Variational Mode Decomposition Weighted Multiscale Support Vector Regression for Spectral Determination of Rapeseed Oil and Rhizoma Alpiniae Offcinarum Adulterants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
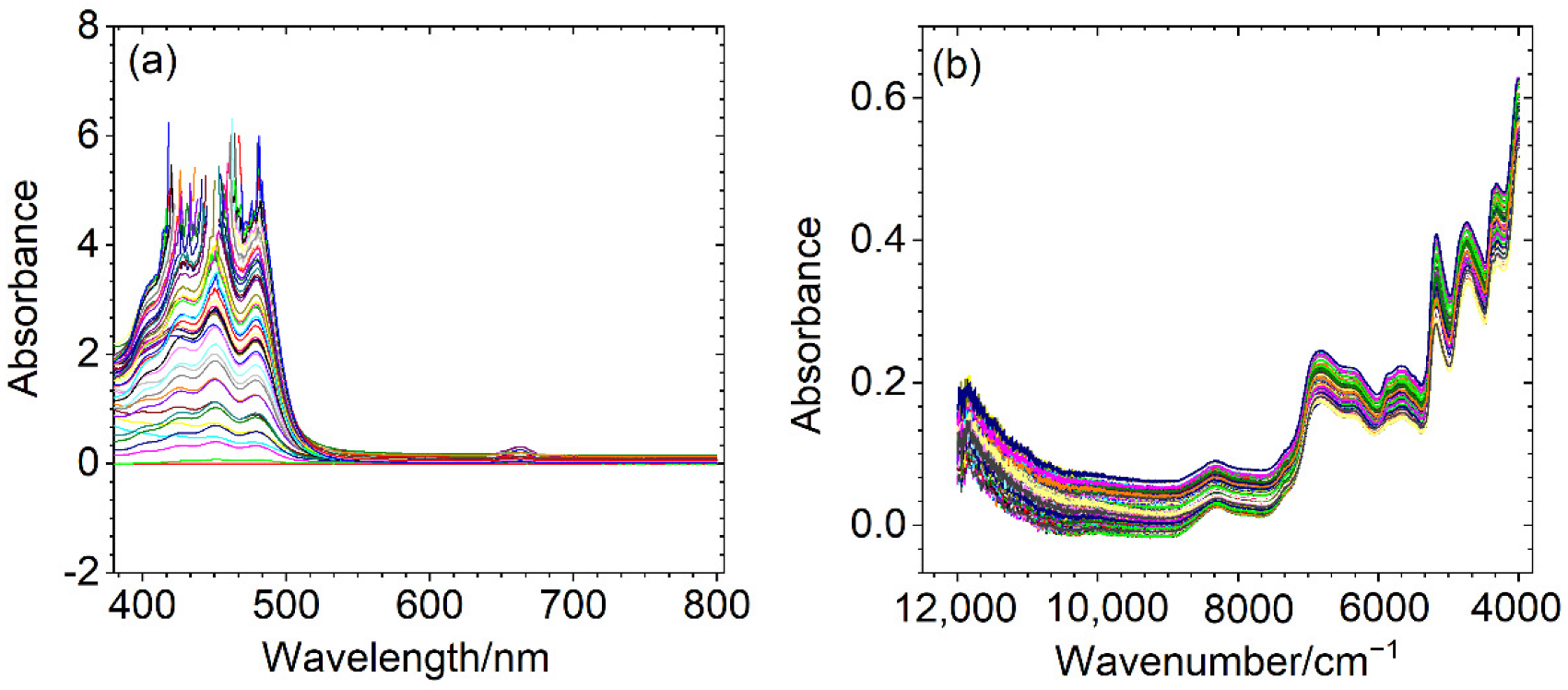
2.2. Spectral Collection
2.3. Variational Mode Decomposition (VMD)
2.4. Support Vector Regression (SVR)
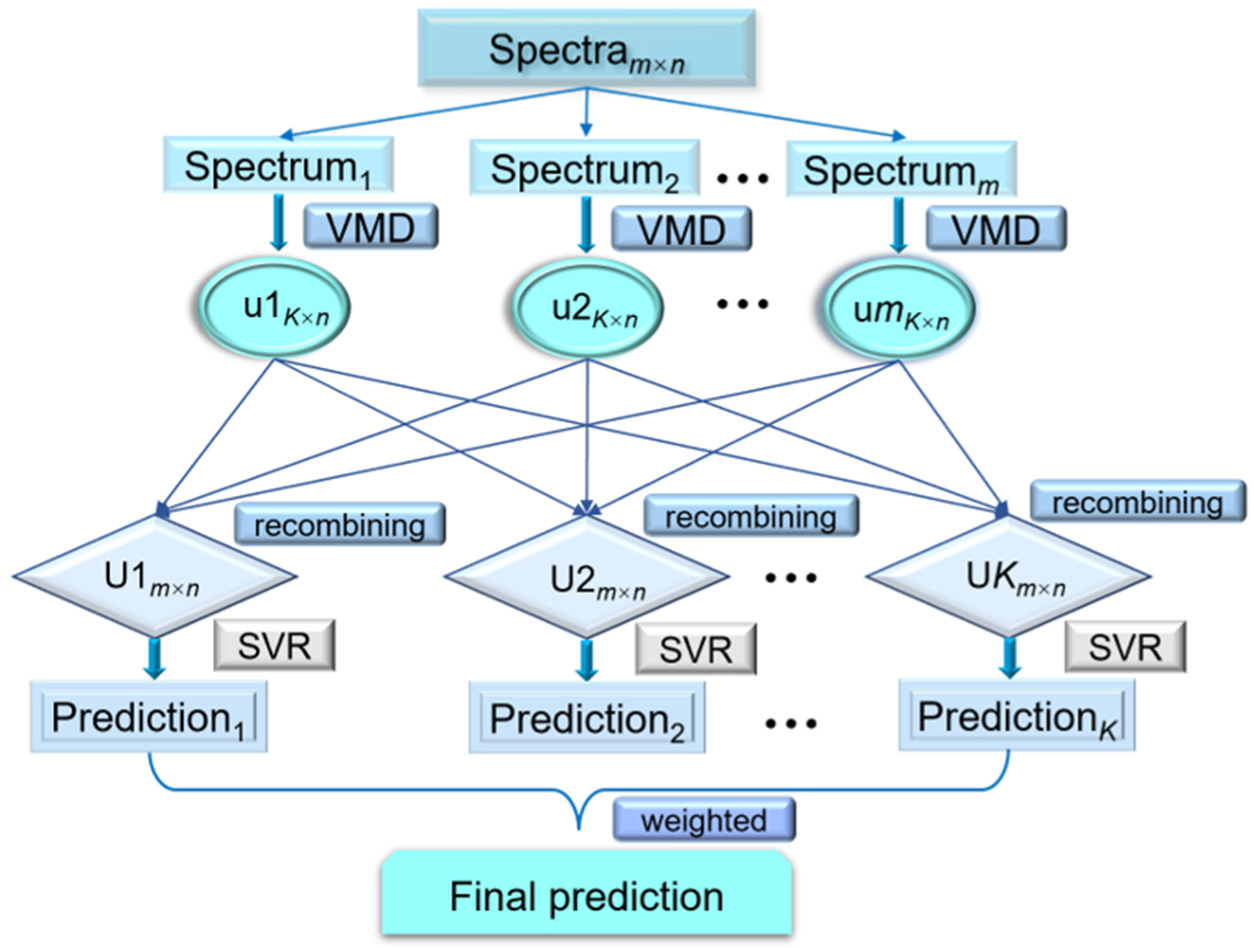
2.5. Variational Mode Decomposition Weighted Multiscale Support Vector Regression (VMD-WMSVR)
3. Results and Discussion
3.1. The Mode Number K
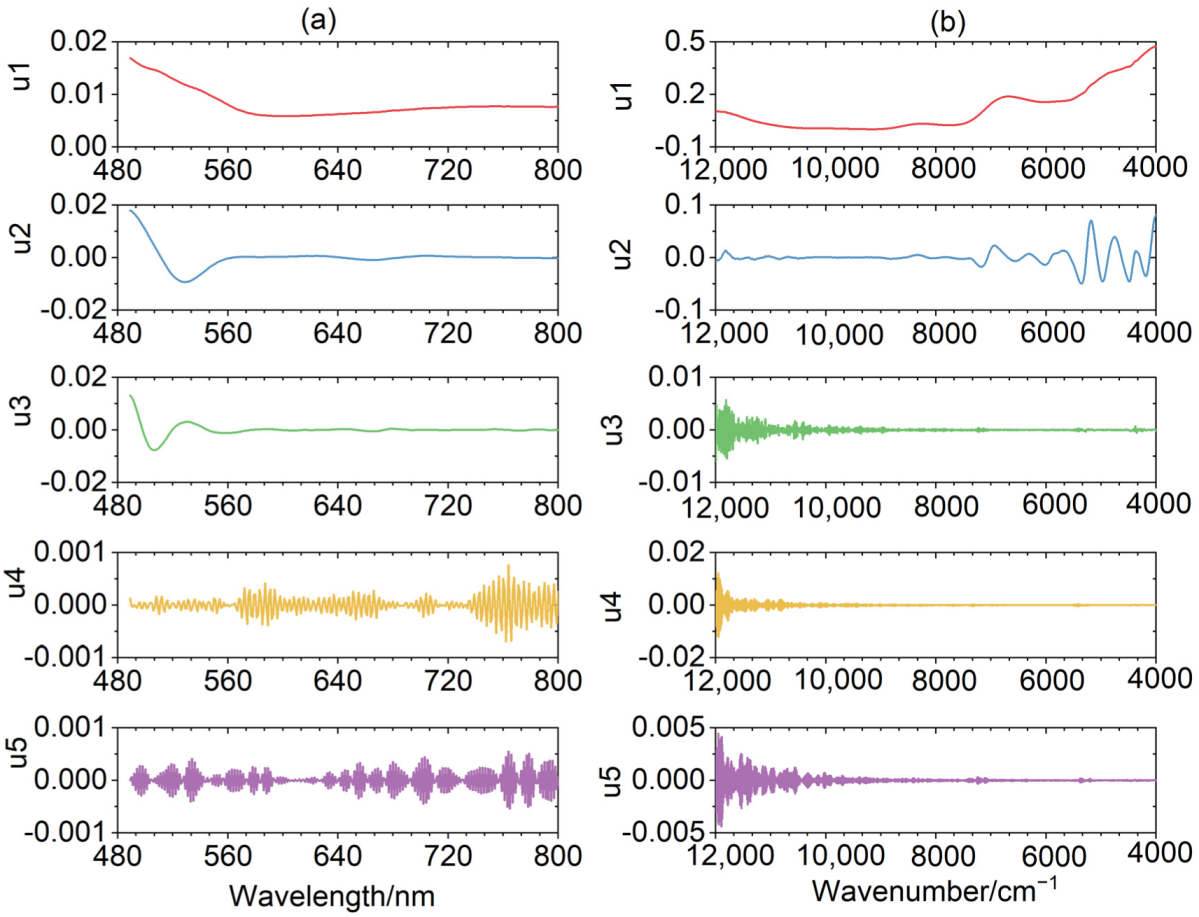
3.2. The Spectral Decomposition of VMD
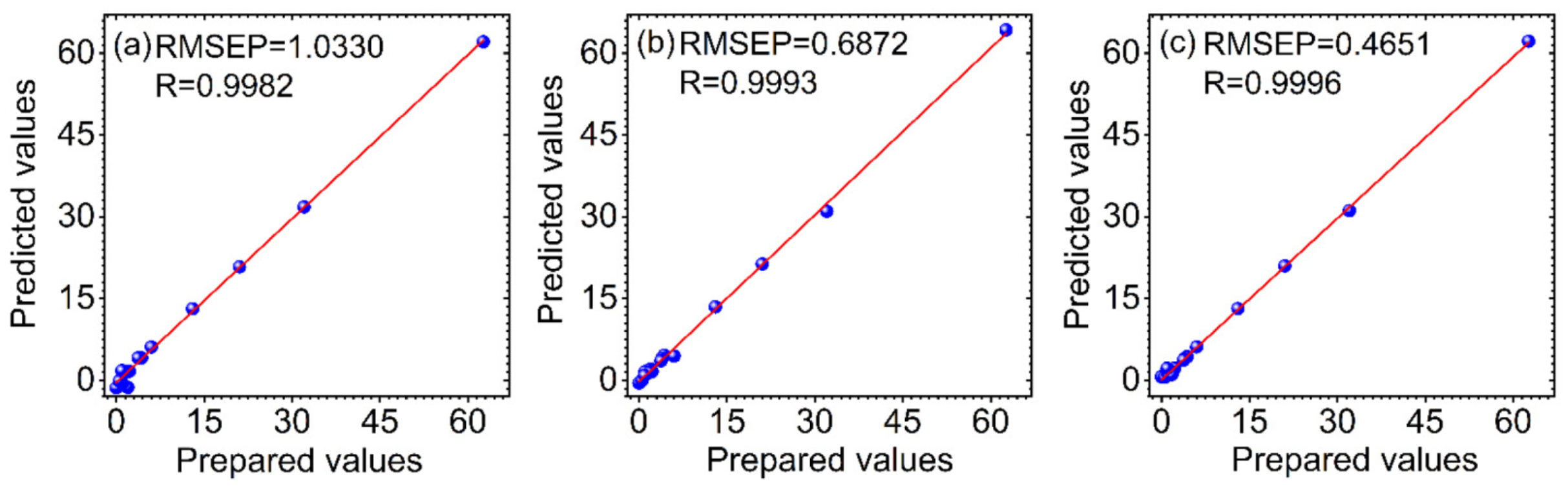
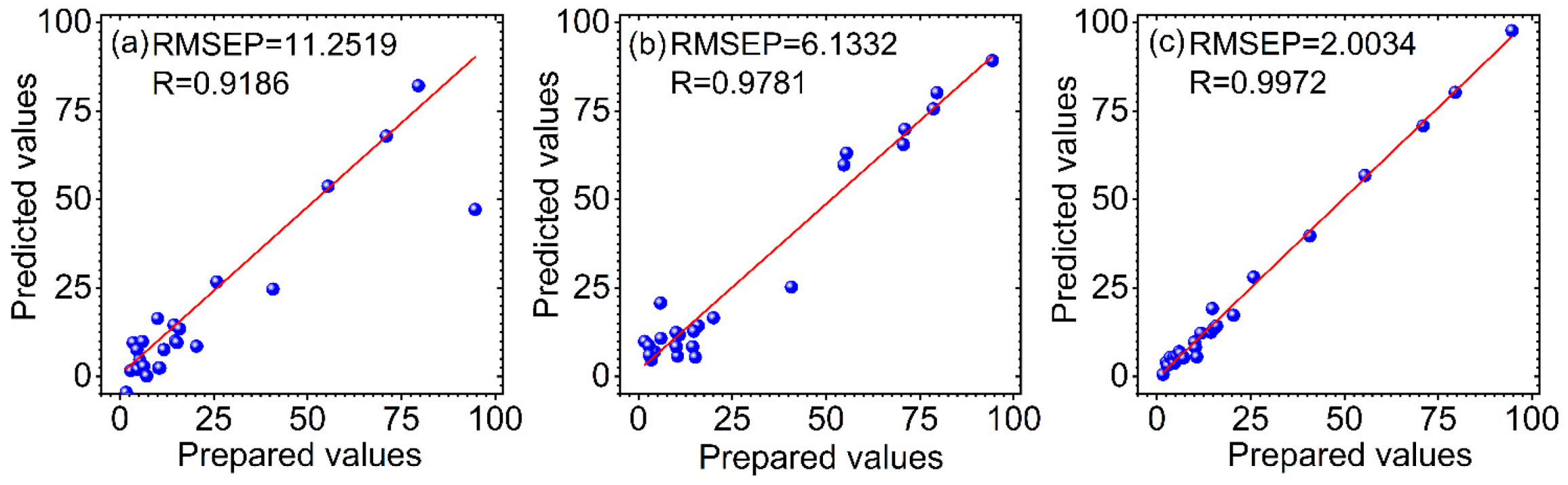
3.3. Comparison of the Predicted Results
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nehal, N.; Choudhary, B.; Nagpure, A.; Gupta, R.K. DNA barcoding: A modern age tool for detection of adulteration in food. Crit. Rev. Biotechnol. 2021, 41, 767–791. [Google Scholar] [CrossRef]
- He, Y.; Bai, X.L.; Xiao, Q.L.; Liu, F.; Zhou, L.; Zhang, C. Detection of adulteration in food based on nondestructive analysis techniques: A review. Crit. Rev. Food Sci. Nutr. 2020, 61, 2351–2371. [Google Scholar] [CrossRef] [PubMed]
- von Wuthenau, K.; Muller, M.-S.; Lina, C.; Marie, O.; Markus, F. Food authentication of almonds (Prunus dulcis Mill.). Fast origin analysis with laser ablation inductively coupled plasma mass spectrometry and chemometrics. J. Agric. Food Chem. 2022, 70, 5237–5244. [Google Scholar] [CrossRef]
