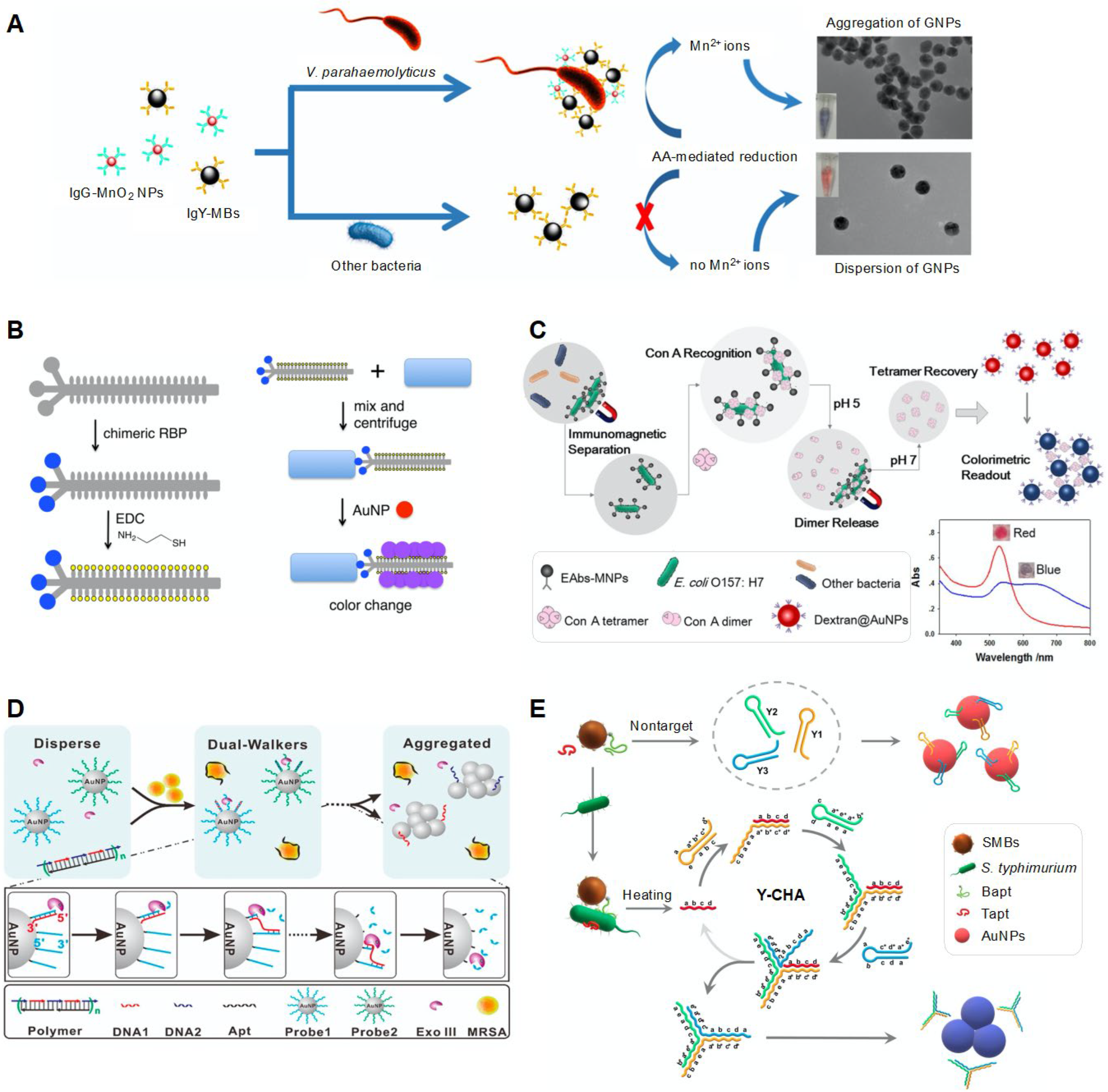
| AuNPs, magnetic nanoparticles (MNPs) | Antibody | V. parahaemolyticus | 1 × 101–1 × 106 CFU/mL | 10 CFU/mL | Oyster | Mn2+-induced AuNP aggregation | Combination with the signal amplification method based on ascorbic acid-mediated Mn2+ reduction and a sandwich assay using IgG-MnO2 NPs and IgY-MNPs. | [51] | |
| AuNPs | Chimeric phage | E. coli, V. cholerae, Pseudomonas aeruginosa, Xanthomonas campestris | 1 × 102 CFU/mL | <1 h | Sea water, tap water | AuNP aggregation | Use of thiolated chimeric phages that can bind to both bacterial cells and AuNPs | [52] | |
| AuNPs | Chimeric phage | P. aeruginosa | 1 × 101–1 × 106 CFU/mL | 1 × 102 CFU/mL | ~30 min | Drinking water, non-fat bovine milk | AuNP aggregation | Detection of antibiotic resistance/susceptibility of bacterial cells | [53] |
| Dextran-coated AuNPs, MNPs | Antibody | E. coli | 1 × 103–1 × 106 CFU/mL | 41 CFU/mL | 95 min | Milk | ConA-driven aggregation of dextran-coated AuNPs | Use of ConA with pH-regulated transformation ability of dimers/tetramers | [54] |
| AuNPs | Fumonisin B1 (FB1) | 2–8 mg/kg | 0.9 mg/kg | Corn | Hydrolyzed FB1-induced AgNP aggregation | Use of cysteamine-functionalised AuNPs (Cys-AuNPs). Need for NaOH treatment to obtain hydrolyzed FB1 with a high affinity towards Cys-AuNPs. | [55] | ||
| AuNPs | DNA | S. aureus | 1–1 × 105 CFU/mL | 1 CFU/mL | 15 min | CSF, urine, spit, serum | Enzyme-driven DNA walker-induced AgNP aggregation | Use of an exonuclease III-driven DNA walker system for signal amplification. | [56] |
