Electrochemical Biosensor for SARS-CoV-2 cDNA Detection Using AuPs-Modified 3D-Printed Graphene Electrodes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Apparatus, Electrochemical Cell, and Electrodes
2.2. Morphological and Electrochemical Characterization
2.3. Production of the G-PLA Electrode
2.4. Optimization and Electrodeposition of Au
2.5. Optimization of SWV Variables for CNN
2.6. Biosensor Preparation
3. Results and Discussion
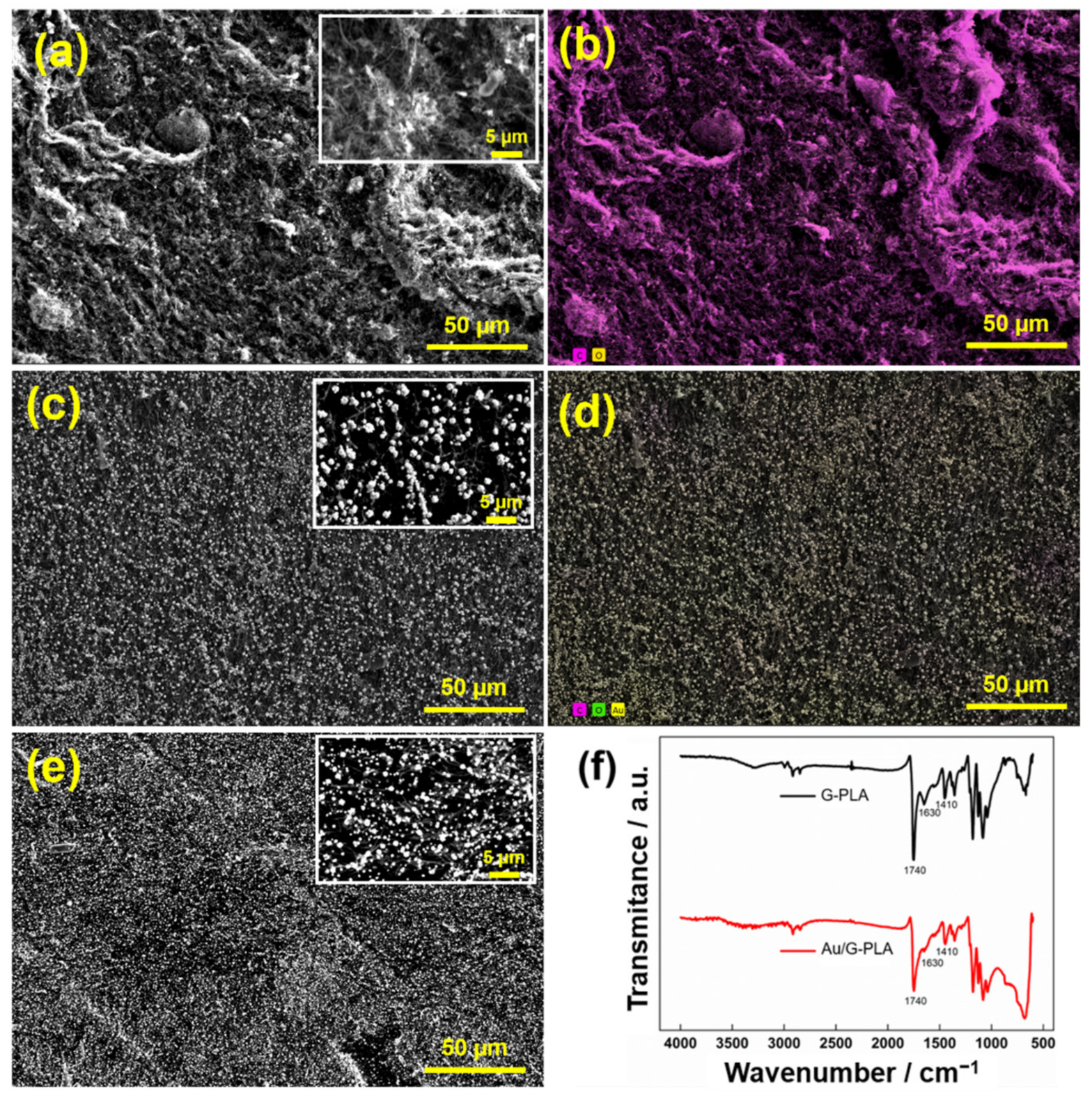
3.1. Morphological and Electrochemical Characterization
3.2. 3D Printed Au/G-PLA Sensor for the Detection of CNN
3.2.1. Cyclic Voltammetry
3.2.2. Optimization and Electrodeposition of Au
3.2.3. Optimization of SWV Variables
3.2.4. Analytical Curve
3.2.5. Interference and Recovery Test
3.3. Biosensor for COVID-19
3.3.1. Biosensor Production
3.3.2. Voltammetric Profile of the Proposed Biosensor
3.3.3. Optimization of Biosensor Parameters
3.3.4. Analytical Performance of the Signal-Off Voltammetric Biosensor
3.3.5. Interfering Study
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stefano, J.S.; Kalinke, C.; da Rocha, R.G.; Rocha, D.P.; da Silva, V.A.O.P.; Bonacin, J.A.; Angnes, L.; Richter, E.M.; Janegitz, B.C.; Muñoz, R.A.A. Electrochemical (bio)sensors enabled by fused deposition modeling-based 3D printing: A guide to selecting designs, printing parameters, and post-treatment protocols. Anal. Chem. 2022, 94, 6417–6429. [Google Scholar] [CrossRef]
- Liyarita, B.R.; Ambrosi, A.; Pumera, M. 3D-printed electrodes for sensing of biologically active molecules. Wiley Online Libr. 2018, 30, 1319–1326. [Google Scholar] [CrossRef]
- Cardoso, R.M.; Kalinke, C.; Rocha, R.G.; dos Santos, P.L.; Rocha, D.P.; Oliveira, P.R.; Janegitz, B.C.; Bonacin, J.A.; Richter, E.M.; Munoz, R.A.A. Additive-manufactured (3D-printed) electrochemical sensors: A critical review. Anal. Chim. Acta 2020, 1118, 73–91. [Google Scholar] [CrossRef] [PubMed]
- Whittingham, M.J.; Crapnell, R.D.; Rothwell, E.J.; Hurst, N.J.; Banks, C.E. Additive manufacturing for electrochemical labs: An overview and tutorial note on the production of cells, electrodes and accessories. Talanta Open 2021, 4, 100051. [Google Scholar] [CrossRef]
- Jayaprakash, G.K. Pre-post redox electron transfer regioselectivity at the alanine modified nano graphene electrode interface. Chem. Phys. Lett. 2022, 789, 139295. [Google Scholar] [CrossRef]
- Coroş, M.; Pruneanu, S.; Stefan-van Staden, R.-I. Recent progress in the graphene-based electrochemical sensors and biosensors. J. Electrochem. Soc. 2020, 167, 037528. [Google Scholar] [CrossRef]
- Wisitsoraat, A.; Tuantranont, A. Graphene-Based Chemical and Biosensors; Springer: Berlin/Heidelberg, Germany, 2013; pp. 103–141. [Google Scholar]
- Foster, C.W.; Down, M.P.; Zhang, Y.; Ji, X.; Rowley-Neale, S.J.; Smith, G.C.; Kelly, P.J.; Banks, C.E. 3D Printed graphene based energy storage devices. Sci. Rep. 2017, 7, 42233. [Google Scholar] [CrossRef]