- Li, D.; Zang, M.W.; Wang, S.W.; Zhang, K.H.; Zhang, Z.Q.; Li, X.M.; Li, J.C.; Guo, W.P. Food fraud of rejected imported foods in China in 2009–2019. Food Control 2022, 133, 108619. [Google Scholar] [CrossRef]
- Lim, K.; Pan, K.; Yu, Z.; Xiao, R.H. Pattern recognition based on machine learning identifies oil adulteration and edible oil mixtures. Nat. Commun. 2020, 11, 5353. [Google Scholar] [CrossRef]
- Negi, A.; Pare, A.; Meenatchi, R. Emerging techniques for adulterant authentication in spices and spice products. Food Control 2021, 127, 108113. [Google Scholar] [CrossRef]
- Czerwenka, C.; Muller, L.; Lindner, W. Detection of the adulteration of water buffalo milk and mozzarella with cow’s milk by liquid chromatography-mass spectrometry analysis of beta-lactoglobulin variants. Food Chem. 2010, 122, 901–908. [Google Scholar] [CrossRef]
- Acosta, G.; Arce, S.; Martinez, L.D.; Llabot, J.; Gomez, M.R. Monitoring of phenolic compounds for the quality control of melissa officinalis products by capillary electrophoresis. Phytochem. Anal. 2012, 23, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Hong, E.; Lee, S.Y.; Jeong, J.Y.; Park, J.M.; Kim, B.H.; Kwon, K.; Chun, H.S. Modern analytical methods for the detection of food fraud and adulteration by food category. J. Sci. Food Agric. 2017, 97, 3877–3896. [Google Scholar] [CrossRef]
- Bian, X.H.; Lu, Z.K.; van Kollenburg, G. Ultraviolet-visible diffuse reflectance spectroscopy combined with chemometrics for rapid discrimination of Angelicae Sinensis Radix from its four similar herbs. Anal. Methods 2020, 12, 3499–3507. [Google Scholar] [CrossRef]