| AuNPs, MNPs | Aptamer | S. typhimurium | 1 × 102–1 × 106 CFU/mL | 2.4 × 102 CFU/mL | Milk | Catalytic hairpin assembly (CHA)-driven AuNP aggregation | Use of Y-shaped CHA for signal amplification. | [57] | |
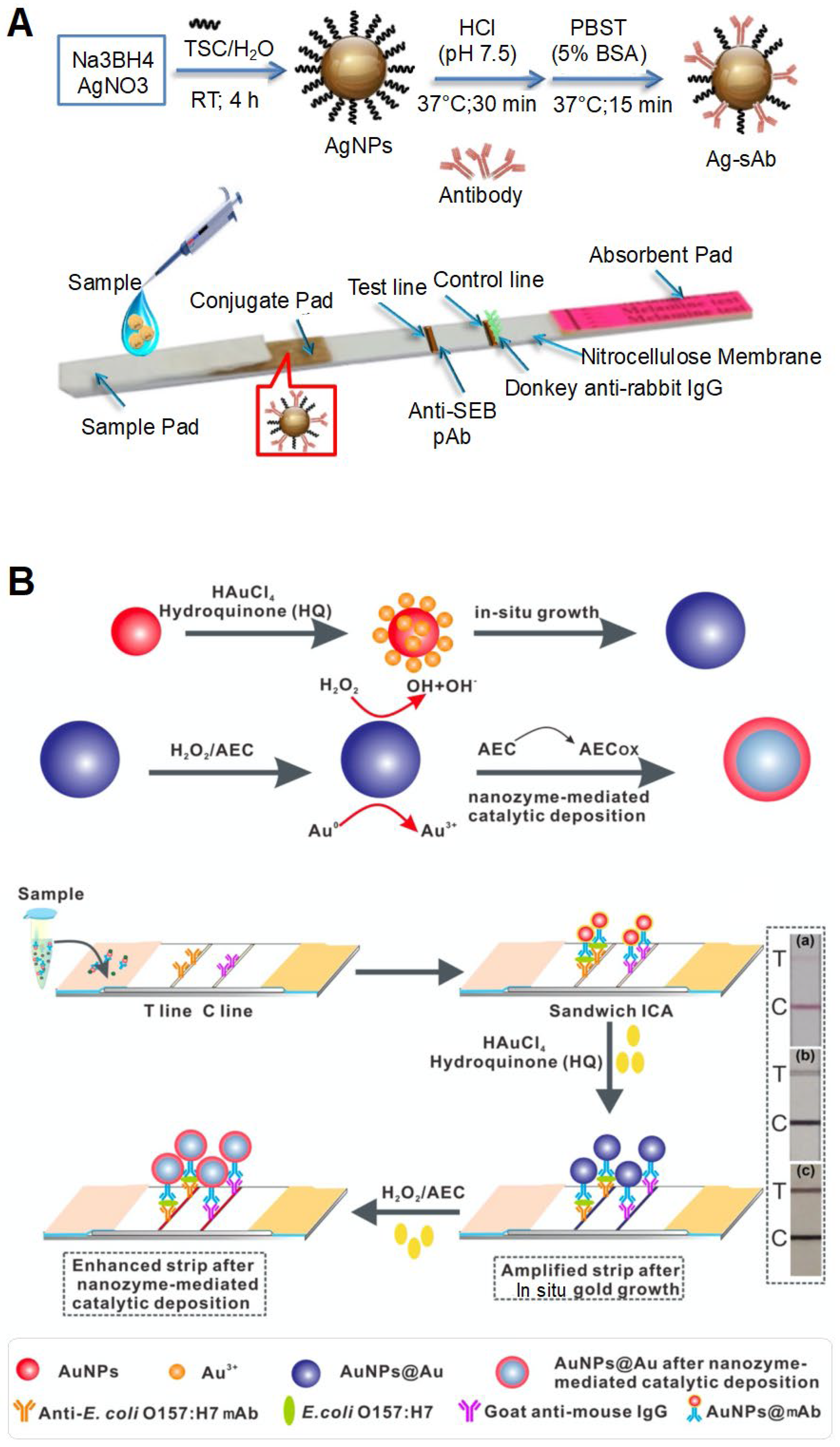
| AgNPs | Antibody | Staphylococcal enterotoxin B | 0–2 ppm | 0.5 ppm | 15 min | Milk, honey | AgNP accumulation | Use of AgNP-based sandwich-type lateral flow immunoassay (LFIA). | [65] |
| AuNPs | Antibody | S. enteritidis | 1 × 105–1 × 108 CFU/mL | 1 × 104 CFU/mL | 20 min | Milk | AuNP accumulation | Use of LFIA. | [66] |
| Use of a signal enhancer, HAuCl4 and NH2OH·HCl for in situ AuNP growth. | |||||||||
| AuNPs | Antibody | V. parahaemolyticus | 4.66 × 105 CFU/mL | 2 h | Oyster hemolymph | AuNP accumulation | Use of a dipstick. | [67] | |
| AuNPs | Aptamer | S. typhimurium, E. coli O157:H7, S. aureus | 1 × 103 CFU/mL for S. typhimurium and 1 × 104 CFU/mL for E. coli O157:H7 and S. aureus | 10 min | Milk, chicken, food | AuNP accumulation | Use of LFA. | [68] | |
| AuNPs, MNPs | Aptamer | V. parahaemolyticus | 1 × 103–1 × 108 CFU/mL | 2.6 × 103 CFU/mL | 67 min | Shrimp | AuNP accumulation | Combination of HCR-mediated signal amplification methods. | [69] |
| AuNPs | Antibody | E. coli O157:H7 | 1.25 × 101–1.25 × 105 CFU/mL | 1.25 × 101 CFU/mL | Milk | AuNP accumulation | Use of LFA. | [70]; | |
| Combination of two signal amplification strategies; use of a signal enhancer (hydroquinone) for in situ AuNP growth and nanozyme-mediated catalytic deposition. | |||||||||
