Nanotechnology-Assisted Biosensors for the Detection of Viral Nucleic Acids: An Overview
Abstract
1. Introduction
2. Nanotechnology Applied for Biosensors
2.1. Nanomaterials as Components of Biosensors
2.1.1. Metal-Based Nanomaterials
2.1.2. Carbon-Based Nanomaterials
2.1.3. Other Nanomaterials
2.2. Nano-Fabrication Techniques and Nano-Biotechnology for Biosensors
2.2.1. Nano-Fabrication Techniques
2.2.2. CRISPR/Cas System-Based Nanotechnology
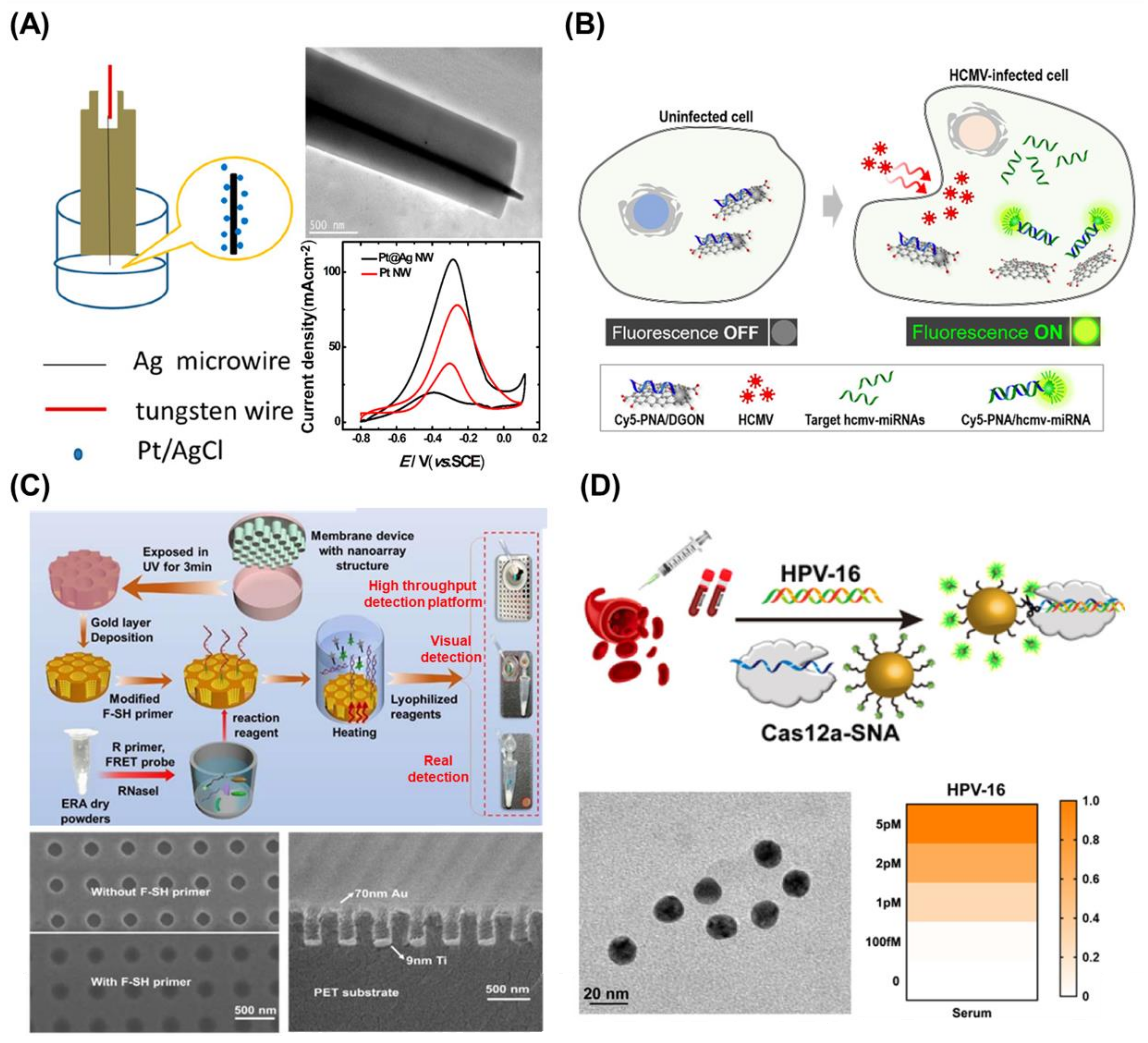
2.2.3. DNA/RNA Structure-Based Nanotechnology
2.2.4. Target Nucleic Acids Amplification Techniques
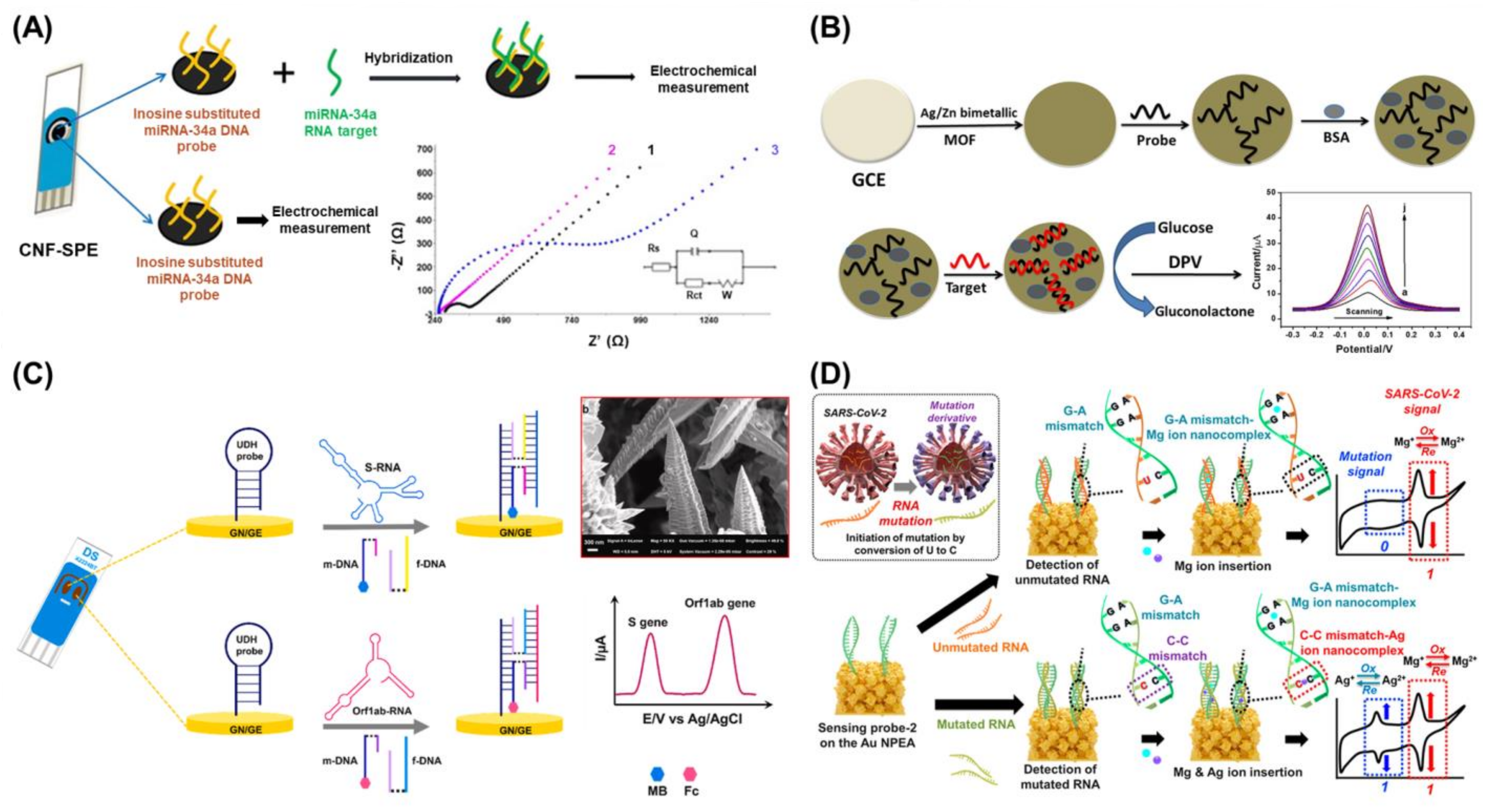
3. Electrochemistry-Based Biosensors for the Detection of Viral Nucleic Acids
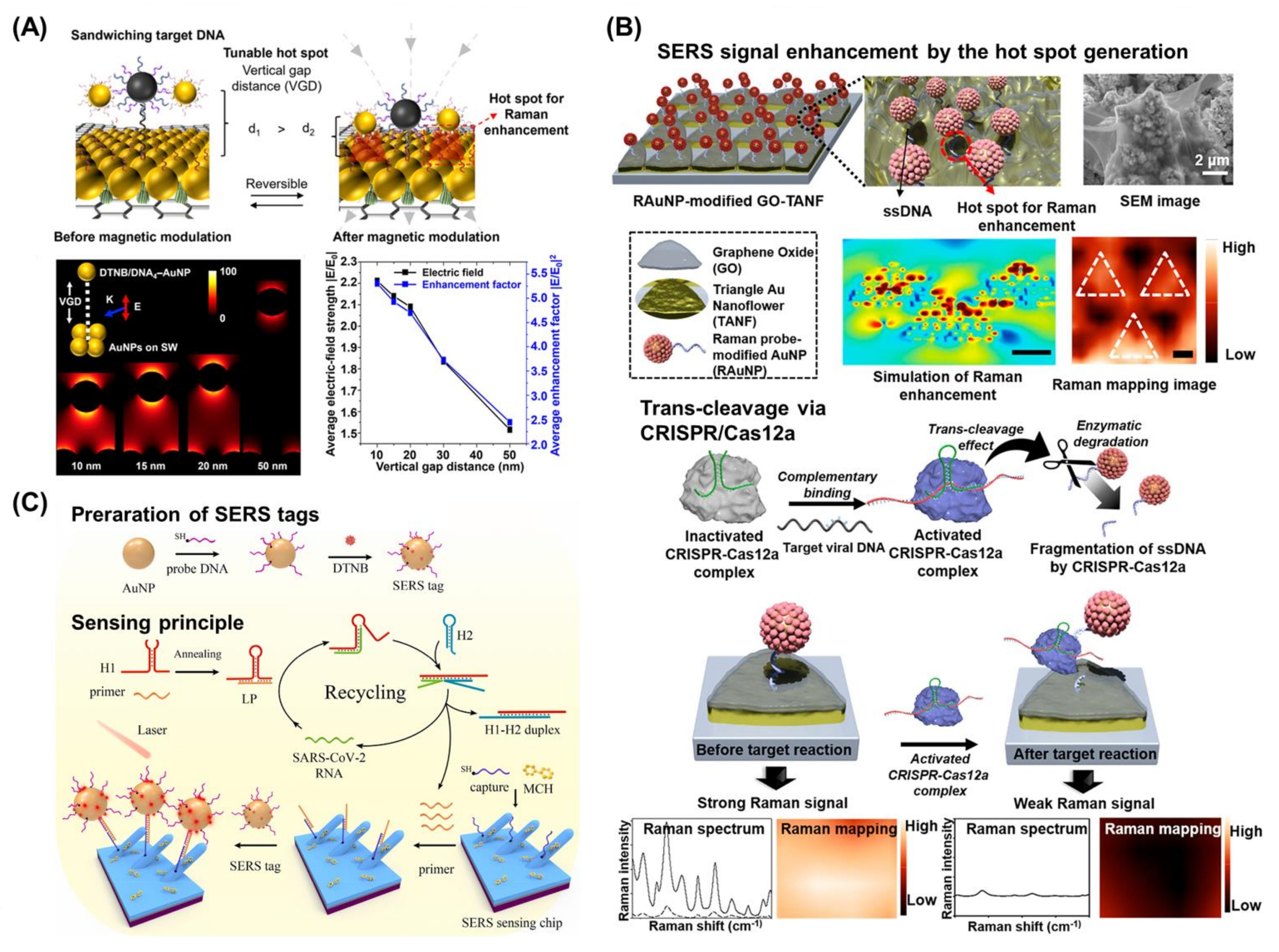
4. Surface-Enhanced Raman Scattering-Based Biosensors for the Detection of Viral Nucleic Acids
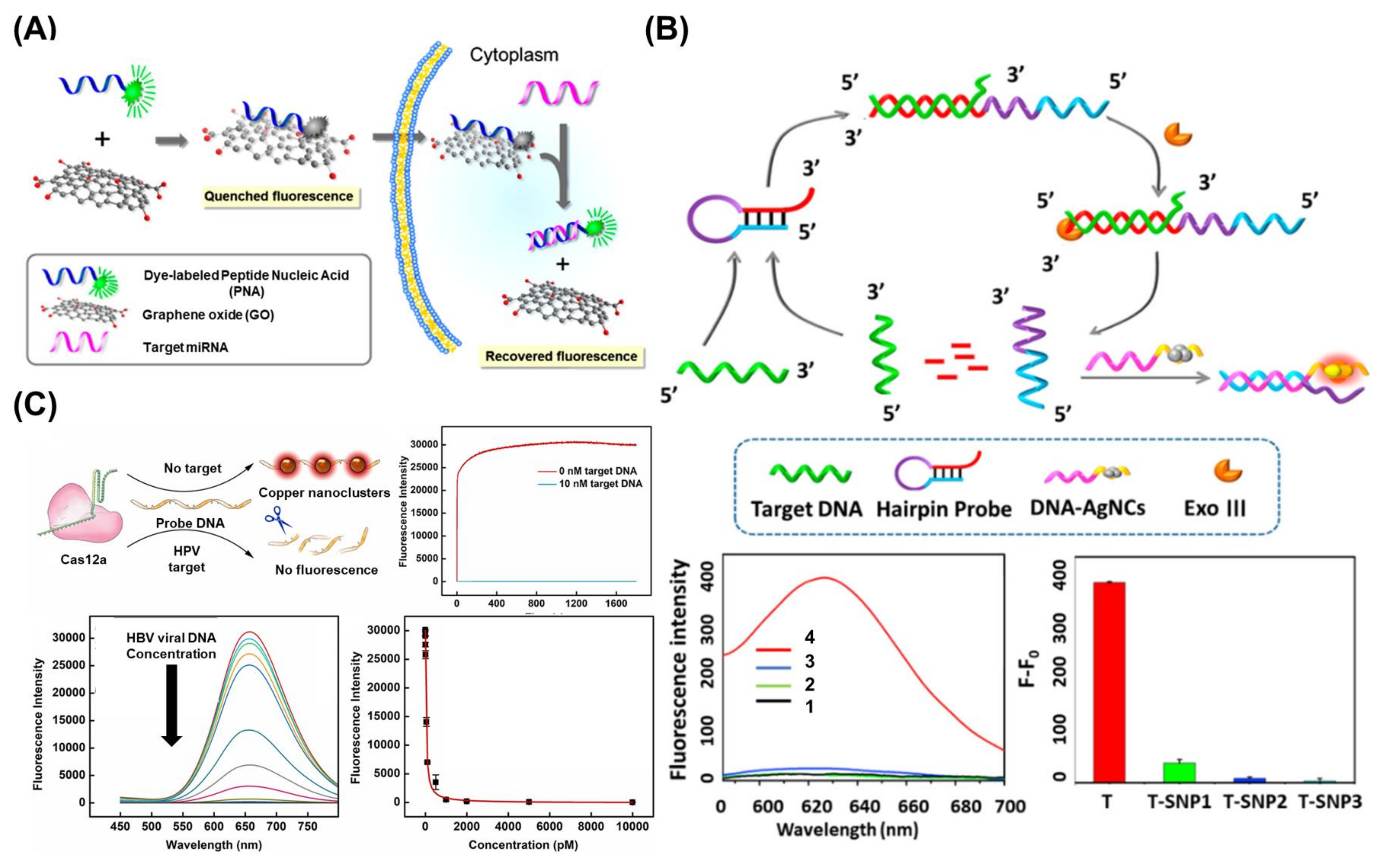
5. Fluorescence-Based Biosensors for the Detection of Viral Nucleic Acids
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shrivastav, A.M.; Cvelbar, U.; Abdulhalim, I. A comprehensive review on plasmonic-based biosensors used in viral diagnostics. Commun. Biol. 2021, 4, 70. [Google Scholar] [CrossRef] [PubMed]
- Narita, F.; Wang, Z.; Kurita, H.; Li, Z.; Shi, Y.; Jia, Y.; Soutis, C. A Review of Piezoelectric and Magnetostrictive Biosensor Materials for Detection of COVID-19 and Other Viruses. Adv. Mater. 2021, 33, 2005448. [Google Scholar] [CrossRef]
- V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Pastula, D.M.; Tyler, K.L. An Overview of Monkeypox Virus and Its Neuroinvasive Potential. Ann. Neurol. 2022, 92, 527–531. [Google Scholar] [CrossRef]
- Isidro, J.; Borges, V.; Pinto, M.; Sobral, D.; Santos, J.D.; Nunes, A.; Mixão, V.; Ferreira, R.; Santos, D.; Duarte, S.; et al. Phylogenomic characterization and signs of microevolution in the 2022 multi-country outbreak of monkeypox virus. Nat. Med. 2022, 28, 1569–1572. [Google Scholar] [CrossRef]
- Baker, R.E.; Mahmud, A.S.; Miller, I.F.; Rajeev, M.; Rasambainarivo, F.; Rice, B.L.; Takahashi, S.; Tatem, A.J.; Wagner, C.E.; Wang, L.-F.; et al. Infectious disease in an era of global change. Nat. Rev. Microbiol. 2022, 20, 193–205. [Google Scholar] [CrossRef]
- Ilkhani, H.; Farhad, S. A novel electrochemical DNA biosensor for Ebola virus detection. Anal. Biochem. 2018, 557, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, H.; Woo, K.; Kim, J.-M.; Jo, H.-J.; Jeong, Y.; Lee, K.H. SARS-CoV-2 Variant Screening Using a Virus-Receptor-Based Electrical Biosensor. Nano Lett. 2022, 22, 50–57. [Google Scholar] [CrossRef]
- Nidzworski, D.; Siuzdak, K.; Niedziałkowski, P.; Bogdanowicz, R.; Sobaszek, M.; Ryl, J.; Weiher, P.; Sawczak, M.; Wnuk, E.; Goddard, W.A.; et al. A rapid-response ultrasensitive biosensor for influenza virus detection using antibody modified boron-doped diamond. Sci. Rep. 2017, 7, 15707. [Google Scholar] [CrossRef] [PubMed]
