Electrical Characterization and Analysis of Single Cells and Related Applications
Abstract
1. Introduction
2. Single-Cell Sorting and Separation
3. Single-Cell Manipulation
| Category | First Author (Year) | Target Cells | Application | Ref. |
|---|---|---|---|---|
| Tumor cells | Desai (2019) | Thyroid, Breast, Lung, and Ovarian cancer cells | Cell recognition | [41] |
| Ren (2019) | MDA-MB-231 cells | Cell recognition | [42] | |
| McGrath (2020) | Six types of pancreatic ductal | Cell screening | [43] | |
| Ostermann (2020) | Adenocarcinoma cell U937 cells | Viability assay | [44] | |
| Zhang (2020) | A549 and Hep G2 cells | Cell screening | [45] | |
| Plant cells | Impe (2019) | Wheat pollen Hazelnut pollen | Viability assay | [46] |
| Ascari (2020) | Wheat microspore | Viability assay | [47] | |
| Canonge (2020) | Herbaceous | Monitoring androgenesis process | [48] | |
| Han (2020) | Arabidopsis thaliana and woody Populus trichocarpa | Cell screening | [49] | |
| Microbes | Xie (2019) | S. cerevisiae | performance assessment | [50] |
| Opitz (2019) | S. cerevisiae | Viability assay | [51] | |
| Bertelsen (2020) | E. coli | Determination of the viability of E. coli | [52] | |
| Spencer (2020) | K. pneumoniae | Antimicrobial susceptibility tests | [53] | |
| Stem cells | Song (2016) | Mesenchymal stem cells | Monitoring differentiation process | [54] |
| Xavier (2017) | Skeletal stem cells | Monitoring differentiation process | [55] |
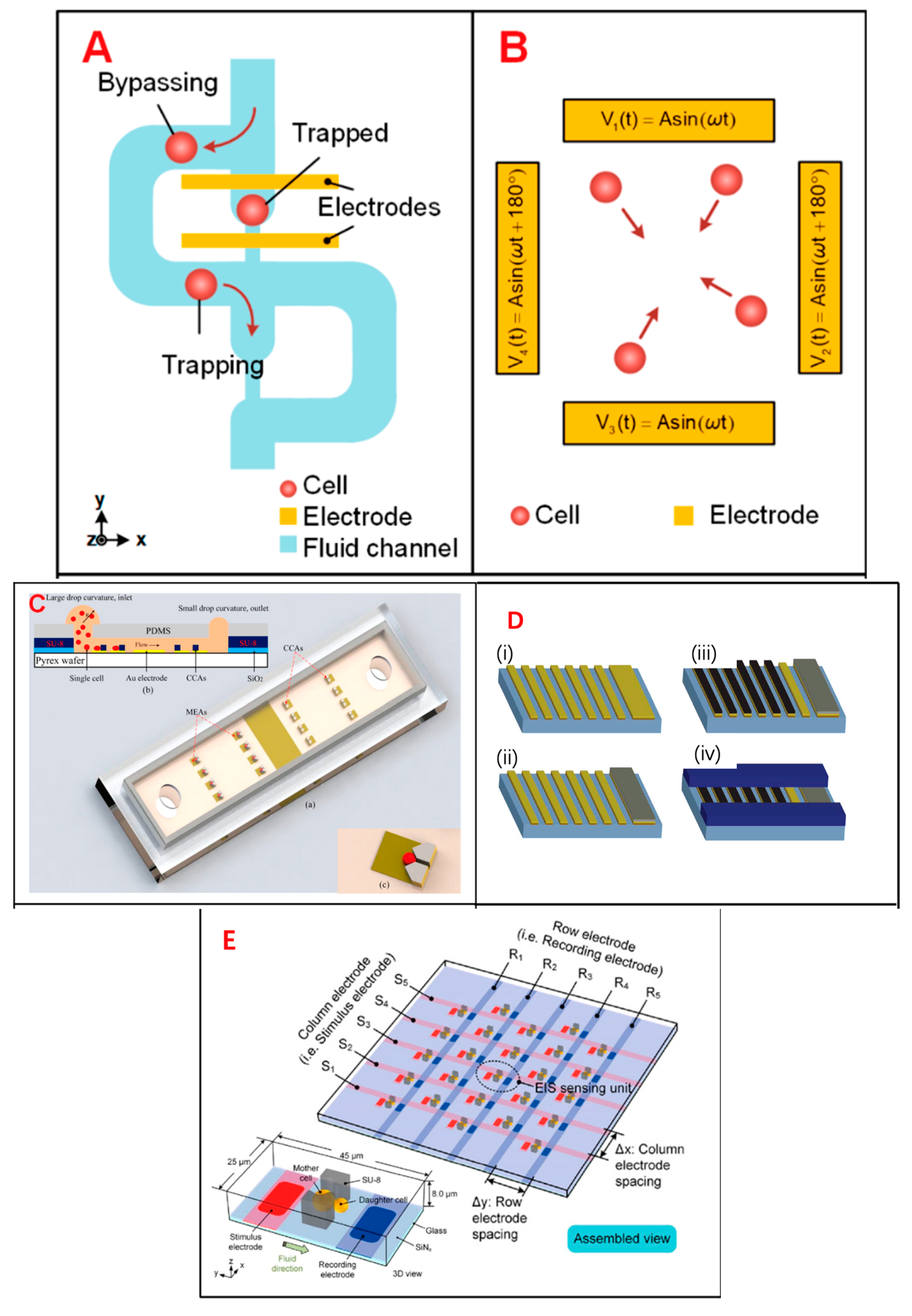


4. Single-Cell Analysis and Application


5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, L.; Wan, S.; Jiang, Y.; Wang, Y.; Fu, T.; Liu, Q.; Cao, Z.; Qiu, L.; Tan, W. Molecular Elucidation of Disease Biomarkers at the Interface of Chemistry and Biology. J. Am. Chem. Soc. 2017, 139, 2532–2540. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, M.; Jendrusch, M.; Constantinou, I. Spatially resolved electrical impedance methods for cell and particle characterization. Electrophoresis 2020, 41, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Adan, A.; Alizada, G.; Kiraz, Y.; Baran, Y.; Nalbant, A. Flow cytometry: Basic principles and applications. Crit. Rev. Biotechnol. 2017, 37, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, Z.R. Cellular dielectrophoresis: Applications to the characterization, manipulation, separation and patterning of cells. Electrophoresis 2011, 32, 2466–2487. [Google Scholar] [CrossRef]
- Xu, J.; Liao, K.; Yang, X.; Wu, C.; Wu, W. Correction to: Using single-cell sequencing technology to detect circulating tumor cells in solid tumors. Mol. Cancer 2022, 21, 100. [Google Scholar] [CrossRef]
- Ramser, K.; Hanstorp, D. Optical manipulation for single-cell studies. J. Biophotonics 2010, 3, 187–206. [Google Scholar] [CrossRef]
- Kirkness, E.F.; Grindberg, R.V.; Yee-Greenbaum, J.; Marshall, C.R.; Scherer, S.W.; Lasken, R.S.; Venter, J.C. Sequencing of isolated sperm cells for direct haplotyping of a human genome. Genome Res. 2013, 23, 826–832. [Google Scholar] [CrossRef]