- Abdalla, A.; Patel, B.A. 3D-printed electrochemical sensors: A new horizon for measurement of biomolecules. Curr. Opin. Electrochem. 2020, 20, 78–81. [Google Scholar] [CrossRef]
- Muñoz, J.; Pumera, M. 3D-Printed COVID-19 immunosensors with electronic readout. Chem. Eng. J. 2021, 425, 131433. [Google Scholar] [CrossRef]
- Khan, M.Z.H.; Hasan, M.R.; Hossain, S.I.; Ahommed, M.S.; Daizy, M. Ultrasensitive detection of pathogenic viruses with electrochemical biosensor: State of the art. Biosens. Bioelectron. 2020, 166, 112431. [Google Scholar] [CrossRef]
- Malik, Y.S.; Kumar, N.; Sircar, S.; Kaushik, R.; Bhat, S.; Dhama, K.; Gupta, P.; Goyal, K.; Singh, M.P.; Ghoshal, U.; et al. Coronavirus disease pandemic (COVID-19): Challenges and a global perspective. Pathogens 2020, 9, 519. [Google Scholar] [CrossRef]
- Vandenberg, O.; Martiny, D.; Rochas, O.; van Belkum, A.; Kozlakidis, Z. Considerations for diagnostic COVID-19 tests. Nat. Rev. Microbiol. 2021, 19, 171–183. [Google Scholar] [CrossRef]
- Giri, B.; Pandey, S.; Shrestha, R.; Pokharel, K.; Ligler, F.S.; Neupane, B.B. Review of analytical performance of COVID-19 detection methods. Anal. Bioanal. Chem. 2021, 413, 35–48. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.W.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef]
- Hwang, C.; Park, N.; Kim, E.S.; Kim, M.; Kim, S.D.; Park, S.; Kim, N.Y.; Kim, J.H. Ultra-fast and recyclable DNA biosensor for point-of-care detection of SARS-CoV-2 (COVID-19). Biosens. Bioelectron. 2021, 185, 113177. [Google Scholar] [CrossRef]
- Brazaca, L.C.; Bramorski, C.B.; Cancino-Bernardi, J.; Janegitz, B.C.; Zucolotto, V. A genosensor for sickle cell anemia trait determination. Wiley Online Libr. 2017, 29, 773–777. [Google Scholar] [CrossRef]
- Beduk, T.; Beduk, D.; de Oliveira Filho, J.I.; Zihnioglu, F.; Cicek, C.; Sertoz, R.; Arda, B.; Goksel, T.; Turhan, K.; Salama, K.N.; et al. Rapid point-of-care COVID-19 diagnosis with a gold-nanoarchitecture-assisted laser-scribed graphene biosensor. Anal. Chem. 2021, 93, 8585–8594. [Google Scholar] [CrossRef]
- Peng, Y.; Pan, Y.; Sun, Z.; Li, J.; Yi, Y.; Yang, J.; Li, G. An electrochemical biosensor for sensitive analysis of the SARS-CoV-2 RNA. Biosens. Bioelectron. 2021, 186, 113309. [Google Scholar] [CrossRef]
- Wang, M.; Lin, Y.; Lu, J.; Sun, Z.; Deng, Y.; Wang, L.; Yi, Y.; Li, J.; Yang, J.; Li, G. Visual naked-eye detection of SARS-CoV-2 RNA based on covalent organic framework capsules. Chem. Eng. J. 2022, 429, 132332. [Google Scholar] [CrossRef]
- Ghanbari, K.; Roushani, M.; Azadbakht, A. Ultra-sensitive aptasensor based on a GQD nanocomposite for detection of hepatitis C virus core antigen. Anal. Biochem. 2017, 534, 64–69. [Google Scholar] [CrossRef]