- Courtney, S.J.; Stromberg, Z.R.; Kubicek-Sutherland, J.Z. Nucleic Acid-Based Sensing Techniques for Diagnostics and Surveillance of Influenza. Biosensors 2021, 11, 47. [Google Scholar] [CrossRef]
- Sanjuán, R.; Domingo-Calap, P. Mechanisms of viral mutation. Cell. Mol. Life Sci. 2016, 73, 4433–4448. [Google Scholar] [CrossRef]
- Ozer, T.; Henry, C.S. Paper-based analytical devices for virus detection: Recent strategies for current and future pandemics. TrAC Trends Anal. Chem. 2021, 144, 116424. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.; Lu, J.; Yu, T.; Long, Y.; Liu, G. Advances in nucleic acid amplification techniques (NAATs): COVID-19 point-of-care diagnostics as an example. Biosens. Bioelectron. 2022, 206, 114109. [Google Scholar] [CrossRef]
- Choi, J.R. Development of Point-of-Care Biosensors for COVID-19. Front. Chem. 2020, 8, 517. [Google Scholar] [CrossRef]
- Lee, E.; Lee, M.; Kwon, S.; Kim, J.; Kwon, Y. Systematic and mechanistic analysis of AuNP-induced nanotoxicity for risk assessment of nanomedicine. Nano Converg. 2022, 9, 27. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kim, T.-H.; Kwon, O.; Ismail, M.; Mahata, C.; Kim, Y.; Kim, S.; Kim, S. Implementation of convolutional neural network and 8-bit reservoir computing in CMOS compatible VRRAM. Nano Energy 2022, 104, 107886. [Google Scholar] [CrossRef]
- Yoon, J.; Shin, M.; Kim, D.; Lim, J.; Kim, H.-W.; Kang, T.; Choi, J.-W. Bionanohybrid composed of metalloprotein/DNA/MoS2/peptides to control the intracellular redox states of living cells and its applicability as a cell-based biomemory device. Biosens. Bioelectron. 2022, 196, 113725. [Google Scholar] [CrossRef]
- Yin, B.; Ho, W.K.H.; Zhang, Q.; Li, C.; Huang, Y.; Yan, J.; Yang, H.; Hao, J.; Wong, S.H.D.; Yang, M. Magnetic-Responsive Surface-Enhanced Raman Scattering Platform with Tunable Hot Spot for Ultrasensitive Virus Nucleic Acid Detection. ACS Appl. Mater. Interfaces 2022, 14, 4714–4724. [Google Scholar] [CrossRef]
- Maddali, H.; Miles, C.E.; Kohn, J.; O’Carroll, D.M. Optical Biosensors for Virus Detection: Prospects for SARS-CoV-2/COVID-19. ChemBioChem 2021, 22, 1176–1189. [Google Scholar] [CrossRef]
- Yoon, J.; Conley, B.M.; Shin, M.; Choi, J.-H.; Bektas, C.K.; Choi, J.-W.; Lee, K.-B. Ultrasensitive Electrochemical Detection of Mutated Viral RNAs with Single-Nucleotide Resolution Using a Nanoporous Electrode Array (NPEA). ACS Nano 2022, 16, 5764–5777. [Google Scholar] [CrossRef]
- Arunadevi, N. Metal nanocomposites for advanced futuristic biosensing applications. Mater. Lett. 2022, 309, 131320. [Google Scholar] [CrossRef]