- Peeters, D.J.; De Laere, B.; Van den Eynden, G.G.; Van Laere, S.J.; Rothé, F.; Ignatiadis, M.; Sieuwerts, A.M.; Lambrechts, D.; Rutten, A.; van Dam, P.A.; et al. Semiautomated isolation and molecular characterisation of single or highly purified tumour cells from CellSearch enriched blood samples using dielectrophoretic cell sorting. Br. J. Cancer 2013, 108, 1358–1367. [Google Scholar] [CrossRef]
- Choi, J.H.; Ogunniyi, A.O.; Du, M.; Du, M.; Kretschmann, M.; Eberhardt, J.; Love, J.C. Development and optimization of a process for automated recovery of single cells identified by microengraving. Biotechnol. Prog. 2010, 26, 888–895. [Google Scholar] [CrossRef]
- Fuchs, A.B.; Romani, A.; Freida, D.; Medoro, G.; Abonnenc, M.; Altomare, L.; Chartier, I.; Guergour, D.; Villiers, C.; Marche, P.N.; et al. Electronic sorting and recovery of single live cells from microlitre sized samples. Lab Chip 2006, 6, 121–126. [Google Scholar] [CrossRef]
- Kolodziejczyk, A.A.; Kim, J.K.; Svensson, V.; Marioni, J.C.; Teichmann, S.A. The technology and biology of single-cell RNA sequencing. Mol. Cell 2015, 58, 610–620. [Google Scholar] [CrossRef] [PubMed]
- Lan, F.; Demaree, B.; Ahmed, N.; Abate, A.R. Single-cell genome sequencing at ultra-high-throughput with microfluidic droplet barcoding. Nat. Biotechnol. 2017, 35, 640–646. [Google Scholar] [CrossRef]
- Klein, A.M.; Mazutis, L.; Akartuna, I.; Tallapragada, N.; Veres, A.; Li, V.; Peshkin, L.; Weitz, D.A.; Kirschner, M.W. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 2015, 161, 1187–1201. [Google Scholar] [CrossRef] [PubMed]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.C.; Wang, J.; Potanina, A.; Quake, S.R. Whole-genome molecular haplotyping of single cells. Nat. Biotechnol. 2011, 29, 51–57. [Google Scholar] [CrossRef]
- Luo, T.; Fan, L.; Zhu, R.; Sun, D. Microfluidic Single-Cell Manipulation and Analysis: Methods and Applications. Micromachines 2019, 10, 104. [Google Scholar] [CrossRef]
- Altschuler, S.J.; Wu, L.F. Cellular heterogeneity: Do differences make a difference? Cell 2010, 141, 559–563. [Google Scholar] [CrossRef]
- Turner, N.C.; Reis-Filho, J.S. Genetic heterogeneity and cancer drug resistance. Lancet Oncol. 2012, 13, e178–e185. [Google Scholar] [CrossRef]
- Bauwens, C.L.; Peerani, R.; Niebruegge, S.; Woodhouse, K.A.; Kumacheva, E.; Husain, M.; Zandstra, P.W. Control of human embryonic stem cell colony and aggregate size heterogeneity influences differentiation trajectories. Stem. Cells 2008, 26, 2300–2310. [Google Scholar] [CrossRef]
- Gao, D.; Jin, F.; Zhou, M.; Jiang, Y. Recent advances in single cell manipulation and biochemical analysis on microfluidics. Analyst 2019, 144, 766–781. [Google Scholar] [CrossRef]
- Nagrath, S.; Sequist, L.V.; Maheswaran, S.; Bell, D.W.; Irimia, D.; Ulkus, L.; Smith, M.R.; Kwak, E.L.; Digumarthy, S.; Muzikansky, A. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007, 450, 1235–1239. [Google Scholar] [CrossRef]
