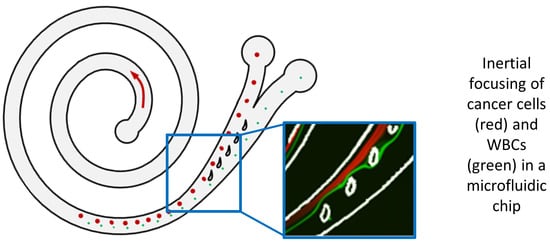
Analytical Validation of a Spiral Microfluidic Chip with Hydrofoil-Shaped Pillars for the Enrichment of Circulating Tumor Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microfluidic Design
2.2. Microfluidic Chip Fabrication
2.3. Cell Culture, Blood Collection, and Sample Preparation
2.4. Experimental Setup
2.5. Chip Conditioning and Sample Processing
2.6. Chip Characterization Using Fluorescent Microbeads and Cells
2.7. Analytical Studies for Design Validation and Performance Characterization
3. Results
3.1. Chip Characterization Using Fluorescent Microbeads and Cells
3.2. Analytical Studies for Design Validation and Performance Characterization
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Micalizzi, D.S.; Maheswaran, S.; Haber, D.A. A Conduit to Metastasis: Circulating Tumor Cell Biology. Genes Dev. 2017, 31, 1827–1840. [Google Scholar] [CrossRef]
- Agashe, R.; Kurzrock, R. Circulating Tumor Cells: From the Laboratory to the Cancer Clinic. Cancers 2020, 12, 2361. [Google Scholar] [CrossRef] [PubMed]
- Bardia, A.; Haber, D.A. Solidifying Liquid Biopsies: Can Circulating Tumor Cell Monitoring Guide Treatment Selection in Breast Cancer? J. Clin. Oncol. 2014, 32, 3470–3471. [Google Scholar] [CrossRef] [PubMed]
- Masuda, T.; Hayashi, N.; Iguchi, T.; Ito, S.; Eguchi, H.; Mimori, K. Clinical and Biological Significance of Circulating Tumor Cells in Cancer. Mol. Oncol. 2016, 10, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Keller, L.; Pantel, K. Unravelling Tumour Heterogeneity by Single-Cell Profiling of Circulating Tumour Cells. Nat. Rev. Cancer 2019, 19, 553–567. [Google Scholar] [CrossRef]
- Powell, A.A.; Talasaz, A.A.H.; Zhang, H.; Coram, M.A.; Reddy, A.; Deng, G.; Telli, M.L.; Advani, R.H.; Carlson, R.W.; Mollick, J.A.; et al. Single Cell Profiling of Circulating Tumor Cells: Transcriptional Heterogeneity and Diversity from Breast Cancer Cell Lines. PLoS ONE 2012, 7, e33788. [Google Scholar] [CrossRef]
- Bin Lim, S.; di Lee, W.; Vasudevan, J.; Lim, W.T.; Lim, C.T. Liquid Biopsy: One Cell at a Time. NPJ Precis. Oncol. 2019, 3, 23. [Google Scholar]
- CTC Studies That Are Actively Recruiting. Available online: https://clinicaltrials.gov/ct2/results?term=circulating+tumor+cells&cond=cancer&Search=Apply&recrs=a&age_v=&gndr=&type=&rslt= (accessed on 30 November 2022).