- Fritea, L.; Banica, F.; Costea, T.O.; Moldovan, L.; Dobjanschi, L.; Muresan, M.; Cavalu, S. Metal Nanoparticles and Carbon-Based Nanomaterials for Improved Performances of Electrochemical (Bio)Sensors with Biomedical Applications. Materials 2021, 14, 6319. [Google Scholar] [CrossRef]
- Fujiwara, H.; Yamauchi, K.; Wada, T.; Ishihara, H.; Sasaki, K. Optical selection and sorting of nanoparticles according to quantum mechanical properties. Sci. Adv. 2021, 7, eabd9551. [Google Scholar] [CrossRef] [PubMed]
- Cava, R.; de Leon, N.; Xie, W. Introduction: Quantum Materials. Chem. Rev. 2021, 121, 2777–2779. [Google Scholar] [CrossRef] [PubMed]
- Pore, O.C.; Fulari, A.V.; Velhal, N.B.; Parale, V.G.; Park, H.H.; Shejwal, R.V.; Fulari, V.J.; Lohar, G.M. Hydrothermally synthesized urchinlike NiO nanostructures for supercapacitor and nonenzymatic glucose biosensing application. Mater. Sci. Semicond. Process. 2021, 134, 105980. [Google Scholar] [CrossRef]
- Reddy, Y.V.M.; Shin, J.H.; Hwang, J.; Kweon, D.-H.; Choi, C.-H.; Park, K.; Kim, S.-K.; Madhavi, G.; Yi, H.; Park, J.P. Fine-tuning of MXene-nickel oxide-reduced graphene oxide nanocomposite bioelectrode: Sensor for the detection of influenza virus and viral protein. Biosens. Bioelectron. 2022, 214, 114511. [Google Scholar] [CrossRef] [PubMed]
- Sehit, E.; Altintas, Z. Significance of nanomaterials in electrochemical glucose sensors: An updated review (2016-2020). Biosens. Bioelectron. 2020, 159, 112165. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Z.; Zhang, L.; Liu, Y.; Xie, L.; Ge, S.; Yu, J. Photoswitchable CRISPR/Cas12a-Amplified and Co3O4@Au Nanoemitter Based Triple-Amplified Diagnostic Electrochemiluminescence Biosensor for Detection of miRNA-141. ACS Appl. Mater. Interfaces 2022, 14, 32960–32969. [Google Scholar] [CrossRef]
- Wu, Z.-L.; Li, C.-K.; Yu, J.-G.; Chen, X.-Q. MnO2/reduced graphene oxide nanoribbons: Facile hydrothermal preparation and their application in amperometric detection of hydrogen peroxide. Sens. Actuators B Chem. 2017, 239, 544–552. [Google Scholar] [CrossRef]
- Rao, D.; Zhang, X.; Sheng, Q.; Zheng, J. Highly improved sensing of dopamine by using glassy carbon electrode modified with MnO2, graphene oxide, carbon nanotubes and gold nanoparticles. Microchim. Acta 2016, 183, 2597–2604. [Google Scholar] [CrossRef]
- Choi, H.K.; Lee, M.-J.; Lee, S.N.; Kim, T.-H.; Oh, B.-K. Noble Metal Nanomaterial-Based Biosensors for Electrochemical and Optical Detection of Viruses Causing Respiratory Illnesses. Front. Chem. 2021, 9, 672739. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Shu, Y.; Wu, Y.; Gao, Q. Co-tuning composition and channel-rich structure of Ag-Pd alloys toward sensitive electrochemical biosensing. Chem. Eng. J. 2021, 425, 131858. [Google Scholar] [CrossRef]