- Zhou, J.; Jiang, D.; Chen, H. Nanoelectrochemical architectures for high-spatial-resolution single cell analysis. Sci. China 2017, 60, 1277–1284. [Google Scholar] [CrossRef]
- Zhang, X.W.; Qiu, Q.F.; Jiang, H.; Zhang, F.L.; Liu, Y.L.; Amatore, C.; Huang, W.H. Real-Time Intracellular Measurements of ROS and RNS in Living Cells with Single Core-Shell Nanowire Electrodes. Angew. Chem. Int. Ed. Engl. 2017, 56, 12997–13000. [Google Scholar] [CrossRef] [PubMed]
- Erofeev, A.; Gorelkin, P.; Garanina, A.; Alova, A.; Efremova, M.; Vorobyeva, N.; Edwards, C.; Korchev, Y.; Majouga, A. Novel method for rapid toxicity screening of magnetic nanoparticles. Sci. Rep. 2018, 8, 7462. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Laforge, F.O.; Abeyweera, T.P.; Rotenberg, S.A.; Carpino, J.; Mirkin, M.V. Nanoelectrochemistry of mammalian cells. Proc. Natl. Acad. Sci. USA 2008, 105, 443–448. [Google Scholar] [CrossRef]
- Zheng, X.T.; Hu, W.; Wang, H.; Yang, H.; Zhou, W.; Li, C.M. Bifunctional electro-optical nanoprobe to real-time detect local biochemical processes in single cells. Biosens. Bioelectron. 2011, 26, 4484–4490. [Google Scholar] [CrossRef]
- Ding, S.; Li, M.; Gong, H.; Zhu, Q.; Shi, G.; Zhu, A. Sensitive and Selective Measurement of Hydroxyl Radicals at Subcellular Level with Tungsten Nanoelectrodes. Anal. Chem. 2020, 92, 2543–2549. [Google Scholar] [CrossRef]
- Li, Y.; Hu, K.; Yu, Y.; Rotenberg, S.A.; Amatore, C.; Mirkin, M.V. Direct Electrochemical Measurements of Reactive Oxygen and Nitrogen Species in Nontransformed and Metastatic Human Breast Cells. J. Am. Chem. Soc. 2017, 139, 13055–13062. [Google Scholar] [CrossRef]
- Vaneev, A.N.; Gorelkin, P.V.; Garanina, A.S.; Lopatukhina, H.V.; Vodopyanov, S.S.; Alova, A.V.; Ryabaya, O.O.; Akasov, R.A.; Zhang, Y.; Novak, P.; et al. In Vitro and In Vivo Electrochemical Measurement of Reactive Oxygen Species After Treatment with Anticancer Drugs. Anal. Chem. 2020, 92, 8010–8014. [Google Scholar] [CrossRef]
- Marquitan, M.; Clausmeyer, J.; Actis, P.; Cόrdoba, A.L.; Korchev, Y.; Mark, M.D.; Herlitze, S.; Schuhmann, W. Intracellular hydrogen peroxide detection with functionalised nanoelectrodes. ChemElectroChem 2016, 3, 2125–2129. [Google Scholar] [CrossRef]
- Ying, Y.L.; Hu, Y.X.; Gao, R.; Yu, R.J.; Gu, Z.; Lee, L.P.; Long, Y.T. Asymmetric Nanopore Electrode-Based Amplification for Electron Transfer Imaging in Live Cells. J. Am. Chem. Soc. 2018, 140, 5385–5392. [Google Scholar] [CrossRef] [PubMed]
- Thein, M.; Asphahani, F.; Cheng, A.; Buckmaster, R.; Zhang, M.; Xu, J. Response characteristics of single-cell impedance sensors employed with surface-modified microelectrodes. Biosens. Bioelectron. 2010, 25, 1963–1969. [Google Scholar] [CrossRef]