- Li, X.; Scida, K.; Crooks, R.M. Detection of hepatitis B virus DNA with a paper electrochemical sensor. Anal. Chem. 2015, 87, 9009–9015. [Google Scholar] [CrossRef]
- Brazaca, L.C.; Imamura, A.H.; Gomes, N.O.; Almeida, M.B.; Scheidt, D.T.; Raymundo-Pereira, P.A.; Oliveira, O.N.; Janegitz, B.C.; Machado, S.A.S.; Carrilho, E. Electrochemical immunosensors using electrodeposited gold nanostructures for detecting the S proteins from SARS-CoV and SARS-CoV-2. Anal. Bioanal. Chem. 2022, 414, 5507–5517. [Google Scholar] [CrossRef]
- Stefano, J.S.; Guterres e Silva, L.R.; Rocha, R.G.; Brazaca, L.C.; Richter, E.M.; Abarza Muñoz, R.A.; Janegitz, B.C. New conductive filament ready-to-use for 3D-printing electrochemical (bio)sensors: Towards the detection of SARS-CoV-2. Anal. Chim. Acta 2021, 1191, 339372. [Google Scholar] [CrossRef]
- Brazaca, L.C.; dos Santos, P.L.; de Oliveira, P.R.; Rocha, D.P.; Stefano, J.S.; Kalinke, C.; Abarza Muñoz, R.A.; Bonacin, J.A.; Janegitz, B.C.; Carrilho, E. Biosensing strategies for the electrochemical detection of viruses and viral diseases—A review. Anal. Chim. Acta 2021, 1159, 338384. [Google Scholar] [CrossRef]
- Tripathy, S.; Singh, S.G. Label-free electrochemical detection of DNA hybridization: A method for COVID-19 diagnosis. Trans. Indian Natl. Acad. Eng. 2020, 5, 205–209. [Google Scholar] [CrossRef]
- Shi, L.; Wang, L.; Ma, X.; Fang, X.; Xiang, L.; Yi, Y.; Li, J.; Luo, Z.; Li, G. Aptamer-functionalized nanochannels for one-step detection of SARS-CoV-2 in samples from COVID-19 patients. Anal. Chem. 2021, 93, 16646–16654. [Google Scholar] [CrossRef]
- Crevillen, A.G.; Mayorga-Martinez, C.C.; Vaghasiya, J.V.; Pumera, M. 3D-printed SARS-CoV-2 RNA genosensing microfluidic system. Adv. Mater. Technol. 2022, 7, 2101121. [Google Scholar] [CrossRef]
- Werion, A.; Belkhir, L.; Perrot, M.; Schmit, G.; Aydin, S.; Chen, Z.; Penaloza, A.; De Greef, J.; Yildiz, H.; Pothen, L.; et al. SARS-CoV-2 causes a specific dysfunction of the kidney proximal tubule. Kidney Int. 2020, 98, 1296–1307. [Google Scholar] [CrossRef]
- Xiang, J.; Wen, J.; Yuan, X.; Xiong, S.; Zhou, X.; Liu, C.; Min, X. Potential biochemical markers to identify severe cases among COVID-19 patients. medRxiv 2020. [Google Scholar] [CrossRef]
- Li, T.J.; Chen, P.Y.; Nien, P.C.; Lin, C.Y.; Vittal, R.; Ling, T.R.; Ho, K.C. Preparation of a novel molecularly imprinted polymer by the sol-gel process for sensing creatinine. Anal. Chim. Acta 2012, 711, 83–90. [Google Scholar] [CrossRef]
- Jacobi, D.; Lavigne, C.; Halimi, J.M.; Fierrard, H.; Andres, C.; Couet, C.; Maillot, F. Variability in creatinine excretion in adult diabetic, overweight men and women: Consequences on creatinine-based classification of renal disease. Diabetes Res. Clin. Pract. 2008, 80, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Lad, U.; Khokhar, S.; Kale, G.M. Electrochemical creatinine biosensors. Anal. Chem. 2008, 80, 7910–7917. [Google Scholar] [CrossRef] [PubMed]