- Maduraiveeran, G.; Sasidharan, M.; Ganesan, V. Electrochemical sensor and biosensor platforms based on advanced nanomaterials for biological and biomedical applications. Biosens. Bioelectron. 2018, 103, 113–129. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, H.; Liu, J.; He, J.; Liu, J.; Yang, W. Electrochemical detection of DNA by formation of efficient electron transfer pathways through adsorbing gold nanoparticles to DNA modified electrodes. Microchem. J. 2021, 169, 106581. [Google Scholar] [CrossRef]
- Wang, D.; Hua, H.; Liu, Y.; Tang, H.; Li, Y. Single Ag Nanowire Electrodes and Single Pt@Ag Nanowire Electrodes: Fabrication, Electrocatalysis, and Surface-Enhanced Raman Scattering Applications. Anal. Chem. 2019, 91, 4291–4295. [Google Scholar] [CrossRef] [PubMed]
- Yaraki, M.T.; Tan, Y.N. Metal Nanoparticles-Enhanced Biosensors: Synthesis, Design and Applications in Fluorescence Enhancement and Surface-enhanced Raman Scattering. Chem. Asian J. 2020, 15, 3180–3208. [Google Scholar] [CrossRef]
- Elahi, N.; Kamali, M.; Baghersad, M.H. Recent biomedical applications of gold nanoparticles: A review. Talanta 2018, 184, 537–556. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kim, J.; Park, C.; Lim, J.-W.; Yeom, M.; Song, D.; Kim, E.; Haam, S. A flap endonuclease 1-assisted universal viral nucleic acid sensing system using surface-enhanced Raman scattering. Analyst 2022, 147, 5028–5037. [Google Scholar] [CrossRef]
- Xie, P.; Yuan, W.; Liu, X.; Peng, Y.; Yin, Y.; Li, Y.; Wu, Z. Advanced carbon nanomaterials for state-of-the-art flexible supercapacitors. Energy Storage Mater. 2021, 36, 56–76. [Google Scholar] [CrossRef]
- Speranza, G. Carbon Nanomaterials: Synthesis, Functionalization and Sensing Applications. Nanomaterials 2021, 11, 967. [Google Scholar] [CrossRef]
- Xu, J.; Tao, J.; Su, L.; Wang, J.; Jiao, T. A Critical Review of Carbon Quantum Dots: From Synthesis toward Applications in Electrochemical Biosensors for the Determination of a Depression-Related Neurotransmitter. Materials 2021, 14, 3987. [Google Scholar] [CrossRef]
- Li, Y.; Li, J.; Cao, Y.; Jiang, P.; Tang, Y.; Chen, Z.; Han, K. A visual method for determination of hepatitis C virus RNAs based on a 3D nanocomposite prepared from graphene quantum dots. Anal. Chim. Acta 2022, 1203, 339693. [Google Scholar] [CrossRef]
- Ferrier, D.C.; Honeychurch, K.C. Carbon Nanotube (CNT)-Based Biosensors. Biosensors 2021, 11, 486. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.; Pint, C.L.; Islam, A.E.; Weatherup, R.S.; Hofmann, S.; Meshot, E.R.; Wu, F.; Zhou, C.; Dee, N.; Amama, P.B.; et al. Carbon Nanotubes and Related Nanomaterials: Critical Advances and Challenges for Synthesis toward Mainstream Commercial Applications. ACS Nano 2018, 12, 11756–11784. [Google Scholar] [CrossRef] [PubMed]