- Hatamie, A.; Ren, L.; Dou, H.; Gandasi, N.R.; Rorsman, P.; Ewing, A. Nanoscale Amperometry Reveals that Only a Fraction of Vesicular Serotonin Content is Released During Exocytosis from Beta Cells. Angew. Chem. Int. Ed. Engl. 2021, 60, 7593–7596. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Huang, X.; Liu, K.; Lan, T.; Wang, Z.; Zhu, Z. Recent Advances in Electrical Impedance Sensing Technology for Single-Cell Analysis. Biosensors 2021, 11, 470. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Song, H.; Ahn, H.; Kim, T.; Jung, J.; Cho, S.K.; Shin, D.M.; Choi, J.R.; Hwang, Y.H.; Kim, K. A Review of Advanced Impedance Biosensors with Microfluidic Chips for Single-Cell Analysis. Biosensors 2021, 11, 412. [Google Scholar] [CrossRef]
- Sun, T.; Morgan, H. Single-cell microfluidic impedance cytometry: A review. Microfluid. Nanofluid. 2010, 8, 423–443. [Google Scholar] [CrossRef]
- Sun, T.; Green, N.G.; Nicolas, G.G.; Hywel, M. Analysis and numerical modeling methods for impedance analysis of single cells on-chip. Nano 2008, 3, 55–63. [Google Scholar] [CrossRef]
- Giana, F.E.; Bonetto, F.J.; Bellotti, M.I. Assay based on electrical impedance spectroscopy to discriminate between normal and cancerous mammalian cells. Phys. Rev. E 2018, 97, 032410. [Google Scholar] [CrossRef]
- Cheung, K.C.; Di Berardino, M.; Schade-Kampmann, G.; Hebeisen, M.; Pierzchalski, A.; Bocsi, J.; Mittag, A.; Tárnok, A. Microfluidic impedance-based flow cytometry. Cytom. A 2010, 77, 648–666. [Google Scholar] [CrossRef]
- Xu, Y.; Xie, X.; Duan, Y.; Wang, L.; Cheng, Z.; Cheng, J. A review of impedance measurements of whole cells. Biosens. Bioelectron. 2016, 77, 824–836. [Google Scholar] [CrossRef]
- Desai, S.P.; Coston, A.; Berlin, A. Micro-electrical impedance spectroscopy and identification of patient-derived, dissociated tumor cells. IEEE Trans. Nanobiosci. 2019, 18, 369–372. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Ghassemi, P.; Strobl, J.S.; Agah, M. Biophysical phenotyping of cells via impedance spectroscopy in parallel cyclic deformability channels. Biomicrofluidics 2019, 13, 044103. [Google Scholar] [CrossRef] [PubMed]
- McGrath, J.S.; Honrado, C.; Moore, J.H.; Adair, S.J.; Varhue, W.B.; Salahi, A.; Farmehini, V.; Goudreau, B.J.; Nagdas, S.; Blais, E.M.; et al. Electrophysiology-based stratification of pancreatic tumorigenicity by label-free single-cell impedance cytometry. Anal. Chim. Acta 2020, 1101, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Ostermann, M.; Sauter, A.; Xue, Y.; Birkeland, E.; Schoelermann, J.; Holst, B.; Cimpan, M.R. Label-free impedance flow cytometry for nanotoxicity screening. Sci. Rep. 2020, 10, 142. [Google Scholar] [CrossRef]
- Zhang, Y.; Liang, H.; Tan, H.; Chen, D.; Wang, Y.; Xu, Y.; Wang, J.; Chen, J. Development of microfluidic platform to high- throughput quantify single-cell intrinsic bioelectrical markers of tumor cell lines, subtypes and patient tumor cells. Sens. Actuators B Chem. 2020, 317, 128231. [Google Scholar] [CrossRef]