- Bishop, M.L.; Fody, E.P.; Schoeff, L.E. Clinical Chemistry: Principles, Techniques, and Correlations, 7th ed.; Wolters Kluwer Health/Hippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; ISBN 9781469837130. [Google Scholar]
- Brutis, C.A.; Bruns, D.E. Tietz Fundamentals of Clinical Chemistry and Molecular Diagnostics, 7th ed.; Elsevier-Health Sciences Division: St. Louis, MO, USA, 2014; ISBN 9780323292061. [Google Scholar]
- Vella, F. Textbook of clinical chemistry. Biochem. Educ. 1986, 14, 146. [Google Scholar] [CrossRef]
- Chan, K.H.; Farouji, I.; Abu Hanoud, A.; Slim, J. Weakness and elevated creatinine kinase as the initial presentation of coronavirus disease 2019 (COVID-19). Am. J. Emerg. Med. 2020, 38, 1548.e1–1548.e3. [Google Scholar] [CrossRef]
- Stradiotto, N.R.; Yamanaka, H.; Zanoni, M.V.B. Electrochemical sensors: A powerful tool in analytical chemistry. J. Braz. Chem. Soc. 2003, 14, 159–173. [Google Scholar] [CrossRef]
- Do, J.S.; Chang, Y.H.; Tsai, M.L. Highly sensitive amperometric creatinine biosensor based on creatinine deiminase/Nafion®-nanostructured polyaniline composite sensing film prepared with cyclic voltammetry. Mater. Chem. Phys. 2018, 219, 1–12. [Google Scholar] [CrossRef]
- Fava, E.L.; Prado, T.M.D.; Garcia-Filho, A.; Silva, T.A.; Cincotto, F.H.; Cruz de Moraes, F.; Faria, R.C.; Fatibello-Filho, O. Non-enzymatic electrochemical determination of creatinine using a novel screen-printed microcell. Talanta 2020, 207, 120277. [Google Scholar] [CrossRef]
- Kumar, V.; Hebbar, S.; Kalam, R.; Panwar, S.; Prasad, S.; Srikanta, S.S.; Krishnaswamy, P.R.; Bhat, N. Creatinine-iron complex and its use in electrochemical measurement of urine creatinine. IEEE Sens. J. 2018, 18, 830–836. [Google Scholar] [CrossRef]
- Naresh Kumar, T.; Ananthi, A.; Mathiyarasu, J.; Joseph, J.; Lakshminarasimha Phani, K.; Yegnaraman, V. Enzymeless creatinine estimation using poly(3,4-ethylenedioxythiophene)-β-cyclodextrin. J. Electroanal. Chem. 2011, 661, 303–308. [Google Scholar] [CrossRef]
- Hooshmand, S.; Es’haghi, Z. Microfabricated disposable nanosensor based on CdSe quantum dot/ionic liquid-mediated hollow fiber-pencil graphite electrode for simultaneous electrochemical quantification of uric acid and creatinine in human samples. Anal. Chim. Acta 2017, 972, 28–37. [Google Scholar] [CrossRef]
- Randviir, E.P.; Brownson, D.A.C.; Banks, C.E. A decade of graphene research: Production, applications and outlook. Mater. Today 2014, 17, 426–432. [Google Scholar] [CrossRef]
- Raveendran, J.; Resmi, P.E.; Ramachandran, T.; Nair, B.G.; Satheesh Babu, T.G. Fabrication of a disposable non-enzymatic electrochemical creatinine sensor. Sens. Actuators B Chem. 2017, 243, 589–595. [Google Scholar] [CrossRef]