- Mujica, M.L.; Tamborelli, A.; Castellaro, A.; Barcudi, D.; Rubianes, M.D.; Rodríguez, M.C.; Saka, H.A.; Bocco, J.L.; Dalmasso, P.R.; Rivas, G.A. Impedimetric and amperometric genosensors for the highly sensitive quantification of SARS-CoV-2 nucleic acid using an avidin-functionalized multi-walled carbon nanotubes biocapture platform. Biosens. Bioelectron. X 2022, 12, 100222. [Google Scholar] [CrossRef] [PubMed]
- Afroj, S.; Britnell, L.; Hasan, T.; Andreeva, D.V.; Novoselov, K.S.; Karim, N. Graphene-Based Technologies for Tackling COVID-19 and Future Pandemics. Adv. Funct. Mater. 2021, 31, 2107407. [Google Scholar] [CrossRef] [PubMed]
- Yim, Y.; Shin, H.; Ahn, S.M.; Min, D.-H. Graphene oxide-based fluorescent biosensors and their biomedical applications in diagnosis and drug discovery. Chem. Comm. 2021, 57, 9820–9833. [Google Scholar] [CrossRef] [PubMed]
- Hwang, M.T.; Heiranian, M.; Kim, Y.; You, S.; Leem, J.; Taqieddin, A.; Jing, Y.; Park, I.; Van Der Zande, A.M.; Nam, S.; et al. Ultrasensitive detection of nucleic acids using deformed graphene channel field effect biosensors. Nat. Commun. 2020, 11, 1543. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-S.; Kim, S.; Kim, S.; Ahn, K.; Min, D.-H. Fluorometric Viral miRNA Nanosensor for Diagnosis of Productive (Lytic) Human Cytomegalovirus Infection in Living Cells. ACS Sensors 2021, 6, 815–822. [Google Scholar] [CrossRef]
- Liu, X.; He, C.; Huang, Q.; Yu, M.; Qiu, Z.; Cheng, H.; Yang, Y.; Hao, X.; Wang, X. A facile visualized solid-phase detection of virus-specific nucleic acid sequences through an upconversion activated linear luminescence recovery process. Analyst 2022, 147, 2378–2387. [Google Scholar] [CrossRef]
- Gogotsi, Y.; Anasori, B. The Rise of MXenes. ACS Nano 2019, 13, 8491–8494. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-H.; Feng, Y.-G.; Wang, A.-J.; Mei, L.-P.; Luo, X.; Feng, J.-J. A signal-on photoelectrochemical aptasensor for chloramphenicol assay based on 3D self-supporting AgI/Ag/BiOI Z-scheme heterojunction arrays. Biosens. Bioelectron. 2021, 181, 113158. [Google Scholar] [CrossRef] [PubMed]
- Rao, C.N.R.; Gopalakrishnan, K.; Maitra, U. Comparative Study of Potential Applications of Graphene, MoS2, and Other Two-Dimensional Materials in Energy Devices, Sensors, and Related Areas. ACS Appl. Mater. Interfaces 2015, 7, 7809–7832. [Google Scholar] [CrossRef]
- Chekin, F.; Bagga, K.; Subramanian, P.; Jijie, R.; Singh, S.K.; Kurungot, S.; Boukherroub, R.; Szunerits, S. Nucleic aptamer modified porous reduced graphene oxide/MoS2 based electrodes for viral detection: Application to human papillomavirus (HPV). Sens. Actuators B Chem. 2018, 262, 991–1000. [Google Scholar] [CrossRef]