- Impe, D.; Reitz, J.; Kopnick, C.; Rolletschek, H.; Borner, A.; Senula, A.; Nagel, M. Assessment of pollen viability for wheat. Front. Plant Sci. 2019, 10, 1588. [Google Scholar]
- Ascari, L.; Cristofori, V.; Macri, F.; Botta, R.; Silvestri, C.; De Gregorio, T.; Huerta, E.S.; Di Berardino, M.; Kaufmann, S.; Siniscalco, C. Hazelnut pollen phenotyping using label-free impedance flow cytometry. Front. Plant Sci. 2020, 11, 615922. [Google Scholar] [CrossRef]
- Canonge, J.; Philippot, M.; Leblanc, C.; Potin, P.; Bodin, M. Impedance flow cytometry allows the early prediction of embryo yields in wheat (Triticum aestivum L.) microspore cultures. Plant Sci. 2020, 300, 110586. [Google Scholar] [CrossRef]
- Han, Z.; Chen, L.; Zhang, S.; Wang, J.; Duan, X. Label-free and simultaneous mechanical and electrical characterization of single plant cells using microfluidic impedance flow cytometry. Anal. Chem. 2020, 92, 14568–14575. [Google Scholar] [CrossRef]
- Xie, X.; Zhang, Z.; Ge, X.; Zhao, X.; Hao, L.; Cheng, Z.; Zhou, W.; Du, Y.; Wang, L.; Tian, F.; et al. Particle self-aligning, focusing, and electric impedance microcytometer device for label-free single cell morphology discrimination and yeast budding analysis. Anal. Chem. 2019, 91, 13398–13406. [Google Scholar] [CrossRef]
- Opitz, C.; Schade, G.; Kaufmann, S.; Berardino, M.D. Rapid determination of general cell status, cell viability, and optimal harvest time in eukaryotic cell cultures by impedance flow cytometry. Appl. Microbiol. Biotechnol. 2019, 103, 8619–8629. [Google Scholar] [CrossRef] [PubMed]
- Bertelsen, C.V.; Franco, J.C.; Skands, G.E.; Dimaki, M.; Svendsen, W.E. Investigating the use of impedance flow cytometry for classifying the viability state of E. coli. Sensors 2020, 20, 6339. [Google Scholar] [CrossRef] [PubMed]
- Spencer, D.C.; Paton, T.F.; Mulroney, K.T.; Inglis, T.J.J.; Sutton, J.M.; Morgan, H. A fast impedance-based antimicrobial susceptibility test. Nat. Commun. 2020, 11, 5328. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Rosano, J.M.; Wang, Y.; Garson, C.J.; Prabhakarpandian, B.; Pant, K.; Klarmann, G.J.; Perantoni, A.; Alvarez, L.M.; Lai, E. Identification of mesenchymal stem cell differentiation state using dual-micropore microfluidic impedance flow cytometry. Anal. Methods 2016, 8, 7437–7444. [Google Scholar] [CrossRef]
- Xavier, M.; de Andres, M.C.; Spencer, D.; Oreffo, R.O.C.; Morgan, H. Size and dielectric properties of skeletal stem cells change critically after enrichment and expansion from human bone marrow: Consequences for microfluidic cell sorting. J. R. Soc. Interface 2017, 14, 20170233. [Google Scholar] [CrossRef]
- Nguyen, T.A.; Yin, T.I.; Reyes, D.; Urban, G.A. Microfluidic chip with integrated electrical cell-impedance sensing for monitoring single cancer cell migration in three-dimensional matrixes. Anal. Chem. 2013, 85, 11068–11076. [Google Scholar] [CrossRef]