- Rakesh Kumar, R.K.; Shaikh, M.O.; Chuang, C.H. A review of recent advances in non-enzymatic electrochemical creatinine biosensing. Anal. Chim. Acta 2021, 1183, 338748. [Google Scholar] [CrossRef]
- Cardoso, R.M.; Castro, S.V.F.; Silva, M.N.T.; Lima, A.P.; Santana, M.H.P.; Nossol, E.; Silva, R.A.B.; Richter, E.M.; Paixão, T.R.L.C.; Muñoz, R.A.A. 3D-printed flexible device combining sampling and detection of explosives. Sens. Actuators B Chem. 2019, 292, 308–313. [Google Scholar] [CrossRef]
- Ngo, T.D.; Kashani, A.; Imbalzano, G.; Nguyen, K.T.Q.; Hui, D. Additive manufacturing (3D printing): A review of materials, methods, applications and challenges. Compos. Part B Eng. 2018, 143, 172–196. [Google Scholar] [CrossRef]
- Da Silva, V.A.O.P.; Tartare, V.A.P.; Kalinke, C.; Roberto De Oliveira, P.; Cardoso De Souza, D.; Bonacin, J.A.; Janegitz, B.C. Construção de um suporte ajustável lab-made impresso em 3D para medição de ângulo de contato. Quim. Nov. 2020, 43, 1312–1319. [Google Scholar] [CrossRef]
- Guo, S.; Wang, L.; Wang, E. Templateless, surfactantless, simple electrochemical route to rapid synthesis of diameter-controlled 3D flowerlike gold microstructure with “clean” surface. Chem. Commun. 2007, 3163–3165. [Google Scholar] [CrossRef]
- Sun, X.; Dong, S.; Wang, E. Large-scale synthesis of micrometer-scale single-crystalline Au plates of nanometer thickness by a wet-chemical route. Angew. Chem. Int. Ed. 2004, 43, 6360–6363. [Google Scholar] [CrossRef]
- Zhang, J.; Du, J.; Han, B.; Liu, Z.; Jiang, T.; Zhang, Z. Sonochemical formation of single-crystalline gold nanobelts. Wiley Online Libr. 2006, 45, 1116–1119. [Google Scholar] [CrossRef]
- Antonini, S.; Nguyen, P.T.; Arnold, U.; Eichert, T.; Clemens, J. Solar thermal evaporation of human urine for nitrogen and phosphorus recovery in Vietnam. Sci. Total Environ. 2012, 414, 592–599. [Google Scholar] [CrossRef]
- Romonti, D.E.; Gomez Sanchez, A.V.; Milošev, I.; Demetrescu, I.; Ceré, S. Effect of anodization on the surface characteristics and electrochemical behaviour of zirconium in artificial saliva. Mater. Sci. Eng. C 2016, 62, 458–466. [Google Scholar] [CrossRef]
- Barros Neto, B.; Scarminio, I.S.; Bruns, R.E. Como Fazer Experimentos: Pesquisa e Desenvolvimento na Ciência e na Indústria; Editora da Unicamp: Campinas, SP, Brazil, 2001. [Google Scholar]
- Teófilo, R.F.; Ferreira, M.M.C. Chemometrics II: Spreadsheets for experimental design calculations, A tutorial. Quim. Nova 2006, 29, 338–350. [Google Scholar] [CrossRef]
- Yi, M.; Zhang, C. The synthesis of two-dimensional MoS2 nanosheets with enhanced tribological properties as oil additives. RSC Adv. 2018, 8, 9564–9573. [Google Scholar] [CrossRef]
- Yagati, A.K.; Go, A.; Vu, N.H.; Lee, M.H. A MoS2–Au nanoparticle-modified immunosensor for T3 biomarker detection in clinical serum samples. Electrochim. Acta 2020, 342, 136065. [Google Scholar] [CrossRef]