- Zhang, K.; Fan, Z.; Yao, B.; Ding, Y.; Zhao, J.; Xie, M.; Pan, J. Exploring the trans-cleavage activity of CRISPR-Cas12a for the development of a Mxene based electrochemiluminescence biosensor for the detection of Siglec-5. Biosens. Bioelectron. 2021, 178, 113019. [Google Scholar] [CrossRef]
- Zhang, K.; Fan, Z.; Huang, Y.; Ding, Y.; Xie, M. A strategy combining 3D-DNA Walker and CRISPR-Cas12a trans-cleavage activity applied to MXene based electrochemiluminescent sensor for SARS-CoV-2 RdRp gene detection. Talanta 2022, 236, 122868. [Google Scholar] [CrossRef] [PubMed]
- Hatamluyi, B.; Rezayi, M.; Amel Jamehdar, S.; Rizi, K.S.; Mojarrad, M.; Meshkat, Z.; Choobin, H.; Soleimanpour, S.; Boroushaki, M.T. Sensitive and specific clinically diagnosis of SARS-CoV-2 employing a novel biosensor based on boron nitride quantum dots/flower-like gold nanostructures signal amplification. Biosens. Bioelectron. 2022, 207, 114209. [Google Scholar] [CrossRef]
- John, A.; Benny, L.; Cherian, A.R.; Narahari, S.Y.; Varghese, A.; Hegde, G. Electrochemical sensors using conducting polymer/noble metal nanoparticle nanocomposites for the detection of various analytes: A review. J. Nanostruct. Chem. 2021, 11, 1–31. [Google Scholar] [CrossRef]
- Kanwar, R.; Rathee, J.; Salunke, D.B.; Mehta, S.K. Green Nanotechnology-Driven Drug Delivery Assemblies. ACS Omega 2019, 4, 8804–8815. [Google Scholar] [CrossRef]
- Wu, C.-Y.; Hsieh, H.; Lee, Y.-C. Contact Photolithography at Sub-Micrometer Scale Using a Soft Photomask. Micromachines 2019, 10, 547. [Google Scholar] [CrossRef]
- Fruncillo, S.; Su, X.; Liu, H.; Wong, L.S. Lithographic Processes for the Scalable Fabrication of Micro- and Nanostructures for Biochips and Biosensors. ACS Sensors 2021, 6, 2002–2024. [Google Scholar] [CrossRef]
- Li, R.; Zhao, Y.; Fan, H.; Chen, M.; Hu, W.; Zhang, Q.; Jin, M.; Liu, G.L.; Huang, L. Versatile nanorobot hand biosensor for specific capture and ultrasensitive quantification of viral nanoparticles. Mater. Today Bio 2022, 16, 100444. [Google Scholar] [CrossRef]
- Kurt, H.; Pishva, P.; Pehlivan, Z.S.; Arsoy, E.G.; Saleem, Q.; Bayazıt, M.K.; Yüce, M. Nanoplasmonic biosensors: Theory, structure, design, and review of recent applications. Anal. Chim. Acta 2021, 1185, 338842. [Google Scholar] [CrossRef]
- Shariati, M.; Sadeghi, M.; Shojaei, S.H.R. Sensory analysis of hepatitis B virus DNA for medicinal clinical diagnostics based on molybdenum doped ZnO nanowires field effect transistor biosensor; a comparative study to PCR test results. Anal. Chim. Acta 2022, 1195, 339442. [Google Scholar] [CrossRef]