- Dittami, G.M.; Ayliffe, H.E.; King, C.S.; Rabbitt, R.D. A Multilayer MEMS Platform for Single-Cell Electric Impedance Spectroscopy and Electrochemical Analysis. J. Microelectromech. Syst. A Jt. IEEE ASME Publ. Microstruct. Microactuators Microsens. Microsyst. 2008, 17, 850–862. [Google Scholar] [CrossRef]
- Geng, Y.; Zhu, Z.; Zhang, Z.; Xu, F.; Marchisio, M.A.; Wang, Z.; Pan, D.; Zhao, X.; Huang, Q.A. Design and 3D modeling investigation of a microfluidic electrode array for electrical impedance measurement of single yeast cells. Electrophoresis 2021, 42, 1996–2009. [Google Scholar] [CrossRef]
- Doijen, J.; Van Loy, T.; Landuyt, B.; Luyten, W.; Schols, D.; Schoofs, L. Advantages and shortcomings of cell-based electrical impedance measurements as a GPCR drug discovery tool. Biosens. Bioelectron. 2019, 137, 33–44. [Google Scholar] [CrossRef]
- Menotti, J.; Alanio, A.; Sturny-Leclère, A.; Vitry, S.; Sauvage, F.; Barratt, G.; Bretagne, S. A cell impedance-based real-time in vitro assay to assess the toxicity of amphotericin B formulations. Toxicol. Appl. Pharmacol. 2017, 334, 18–23. [Google Scholar] [CrossRef]
- Clausen, C.H.; Skands, G.E.; Bertelsen, C.V.; Svendsen, W.E. Coplanar Electrode Layout Optimized for Increased Sensitivity for Electrical Impedance Spectroscopy. Micromachines. 2015, 6, 110–120. [Google Scholar] [CrossRef]
- De Ninno, A.; Errico, V.; Bertani, F.R.; Businaro, L.; Bisegna, P.; Caselli, F. Coplanar electrode microfluidic chip enabling accurate sheathless impedance cytometry. Lab A Chip 2017, 17, 1158–1166. [Google Scholar] [CrossRef] [PubMed]
- Valero, A.; Braschler, T.; Renaud, P. A unified approach to dielectric single cell analysis: Impedance and dielectrophoretic force spectroscopy. Lab A Chip 2010, 10, 2216–2225. [Google Scholar] [CrossRef] [PubMed]
- Cottingham, K. The single-cell scene. Anal. Chem. 2004, 76, 235A–238A. [Google Scholar] [CrossRef][Green Version]
- Tang, B.; Liu, M.; Dietzel, A. Low-Cost Impedance Camera for Cell Distribution Monitoring. Biosensors 2023, 13, 281. [Google Scholar] [CrossRef]
- Liu, J.; Qiang, Y.; Alvarez, O.; Du, E. Electrical impedance microflow cytometry with oxygen control for detection of sickle cells. Sens. Actuators B Chem. 2018, 255 (Pt 2), 2392–2398. [Google Scholar] [CrossRef]
- Hassan, U.; Watkins, N.N.; Reddy, B.; Jr Damhorst, G.; Bashir, R. Microfluidic differential immunocapture biochip for specific leukocyte counting. Nat. Protoc. 2016, 11, 714–726. [Google Scholar] [CrossRef]
- Gawad, S.; Schild, L.; Renaud, P.H. Micromachined impedance spectroscopy flow cytometer for cell analysis and particle sizing. Lab A Chip 2001, 1, 76–82. [Google Scholar] [CrossRef]
- Honrado, C.; Ciuffreda, L.; Spencer, D.; Ranford-Cartwright, L.; Morgan, H. Dielectric characterization of Plasmodium falciparum-infected red blood cells using microfluidic impedance cytometry. J. R. Soc. Interface 2018, 15, 20180416. [Google Scholar] [CrossRef]