- Torrecilla, J.S.; Rojo, E.; Dominguez, J.C.; Rodriguez, F. A novel method to quantify the adulteration of extra virgin olive oil with low-grade olive oils by UV-Vis. J. Agric. Food Chem. 2010, 58, 1679–1684. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, L.X.; Zhang, Y.; Wang, D.; Wang, X.F.; Yu, L.; Zhang, W.; Li, P.W. Review of NIR spectroscopy methods for nondestructive quality analysis of oilseeds and edible oils. Trends Food Sci. Technol. 2020, 101, 172–181. [Google Scholar] [CrossRef]
- Lohumi, S.; Lee, S.; Lee, H.; Cho, B.K. A review of vibrational spectroscopic techniques for the detection of food authenticity and adulteration. Trends Food Sci. Technol. 2015, 46, 85–98. [Google Scholar] [CrossRef]
- Rivera-Perez, A.; Romero-Gonzalez, R.; Frenich, A.G. Feasibility of applying untargeted metabolomics with GC-Orbitrap-HRMS and chemometrics for authentication of black pepper (Piper nigrum L.) and identification of geographical and processing markers. J. Agric. Food Chem. 2021, 69, 5547–5558. [Google Scholar] [CrossRef] [PubMed]
- Rios-Reina, R.; Garcia-Gonzalez, D.L.; Callejon, R.M.; Amigo, J.M. NIR spectroscopy and chemometrics for the typification of Spanish wine vinegars with a protected designation of origin. Food Control 2018, 89, 108–116. [Google Scholar] [CrossRef]
- Chen, Z.W.; Harrington, P.D. Self-optimizing support vector elastic net. Anal. Chem. 2020, 92, 15306–15316. [Google Scholar] [CrossRef]
- Jia, W.; Dong, X.Y.; Shi, L.; Chu, X.G. Discrimination of milk from different animal species by a foodomics approach based on high-resolution mass spectrometry. J. Agric. Food Chem. 2020, 68, 6638–6645. [Google Scholar] [CrossRef]