- Leitis, A.; Tseng, M.L.; John-Herpin, A.; Kivshar, Y.S.; Altug, H. Wafer-Scale Functional Metasurfaces for Mid-Infrared Photonics and Biosensing. Adv. Mater. 2021, 33, 2102232. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, P.; Hu, X.; Huang, L.; Geng, Z.; Xu, H.; Hu, W.; Wang, L.; Wu, P.; Liu, G.L. An ultra-sensitive and specific nanoplasmonic-enhanced isothermal amplification platform for the ultrafast point-of-care testing of SARS-CoV-2. Chem. Eng. J. 2023, 451, 138822. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, M.M.; Abudayyeh, O.O.; Gootenberg, J.S.; Zhang, F.; Collins, J.J. CRISPR-based diagnostics. Nat. Biomed. Eng. 2021, 5, 643–656. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-Y.; Cheng, Q.-X.; Wang, J.-M.; Li, X.-Y.; Zhang, Z.-L.; Gao, S.; Cao, R.-B.; Zhao, G.-P.; Wang, J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018, 4, 20. [Google Scholar] [CrossRef]
- Patchsung, M.; Jantarug, K.; Pattama, A.; Aphicho, K.; Suraritdechachai, S.; Meesawat, P.; Sappakhaw, K.; Leelahakorn, N.; Ruenkam, T.; Wongsatit, T.; et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1140–1149. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Shi, Y.; Peng, F.; Zhou, M.; Yin, Y.; Tan, Y.; Chen, M.; Yin, X.; Ke, G.; Zhang, X.-B. Exploring the Trans-Cleavage Activity of CRISPR/Cas12a on Gold Nanoparticles for Stable and Sensitive Biosensing. Anal. Chem. 2021, 93, 4967–4974. [Google Scholar] [CrossRef] [PubMed]
- Aquino-Jarquin, G. Recent progress on rapid SARS-CoV-2/COVID-19 detection by CRISPR-Cas13-based platforms. Drug Discov. Today 2021, 26, 2025–2035. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Lai, W.; Man, T.; Chang, B.; Li, L.; Chandrasekaran, A.R.; Pei, H. Rationally engineered nucleic acid architectures for biosensing applications. Chem. Rev. 2019, 119, 11631–11717. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Liu, J. Catalytic Nucleic Acids: Biochemistry, Chemical Biology, Biosensors, and Nanotechnology. iScience 2020, 23, 100815. [Google Scholar] [CrossRef]
- Shen, L.; Wang, P.; Ke, Y. DNA Nanotechnology-Based Biosensors and Therapeutics. Adv. Healthc. Mater. 2021, 10, 2002205. [Google Scholar] [CrossRef]
- Guo, X.; Chen, L.; Li, P.; Li, X.; Wang, J.; Guo, L.; Yang, H. Construction of electrochemiluminescence biosensor via click chemistry and ARGET-ATRP for detecting tobacco mosaic virus RNA. Anal. Biochem. 2022, 655, 114834. [Google Scholar] [CrossRef]
- Kong, D.; Wang, X.; Gu, C.; Guo, M.; Wang, Y.; Ai, Z.; Zhang, S.; Chen, Y.; Liu, W.; Wu, Y.; et al. Direct SARS-CoV-2 Nucleic Acid Detection by Y-Shaped DNA Dual-Probe Transistor Assay. J. Am. Chem. Soc. 2021, 143, 17004–17014. [Google Scholar] [CrossRef]
- Yang, H.; Zhou, Y.; Liu, J. G-quadruplex DNA for construction of biosensors. TrAC Trends Anal. Chem. 2020, 132, 116060. [Google Scholar] [CrossRef]