- Chi, H.; Liu, B.; Guan, G.; Zhang, Z.; Han, M.Y. A simple, reliable and sensitive colorimetric visualization of melamine in milk by unmodified gold nanoparticles. Analyst 2010, 135, 1070–1075. [Google Scholar] [CrossRef]
- Cui, H.; Wang, W.; Duan, C.F.; Dong, Y.P.; Guo, J.Z. Synthesis, characterization, and electrochemiluminescence of luminolreduced gold nanoparticles and their application in a hydrogen peroxide sensor. Chem.—A Eur. J. 2007, 13, 6975–6984. [Google Scholar] [CrossRef]
- He, Y.; Peng, R. Luminol functionalized gold nanoparticles as colorimetric and chemiluminescent probes for visual, label free, highly sensitive and selective detection of minocycline. Nanotechnology 2014, 25, 455502. [Google Scholar] [CrossRef]
- Gittins, D.I.; Caruso, F. Spontaneous phase transfer of nanoparticulate metals from organic to aqueous media. Angew. Chemie—Int. Ed. 2001, 40, 3001–3004. [Google Scholar] [CrossRef]
- He, Y.; Zhang, X.; Yu, H. Gold nanoparticles-based colorimetric and visual creatinine assay. Microchim. Acta 2015, 182, 2037–2043. [Google Scholar] [CrossRef]
- Ziegel, E.R.; Massart, D.L.; Vandeginste, B.G.M.; Buydens, L.M.C.; de Jong, S.; Lewi, P.J.; Verbeke, J.S. Handbook of chemometrics and qualimetrics, part B. Technometrics 2000, 42, 218. [Google Scholar] [CrossRef]
- Carr, O.; Raymundo-Pereira, P.A.; Shimizu, F.M.; Sorroche, B.P.; Melendez, M.E.; de Oliveira Pedro, R.; Miranda, P.B.; Carvalho, A.L.; Reis, R.M.; Arantes, L.M.R.B.; et al. Genosensor made with a self-assembled monolayer matrix to detect MGMT gene methylation in head and neck cancer cell lines. Talanta 2020, 210, 120609. [Google Scholar] [CrossRef] [PubMed]
- Arnaout, R.; Lee, R.A.; Lee, G.R.; Callahan, C.; Yen, C.F.; Smith, K.P.; Arora, R.; Kirby, J.E. SARS-CoV2 testing: The limit of detection matters. bioRxiv 2020. [Google Scholar] [CrossRef]
- Bustin, S.A. How to speed up the polymerase chain reaction. Biomol. Detect. Quantif. 2017, 12, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.L.L.; Maslova, A.; Hsing, I.-M. Rational design of electrochemical DNA biosensors for point-of-care applications. ChemElectroChem 2017, 4, 795–805. [Google Scholar] [CrossRef]
- Dubé, C.; Kelton, D.F.; Dubé, C.; Ribble, C.; Kelton, D.; Mcnab, B. Introduction to network analysis and its implications for animal disease modelling. Rev. Sci. Tech. Off. Int. Epiz. 2011, 30, 425–436. [Google Scholar] [CrossRef]
- Janegitz, B.C.; Cancino, J.; Zucolotto, V. Disposable biosensors for clinical diagnosis. J. Nanosci. Nanotechnol. 2014, 14, 378–389. [Google Scholar] [CrossRef]
- del Caño, R.; García-Mendiola, T.; García-Nieto, D.; Álvaro, R.; Luna, M.; Iniesta, H.A.; Coloma, R.; Diaz, C.R.; Milán-Rois, P.; Castellanos, M.; et al. Amplification-free detection of SARS-CoV-2 using gold nanotriangles functionalized with oligonucleotides. Microchim. Acta 2022, 189, 171. [Google Scholar] [CrossRef]