- Shao, X.G.; Bian, X.H.; Liu, J.J.; Zhang, M.; Cai, W.S. Multivariate calibration methods in near infrared spectroscopic analysis. Anal. Methods 2010, 2, 1662–1666. [Google Scholar] [CrossRef]
- Olivieri, A.C. Analytical advantages of multivariate data processing. One, two, three, infinity? Anal. Chem. 2008, 80, 5713–5720. [Google Scholar] [CrossRef]
- Ying, Y.W.; Jin, W.; Yu, H.X.; Yu, B.W.; Shan, J.; Lv, S.W.; Zhu, D.; Jin, Q.H.; Mu, Y. Development of particle swarm optimization-support vector regression (PSO-SVR) coupled with microwave plasma torch-atomic emission spectrometry for quality control of ginsengs. J. Chemom. 2017, 31, e2862. [Google Scholar] [CrossRef]
- Bian, X.H.; Diwu, P.Y.; Liu, Y.R.; Liu, P.; Li, Q.; Tan, X.Y. Ensemble calibration for the spectral quantitative analysis of complex samples. J. Chemom. 2018, 32, e2940. [Google Scholar] [CrossRef]
- Li, Y.K.; Shao, X.G.; Cai, W.S. A consensus least squares support vector regression (LS-SVR) for analysis of near-infrared spectra of plant samples. Talanta 2007, 72, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Ye, Z.-H.; Yan, S.-M.; Shi, P.-T.; Cui, H.-F.; Fu, X.-S.; Yu, X.-P. Combining local wavelength information and ensemble learning to enhance the specificity of class modeling techniques: Identification of food geographical origins and adulteration. Anal. Chim. Acta 2012, 754, 31–38. [Google Scholar] [CrossRef]
- Hu, Y.; Peng, S.L.; Peng, J.T.; Wei, J.P. An improved ensemble partial least squares for analysis of near-infrared spectra. Talanta 2012, 94, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Granato, D.; Santos, J.S.; Escher, G.B.; Ferreira, B.L.; Maggio, R.M. Use