Fluorescence-Based Measurements of Membrane-Bound Angiotensin Converting Enzyme 2 Activity Using Xenopus Laevis Oocytes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Molecular Biology
2.2. Oocyte Preparation
2.3. ACE2 Activity Assay
2.4. Data Analysis and Statistics
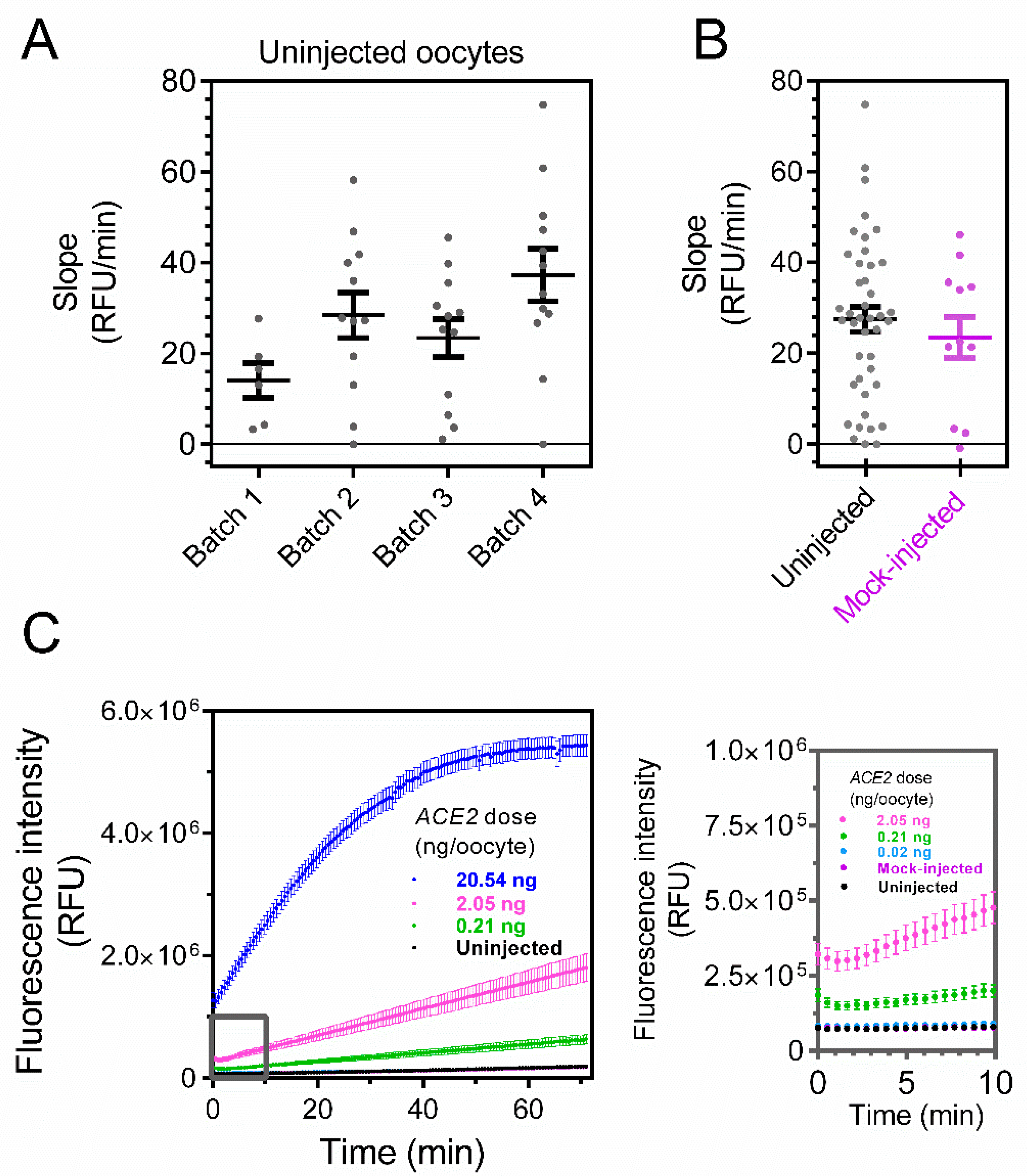
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McDonald, A.G.; Boyce, S.; Tipton, K.F. ExplorEnz: The primary source of the IUBMB enzyme list. Nucleic Acids Res. 2009, 37, D593–D597. [Google Scholar] [CrossRef] [PubMed]
- Bisswanger, H. Enzyme assays. Perspect. Sci. 2014, 1, 41–55. [Google Scholar] [CrossRef] [Green Version]
- Ou, Y.; Wilson, R.E.; Weber, S.G. Methods of Measuring Enzyme Activity Ex Vivo and In Vivo. Ann. Rev. Anal. Chem. 2018, 11, 509–533. [Google Scholar] [CrossRef] [PubMed]
- Boland, M.J.; Hesselink, P.G.; Papamichael, N.; Hustedt, H. Extractive purification of enzymes from animal tissue using aqueous two phase systems: Pilot scale studies. J. Biotechnol. 1991, 19, 19–33. [Google Scholar] [CrossRef]
- Ooi, A.; Wong, A.; Esau, L.; Lemtiri-Chlieh, F.; Gehring, C. A Guide to Transient Expression of Membrane Proteins in HEK-293 Cells for Functional Characterization. Front. Physiol. 2016, 7, 300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.K.; Eberwine, J.H. Mammalian cell transfection: The present and the future. Anal. Bioanal. Chem. 2010, 397, 3173–3178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dascal, N. The use of Xenopus oocytes for the study of ion channels. CRC Crit. Rev. Biochem. 1987, 22, 317–387. [Google Scholar] [CrossRef] [PubMed]
- Ågren, R.; Århem, P.; Nilsson, J.; Sahlholm, K. The Beta-Arrestin-Biased Dopamine D2 Receptor Ligand, UNC9994, Is a Partial Agonist at G-Protein-Mediated Potassium Channel Activation. Int. J. Neuropsychopharmacol. 2018, 21, 1102–1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paucar, M.; Ågren, R.; Li, T.; Lissmats, S.; Bergendal, Å.; Weinberg, J.; Nilsson, D.; Savichetva, I.; Sahlholm, K.; Nilsson, J.; et al. V374A KCND3 Pathogenic Variant Associated with Paroxysmal Ataxia Exacerbations. Neurol. Genet. 2021, 7, e546. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.E.L.; Bezanilla, F. Methodological improvements for fluorescence recordings in Xenopus laevis oocytes. J. Gen. Physiol. 2019, 151, 264–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vickers, C.; Hales, P.; Kaushik, V.; Dick, L.; Gavin, J.; Tang, J.; Godbout, K.; Parsons, T.; Baronas, E.; Hsieh, F.; et al. Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase. J. Biol. Chem. 2002, 277, 14838–14843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rice, G.I.; Thomas, D.A.; Grant, P.J.; Turner, A.J.; Hooper, N.M. Evaluation of angiotensin-converting enzyme (ACE), its homologue ACE2 and neprilysin in angiotensin peptide metabolism. Biochem. J. 2004, 383, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Jespersen, T.; Grunnet, M.; Angelo, K.; Klaerke, D.A.; Olesen, S.P. Dual-function vector for protein expression in both mammalian cells and Xenopus laevis oocytes. Biotechniques 2002, 32, 536–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahlholm, K.; Barchad-Avitzur, O.; Marcellino, D.; Gomez-Soler, M.; Fuxe, K.; Ciruela, F.; Arhem, P. Agonist-specific voltage sensitivity at the dopamine D2S receptor--molecular determinants and relevance to therapeutic ligands. Neuropharmacology 2011, 61, 937–949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graf, S.F.; Madigou, T.; Li, R.; Chesné, C.; Stemmer, A.; Knapp, H.F. Fully automated microinjection system for Xenopus laevis oocytes with integrated sorting and collection. J. Lab. Autom. 2011, 16, 186–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Touret, F.; Gilles, M.; Barral, K.; Nougairède, A.; van Helden, J.; Decroly, E.; de Lamballerie, X.; Coutard, B. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. Sci. Rep. 2020, 10, 13093. [Google Scholar] [CrossRef] [PubMed]
- Bröer, S. Xenopus laevis Oocytes. Methods Mol. Biol. 2010, 637, 295–310. [Google Scholar] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fast, L.; Ågren, R.; Zeberg, H. Fluorescence-Based Measurements of Membrane-Bound Angiotensin Converting Enzyme 2 Activity Using Xenopus Laevis Oocytes. Biosensors 2022, 12, 601. https://doi.org/10.3390/bios12080601
Fast L, Ågren R, Zeberg H. Fluorescence-Based Measurements of Membrane-Bound Angiotensin Converting Enzyme 2 Activity Using Xenopus Laevis Oocytes. Biosensors. 2022; 12(8):601. https://doi.org/10.3390/bios12080601
Chicago/Turabian StyleFast, Luise, Richard Ågren, and Hugo Zeberg. 2022. "Fluorescence-Based Measurements of Membrane-Bound Angiotensin Converting Enzyme 2 Activity Using Xenopus Laevis Oocytes" Biosensors 12, no. 8: 601. https://doi.org/10.3390/bios12080601
APA StyleFast, L., Ågren, R., & Zeberg, H. (2022). Fluorescence-Based Measurements of Membrane-Bound Angiotensin Converting Enzyme 2 Activity Using Xenopus Laevis Oocytes. Biosensors, 12(8), 601. https://doi.org/10.3390/bios12080601