- Lee, J.; Kwak, B. Simultaneous on-chip isolation and characterization of circulating tumor cell sub-populations. Biosens. Bioelectron. 2020, 168, 112564. [Google Scholar] [CrossRef] [PubMed]
- Nagano, T.; Lubling, Y.; Stevens, T.J.; Schoenfelder, S.; Yaffe, E.; Dean, W.; Laue, E.D.; Tanay, A.; Fraser, P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 2013, 502, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.B.; Lim, C.T.; Lim, W.T. Single-Cell Analysis of Circulating Tumor Cells: Why Heterogeneity Matters. Cancers 2019, 11, 1595. [Google Scholar] [CrossRef] [PubMed]
- Lill, M.; Kõks, S.; Soomets, U.; Schalkwyk, L.C.; Fernandes, C.; Lutsar, I.; Taba, P. Peripheral blood RNA gene expression profiling in patients with bacterial meningitis. Front. Neurosci. 2013, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, S.V.; Bingham, C.; Fittipaldi, P.; Austin, L.; Palazzo, J.; Palmer, G.; Alpaugh, K.; Cristofanilli, M. TP53 mutations detected in circulating tumor cells present in the blood of metastatic triple negative breast cancer patients. Breast. Cancer Res. 2014, 16, 445. [Google Scholar] [CrossRef]
- Aceto, N.; Bardia, A.; Miyamoto, D.T.; Donaldson, M.C.; Wittner, B.S.; Spencer, J.A.; Yu, M.; Pely, A.; Engstrom, A.; Zhu, H.; et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 2014, 158, 1110–1122. [Google Scholar] [CrossRef]
- Bedbrook, C.N.; Deverman, B.E.; Gradinaru, V. Viral Strategies for Targeting the Central and Peripheral Nervous Systems. Annu. Rev. Neurosci. 2018, 41, 323–348. [Google Scholar] [CrossRef]
- Mével, M.; Bouzelha, M.; Leray, A.; Pacouret, S.; Guilbaud, M.; Penaud-Budloo, M.; Alvarez-Dorta, D.; Dubreil, L.; Gouin, S.G.; Combal, J.P.; et al. Chemical modification of the adeno-associated virus capsid to improve gene delivery. Chem. Sci. 2019, 11, 1122–1131. [Google Scholar] [CrossRef]
- Paulk, N. Gene Therapy: It Is Time to Talk About High-Dose AAV: The Deaths of Two Children With X-Linked Myotubular Myopathy in the ASPIRO Trial Prompts a Reexamination of Vector Safety. Genet. Eng. Biotechnol. News 2020, 40, 14–16. [Google Scholar] [CrossRef]
- Reemann, P.; Reimann, E.; Ilmjärv, S.; Porosaar, O.; Silm, H.; Jaks, V.; Vasar, E.; Kingo, K.; Kõks, S. Melanocytes in the skin–comparative whole transcriptome analysis of main skin cell types. PLoS ONE 2014, 9, e115717. [Google Scholar] [CrossRef]
- Choi, H.; Jeon, C.S.; Hwang, I.; Ko, J.; Lee, S.; Choo, J.; Boo, J.H.; Kim, H.C.; Chung, T.D. A flow cytometry-based submicron-sized bacterial detection system using a movable virtual wall. Lab Chip 2014, 14, 2327–2333. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Jiang, M.; Chen, D.; Zhao, X.; Xue, C.; Hao, R.; Yue, W.; Wang, J.; Chen, J. Single-Cell Electrical Phenotyping Enabling the Classification of Mouse Tumor Samples. Sci. Rep. 2016, 6, 19487. [Google Scholar] [CrossRef] [PubMed]