of principal component analysis (PCA) and hierarchical cluster analysis (HCA) for multivariate association between bioactive compounds and functional properties in foods: A critical perspective. Trends Food Sci. Tech. 2018, 72, 83–90. [Google Scholar] [CrossRef]
- Jiang, Y.Y.; Ge, H.Y.; Zhang, Y. Quantitative analysis of wheat maltose by combined terahertz spectroscopy and imaging based on Boosting ensemble learning. Food Chem. 2020, 307, 125533. [Google Scholar]
- Bian, X.H.; Li, S.J.; Lin, L.G.; Tan, X.Y.; Fan, Q.J.; Li, M. High and low frequency unfolded partial least squares regression based on empirical mode decomposition for quantitative analysis of fuel oil samples. Anal. Chim. Acta 2016, 925, 16–22. [Google Scholar] [CrossRef]
- Cadet, F.; Fontaine, N.; Vetrivel, I.; Chong, M.N.F.; Savriama, O.; Cadet, X.; Charton, P. Application of fourier transform and proteochemometrics principles to protein engineering. BMC Bioinform. 2018, 19, 382. [Google Scholar] [CrossRef]
- Liu, Z.C.; Cai, W.S.; Shao, X.G. A weighted multiscale regression for multivariate calibration of near infrared spectra. Analyst 2009, 134, 261–266. [Google Scholar] [CrossRef]
- Wu, X.Y.; Bian, X.H.; Lin, E.; Wang, H.T.; Guo, Y.G.; Tan, X.Y. Weighted multiscale support vector regression for fast quantification of vegetable oils in edible blend oil by ultraviolet-visible spectroscopy. Food Chem. 2021, 342, 128245. [Google Scholar] [CrossRef]
- Perez-Canales, D.; Alvarez-Ramirez, J.; Jauregui-Correa, J.C.; Vela-Martinez, L.; Herrera-Ruiz, G. Identification of dynamic instabilities in machining process using the approximate entropy method. Int. J. Mach. Tools Manu. 2011, 51, 556–564. [Google Scholar] [CrossRef]