- Deng, Y.; Peng, Y.; Wang, L.; Wang, M.; Zhou, T.; Xiang, L.; Li, J.; Yang, J.; Li, G. Target-triggered cascade signal amplification for sensitive electrochemical detection of SARS-CoV-2 with clinical application. Anal. Chim. Acta 2022, 1208, 339846. [Google Scholar] [CrossRef]
- Ang, W.L.; Lim, R.R.X.; Ambrosi, A.; Bonanni, A. Rapid electrochemical detection of COVID-19 genomic sequence with dual-function graphene nanocolloids based biosensor. FlatChem 2022, 32, 100336. [Google Scholar] [CrossRef]
- Damiati, S.; Sopstad, S.; Peacock, M.; Akhtar, A.S.; Pinto, I.; Soares, R.R.G.; Russom, A. Flex printed circuit board implemented graphene-based DNA aensor for setection of SARS-CoV-2. IEEE Sens. J. 2021, 21, 13060–13067. [Google Scholar] [CrossRef]
- Kozitsina, A.N.; Shalygina, Z.V.; Dedeneva, S.S.; Rusinov, G.L.; Tolshchina, S.G.; Verbitskiy, E.V.; Brainina, K.Z. Catalytic systems based on the organic nickel (II) complexes in chronoamperometric determination of urea and creatinine. Russ. Chem. Bull. 2009, 58, 1119–1125. [Google Scholar] [CrossRef]
- Chen, J.C.; Kumar, A.S.; Chung, H.H.; Chien, S.H.; Kuo, M.C.; Zen, J.M. An enzymeless electrochemical sensor for the selective determination of creatinine in human urine. Sens. Actuators B Chem. 2006, 115, 473–480. [Google Scholar] [CrossRef]
- Randviir, E.P.; Kampouris, D.K.; Banks, C.E. An improved electrochemical creatinine detection method via a Jaffe-based procedure. Analyst 2013, 138, 6565–6572. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, L.R.G.; Stefano, J.S.; Orzari, L.O.; Brazaca, L.C.; Carrilho, E.; Marcolino-Junior, L.H.; Bergamini, M.F.; Munoz, R.A.A.; Janegitz, B.C. Electrochemical Biosensor for SARS-CoV-2 cDNA Detection Using AuPs-Modified 3D-Printed Graphene Electrodes. Biosensors 2022, 12, 622. https://doi.org/10.3390/bios12080622
Silva LRG, Stefano JS, Orzari LO, Brazaca LC, Carrilho E, Marcolino-Junior LH, Bergamini MF, Munoz RAA, Janegitz BC. Electrochemical Biosensor for SARS-CoV-2 cDNA Detection Using AuPs-Modified 3D-Printed Graphene Electrodes. Biosensors. 2022; 12(8):622. https://doi.org/10.3390/bios12080622
Chicago/Turabian StyleSilva, Luiz R. G., Jéssica S. Stefano, Luiz O. Orzari, Laís C. Brazaca, Emanuel Carrilho, Luiz H. Marcolino-Junior, Marcio F. Bergamini, Rodrigo A. A. Munoz, and Bruno C. Janegitz. 2022. "Electrochemical Biosensor for SARS-CoV-2 cDNA Detection Using AuPs-Modified 3D-Printed Graphene Electrodes" Biosensors 12, no. 8: 622. https://doi.org/10.3390/bios12080622
APA StyleSilva, L. R. G., Stefano, J. S., Orzari, L. O., Brazaca, L. C., Carrilho, E., Marcolino-Junior, L. H., Bergamini, M. F., Munoz, R. A. A., & Janegitz, B. C. (2022). Electrochemical Biosensor for SARS-CoV-2 cDNA Detection Using AuPs-Modified 3D-Printed Graphene Electrodes. Biosensors, 12(8), 622. https://doi.org/10.3390/bios12080622