- Ju, S.; Chen, C.; Zhang, J.; Xu, L.; Zhang, X.; Li, Z.; Chen, Y.; Zhou, J.; Ji, F.; Wang, L. Detection of Circulating Tumor Cells: Opportunities and Challenges. Biomark. Res. 2022, 10, 58. [Google Scholar] [CrossRef]
- Cho, H.; Kim, J.; Song, H.; Sohn, K.Y.; Jeon, M.; Han, K.H. Microfluidic Technologies for Circulating Tumor Cell Isolation. Analyst 2018, 143, 2936–2970. [Google Scholar] [CrossRef]
- Hao, S.J.; Wan, Y.; Xia, Y.Q.; Zou, X.; Zheng, S.Y. Size-Based Separation Methods of Circulating Tumor Cells. Adv. Drug Deliv. Rev. 2018, 125, 3–20. [Google Scholar] [CrossRef]
- Bankó, P.; Lee, S.Y.; Nagygyörgy, V.; Zrínyi, M.; Chae, C.H.; Cho, D.H.; Telekes, A. Technologies for Circulating Tumor Cell Separation from Whole Blood. J. Hematol. Oncol. 2019, 12, 48. [Google Scholar] [CrossRef] [PubMed]
- Pødenphant, M.; Ashley, N.; Koprowska, K.; Mir, K.U.; Zalkovskij, M.; Bilenberg, B.; Bodmer, W.; Kristensen, A.; Marie, R. Separation of Cancer Cells from White Blood Cells by Pinched Flow Fractionation. Lab Chip 2015, 15, 4598–4606. [Google Scholar] [CrossRef] [PubMed]
- Karabacak, N.M.; Spuhler, P.S.; Fachin, F.; Lim, E.J.; Pai, V.; Ozkumur, E.; Martel, J.M.; Kojic, N.; Smith, K.; Chen, P.I.; et al. Microfluidic, Marker-Free Isolation of Circulating Tumor Cells from Blood Samples. Nat. Protoc. 2014, 9, 694–710. [Google Scholar] [CrossRef] [PubMed]
- Fachin, F.; Spuhler, P.; Martel-Foley, J.M.; Edd, J.F.; Barber, T.A.; Walsh, J.; Karabacak, M.; Pai, V.; Yu, M.; Smith, K.; et al. Monolithic Chip for High-Throughput Blood Cell Depletion to Sort Rare Circulating Tumor Cells. Sci. Rep. 2017, 7, 10936. [Google Scholar] [CrossRef]
- Bhagat, A.A.S.; Bow, H.; Hou, H.W.; Tan, S.J.; Han, J.; Lim, C.T. Microfluidics for Cell Separation. Med. Biol. Eng. Comput. 2010, 48, 999–1014. [Google Scholar] [CrossRef]
- di Carlo, D.; Irimia, D.; Tompkins, R.G.; Toner, M. Continuous Inertial Focusing, Ordering, and Separation of Particles in Microchannels. Proc. Natl. Acad. Sci. USA 2007, 104, 18892–18897. [Google Scholar] [CrossRef]
- Nivedita, N.; Papautsky, I. Continuous Separation of Blood Cells in Spiral Microfluidic Devices. Biomicrofluidics 2013, 7, 054101. [Google Scholar] [CrossRef]
- Warkiani, M.E.; Khoo, B.L.; Wu, L.; Tay, A.K.P.; Bhagat, A.A.S.; Han, J.; Lim, C.T. Ultra-Fast, Label-Free Isolation of Circulating Tumor Cells from Blood Using Spiral Microfluidics. Nat. Protoc. 2016, 11, 134–148. [Google Scholar] [CrossRef]
- Warkiani, M.E.; Guan, G.; Luan, K.B.; Lee, W.C.; Bhagat, A.A.S.; Kant Chaudhuri, P.; Tan, D.S.W.; Lim, W.T.; Lee, S.C.; Chen, P.C.Y.; et al. Slanted Spiral Microfluidics for the Ultra-Fast, Label-Free Isolation of Circulating Tumor Cells. Lab Chip 2014, 14, 128–137. [Google Scholar] [CrossRef]
- Sun, J.; Liu, C.; Li, M.; Wang, J.; Xianyu, Y.; Hu, G.; Jiang, X. Size-Based Hydrodynamic Rare Tumor Cell Separation in Curved Microfluidic Channels. Biomicrofluidics 2013, 7, 011802. [Google Scholar] [CrossRef]
- Ookawara, S.; Higashi, R.; Street, D.; Ogawa, K. Feasibility Study on Concentration of Slurry and Classification of Contained Particles by Microchannel. Chem. Eng. J. 2004, 101, 171–178. [Google Scholar] [CrossRef]