- Liu, Y.; Cai, W.S.; Shao, X.G. Intelligent background correction using an adaptive lifting wavelet. Chemom. Intell. Lab. Syst. 2013, 125, 11–17. [Google Scholar] [CrossRef]
- Wu, W.H.; Chen, C.C.; Jhou, J.W.; Lai, G. A rapidly convergent empirical mode decomposition method for analyzing the environmental temperature effects on stay cable force. Comput. Aided. Civ. Infrastruct. Eng. 2018, 33, 672–690. [Google Scholar] [CrossRef]
- Dragomiretskiy, K.; Zosso, D. Variational mode decomposition. IEEE. Trans. Signal Proces. 2014, 62, 531–544. [Google Scholar] [CrossRef]
- Chen, Q.M.; Chen, J.H.; Lang, X.; Xie, L.; Rehman, N.U.; Su, H.Y. Self-tuning variational mode decomposition. J. Franklin Inst. 2021, 358, 7825–7862. [Google Scholar] [CrossRef]
- Lahmiri, S. Intraday stock price forecasting based on variational mode decomposition. J. Comput. Sci. Neth. 2016, 12, 23–27. [Google Scholar] [CrossRef]
- Hu, H.L.; Wang, L.; Tao, R. Wind speed forecasting based on variational mode decomposition and improved echo state network. Renew. Energ. 2021, 164, 729–751. [Google Scholar] [CrossRef]
- Wang, Z.J.; Wang, J.J.; Du, W.H. Research on fault diagnosis of gearbox with improved variational mode decomposition. Sensors 2018, 18, 3510. [Google Scholar] [CrossRef] [Green Version]
- Thissen, U.; Pepers, M.; Ustun, B.; Melssen, W.J.; Buydens, L.M.C. Comparing support vector machines to PLS for spectral regression applications. Chemom. Intell. Lab. Syst. 2004, 73, 169–179. [Google Scholar] [CrossRef]