- Bernabini, C.; Holmes, D.; Morgan, H. Micro-impedance cytometry for detection and analysis of micron-sized particles and bacteria. Lab Chip 2011, 11, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Guler, M.T.; Bilican, I. Capacitive detection of single bacterium from drinking water with a detailed investigation of electrical flow cytometry. Sens. Actuator A Phys. 2018, 269, 454–463. [Google Scholar] [CrossRef]
- Yang, J.; Huang, L.; Qian, K. Nanomaterials-assisted metabolic analysis toward in vitro diagnostics. Exploration 2022, 2, 20210222. [Google Scholar] [CrossRef]
- Feng, D.; Li, H.; Xu, T.; Zheng, F.; Hu, C.; Shi, X.; Xu, G. High-throughput single cell metabolomics and cellular heterogeneity exploration by inertial microfluidics coupled with pulsed electric field-induced electrospray ionization-high resolution mass spectrometry. Anal. Chim. Acta 2022, 1221, 340116. [Google Scholar] [CrossRef]
- Flangea, C.; Serb, A.; Sisu, E.; Zamfir, A.D. Chip-based nanoelectrospray mass spectrometry of brain gangliosides. Biochim. Biophys. Acta 2011, 1811, 513–535. [Google Scholar] [CrossRef]
- Wei, Z.; Xiong, X.; Guo, C.; Si, X.; Zhao, Y.; He, M.; Yang, C.; Xu, W.; Tang, F.; Fang, X.; et al. Pulsed Direct Current Electrospray: Enabling Systematic Analysis of Small Volume Sample by Boosting Sample Economy. Anal. Chem. 2015, 87, 11242–11248. [Google Scholar] [CrossRef]
- Dago, A.E.; Stepansky, A.; Carlsson, A.; Luttgen, M.; Kendall, J.; Baslan, T.; Kolatkar, A.; Wigler, M.; Bethel, K.; Gross, M.E.; et al. Rapid phenotypic and genomic change in response to therapeutic pressure in prostate cancer inferred by high content analysis of single circulating tumor cells. PLoS ONE 2014, 9, e101777. [Google Scholar] [CrossRef]
- Jessri, M.; Farah, C.S. Harnessing massively parallel sequencing in personalized head and neck oncology. J. Dent. Res. 2014, 93, 437–444. [Google Scholar] [CrossRef]
- Shi, X.M.; Liu, F.Q.; Wang, B.; Yu, S.Y.; Xu, Y.T.; Zhao, W.W.; Jiang, D.; Chen, H.Y.; Xu, J.J. Functional nucleic acid engineered double-barreled nanopores for measuring sodium to potassium ratio at single-cell level. Exploration 2022, 2, 20220025. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, W.; Wang, J.; Luo, H.; Luo, B.; Li, X.; Liu, S.; Li, C. Electrical Characterization and Analysis of Single Cells and Related Applications. Biosensors 2023, 13, 907. https://doi.org/10.3390/bios13100907
Zhu W, Wang J, Luo H, Luo B, Li X, Liu S, Li C. Electrical Characterization and Analysis of Single Cells and Related Applications. Biosensors. 2023; 13(10):907. https://doi.org/10.3390/bios13100907
Chicago/Turabian StyleZhu, Weitao, Jiaao Wang, Hongzhi Luo, Binwen Luo, Xue Li, Shan Liu, and Chenzhong Li. 2023. "Electrical Characterization and Analysis of Single Cells and Related Applications" Biosensors 13, no. 10: 907. https://doi.org/10.3390/bios13100907
APA StyleZhu, W., Wang, J., Luo, H., Luo, B., Li, X., Liu, S., & Li, C. (2023). Electrical Characterization and Analysis of Single Cells and Related Applications. Biosensors, 13(10), 907. https://doi.org/10.3390/bios13100907