| Pd-Pt NPs | Antibody | E. coli O157:H7 | 1 × 102–1 × 106 CFU/mL | 0.87 × 102 CFU/mL | 10 min | Milk | Pd-Pt NP accumulation-driven catalytic reaction | Use of LFA. | [71] |
| Signal readout by oxidised TMB through Pd-Pt NP-mediated catalytic reactions. | |||||||||
| Pt-Au NPs | Antibody | E. coli O157:H7 | 1 × 102–1 × 108 CFU/mL | 1 × 102 CFU/mL | 1 min | Pt-Au NP accumulation-driven catalytic reaction | Use of LFA. | [72] | |
| Use of Pt-Au-mediated signal amplification. |
| Challenge | Performance Improvement Strategy | Reference(s) |
|---|---|---|
| Sensitivity |
| [22] [36] [34] [19] [56,57,69,81,82,83,84] [70,71,72] |
| Simple operation |
| [32] [21] [22] |
| Correct signal in complex real samples |
| [37,38,39,40,41,42,43,44] |
| Multiplexing capability |
| [22] [31] |
| Quantification and expansion of analyzed spectra |
| [22,23,34,36] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.-M.; Yoo, S.-M. Colorimetric Systems for the Detection of Bacterial Contamination: Strategy and Applications. Biosensors 2022, 12, 532. https://doi.org/10.3390/bios12070532
Kim D-M, Yoo S-M. Colorimetric Systems for the Detection of Bacterial Contamination: Strategy and Applications. Biosensors. 2022; 12(7):532. https://doi.org/10.3390/bios12070532
Chicago/Turabian StyleKim, Dong-Min, and Seung-Min Yoo. 2022. "Colorimetric Systems for the Detection of Bacterial Contamination: Strategy and Applications" Biosensors 12, no. 7: 532. https://doi.org/10.3390/bios12070532
APA StyleKim, D.-M., & Yoo, S.-M. (2022). Colorimetric Systems for the Detection of Bacterial Contamination: Strategy and Applications. Biosensors, 12(7), 532. https://doi.org/10.3390/bios12070532