- Nazari, M.; Sakhaei, S.M. Successive variational mode decomposition. Signal Process. 2020, 174, 107610. [Google Scholar] [CrossRef]
- Tan, C.; Wang, J.Y.; Wu, T.; Qin, X.; Li, M.L. Determination of nicotine in tobacco samples by near-infrared spectroscopy and boosting partial least squares. Vib. Spectrosc. 2010, 54, 35–41. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bian, X.; Wu, D.; Zhang, K.; Liu, P.; Shi, H.; Tan, X.; Wang, Z. Variational Mode Decomposition Weighted Multiscale Support Vector Regression for Spectral Determination of Rapeseed Oil and Rhizoma Alpiniae Offcinarum Adulterants. Biosensors 2022, 12, 586. https://doi.org/10.3390/bios12080586
Bian X, Wu D, Zhang K, Liu P, Shi H, Tan X, Wang Z. Variational Mode Decomposition Weighted Multiscale Support Vector Regression for Spectral Determination of Rapeseed Oil and Rhizoma Alpiniae Offcinarum Adulterants. Biosensors. 2022; 12(8):586. https://doi.org/10.3390/bios12080586
Chicago/Turabian StyleBian, Xihui, Deyun Wu, Kui Zhang, Peng Liu, Huibing Shi, Xiaoyao Tan, and Zhigang Wang. 2022. "Variational Mode Decomposition Weighted Multiscale Support Vector Regression for Spectral Determination of Rapeseed Oil and Rhizoma Alpiniae Offcinarum Adulterants" Biosensors 12, no. 8: 586. https://doi.org/10.3390/bios12080586
APA StyleBian, X., Wu, D., Zhang, K., Liu, P., Shi, H., Tan, X., & Wang, Z. (2022). Variational Mode Decomposition Weighted Multiscale Support Vector Regression for Spectral Determination of Rapeseed Oil and Rhizoma Alpiniae Offcinarum Adulterants. Biosensors, 12(8), 586. https://doi.org/10.3390/bios12080586