- Mihandoust, A.; Maleki-Jirsaraei, N.; Rouhani, S.; Safi, S.; Alizadeh, M. Improvement of Size-Based Particle Separation Throughput in Slanted Spiral Microchannel by Modifying Outlet Geometry. Electrophoresis 2020, 41, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Martel, J.M.; Toner, M. Particle Focusing in Curved Microfluidic Channels. Sci. Rep. 2013, 3, 3340. [Google Scholar] [CrossRef]
- Nivedita, N.; Ligrani, P.; Papautsky, I. Dean Flow Dynamics in Low-Aspect Ratio Spiral Microchannels. Sci. Rep. 2017, 7, 44072. [Google Scholar] [CrossRef] [PubMed]
- Özkayar, G.; Mutlu, E.; Şahin, Ş.; Demircan Yalçın, Y.; Töral, T.; Külah, H.; Yıldırım, E.; Zorlu, Ö.; Özgür, E. A Novel Microfluidic Method Utilizing a Hydrofoil Structure to Improve Circulating Tumor Cell Enrichment: Design and Analytical Validation. Micromachines 2020, 11, 981. [Google Scholar] [CrossRef]
- Mousapour, B. Measuring Biomechanical Properties of Cancer Cells Using a High-Throughput Microfluidic Platform and Their Correlation with Surface Rheology and Internal Elasticity; University of California: Irvine, CA, USA, 2016. [Google Scholar]
- Hwang, J.Y.; Kim, J.; Park, J.M.; Lee, C.; Jung, H.; Lee, J.; Shung, K.K. Cell Deformation by Single-Beam Acoustic Trapping: A Promising Tool for Measurements of Cell Mechanics. Sci. Rep. 2016, 6, 27238. [Google Scholar] [CrossRef]
- Jhaveri, D. Study of Cell Mechanics and Surface Rheology of Cancer Cells Using High Throughput Microfluidic Device; University of California: Irvine, CA, USA, 2015. [Google Scholar]
- Masaeli, M.; Sollier, E.; Amini, H.; Mao, W.; Camacho, K.; Doshi, N.; Mitragotri, S.; Alexeev, A.; di Carlo, D. Continuous Inertial Focusing and Separation of Particles by Shape. Phys. Rev. X 2012, 2, 031017. [Google Scholar] [CrossRef]











Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sen-Dogan, B.; Demir, M.A.; Sahin, B.; Yildirim, E.; Karayalcin, G.; Sahin, S.; Mutlu, E.; Toral, T.B.; Ozgur, E.; Zorlu, O.; et al. Analytical Validation of a Spiral Microfluidic Chip with Hydrofoil-Shaped Pillars for the Enrichment of Circulating Tumor Cells. Biosensors 2023, 13, 938. https://doi.org/10.3390/bios13100938
Sen-Dogan B, Demir MA, Sahin B, Yildirim E, Karayalcin G, Sahin S, Mutlu E, Toral TB, Ozgur E, Zorlu O, et al. Analytical Validation of a Spiral Microfluidic Chip with Hydrofoil-Shaped Pillars for the Enrichment of Circulating Tumor Cells. Biosensors. 2023; 13(10):938. https://doi.org/10.3390/bios13100938
Chicago/Turabian StyleSen-Dogan, Begum, Mehmet Alper Demir, Buket Sahin, Ender Yildirim, Gizem Karayalcin, Sebnem Sahin, Ege Mutlu, Taylan Berkin Toral, Ebru Ozgur, Ozge Zorlu, and et al. 2023. "Analytical Validation of a Spiral Microfluidic Chip with Hydrofoil-Shaped Pillars for the Enrichment of Circulating Tumor Cells" Biosensors 13, no. 10: 938. https://doi.org/10.3390/bios13100938
APA StyleSen-Dogan, B., Demir, M. A., Sahin, B., Yildirim, E., Karayalcin, G., Sahin, S., Mutlu, E., Toral, T. B., Ozgur, E., Zorlu, O., & Kulah, H. (2023). Analytical Validation of a Spiral Microfluidic Chip with Hydrofoil-Shaped Pillars for the Enrichment of Circulating Tumor Cells. Biosensors, 13(10), 938. https://doi.org/10.3390/bios13100938