- Bialy, R.M.; Mainguy, A.; Li, Y.; Brennan, J.D. Functional nucleic acid biosensors utilizing rolling circle amplification. Chem. Soc. Rev. 2022, 51, 9009–9067. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Zhang, H.-C.; Sun, X.-H.; Guo, F.-N.; Feng, L.-X.; Yang, T.; Wang, J.-H. Detection of HIV/HCV virus DNA with homogeneous DNA machine-triggered in situ formation of silver nanoclusters. Sens. Actuators B Chem. 2022, 352, 131041. [Google Scholar] [CrossRef]
- Thévenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical biosensors: Recommended definitions and classification. Biosens. Bioelectron. 2001, 16, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Diculescu, V.C.; Chiorcea-Paquim, A.-M.; Oliveira-Brett, A.M. Applications of a DNA-electrochemical biosensor. TrAC Trends Anal. Chem. 2016, 79, 23–36. [Google Scholar] [CrossRef]
- Islam, M.N.; Masud, M.K.; Haque, M.H.; Hossain, M.S.A.; Yamauchi, Y.; Nguyen, N.-T.; Shiddiky, M.J.A. RNA Biomarkers: Diagnostic and Prognostic Potentials and Recent Developments of Electrochemical Biosensors. Small Methods 2017, 1, 1700131. [Google Scholar] [CrossRef]
- Erdem, A.; Eksin, E.; Congur, G. Indicator-free electrochemical biosensor for microRNA detection based on carbon nanofibers modified screen printed electrodes. J. Electroanal. Chem. 2015, 755, 167–173. [Google Scholar] [CrossRef]
- Cho, I.-H.; Kim, D.H.; Park, S. Electrochemical biosensors: Perspective on functional nanomaterials for on-site analysis. Biomater. Res. 2020, 24, 6. [Google Scholar] [CrossRef]
- Chowdhury, A.D.; Takemura, K.; Li, T.-C.; Suzuki, T.; Park, E.Y. Electrical pulse-induced electrochemical biosensor for hepatitis E virus detection. Nat. Commun. 2019, 10, 3737. [Google Scholar] [CrossRef] [PubMed]
- Cajigas, S.; Alzate, D.; Orozco, J. Gold nanoparticle/DNA-based nanobioconjugate for electrochemical detection of Zika virus. Microchim. Acta 2020, 187, 594. [Google Scholar] [CrossRef] [PubMed]
- El-Sheikh, S.M.; Osman, D.I.; Ali, O.I.; Shousha, W.G.; Shoeib, M.A.; Shawky, S.M.; Sheta, S.M. A novel Ag/Zn bimetallic MOF as a superior sensitive biosensing platform for HCV-RNA electrochemical detection. Appl. Surf. Sci. 2021, 562, 150202. [Google Scholar] [CrossRef]
- Kashefi-Kheyrabadi, L.; Nguyen, H.V.; Go, A.; Baek, C.; Jang, N.; Lee, J.M.; Cho, N.-H.; Min, J.; Lee, M.-H. Rapid, multiplexed, and nucleic acid amplification-free detection of SARS-CoV-2 RNA using an electrochemical biosensor. Biosens. Bioelectron. 2022, 195, 113649. [Google Scholar] [CrossRef]
- Ji, D.; Guo, M.; Wu, Y.; Liu, W.; Luo, S.; Wang, X.; Kang, H.; Chen, Y.; Dai, C.; Kong, D.; et al. Electrochemical Detection of a Few Copies of Unamplified SARS-CoV-2 Nucleic Acids by a Self-Actuated Molecular System. J. Am. Chem. Soc. 2022, 144, 13526–13537. [Google Scholar] [CrossRef] [PubMed]
- Moço, A.C.R.; Neto, J.A.S.; de Moraes, D.D.; Guedes, P.H.; Brussasco, J.G.; Flauzino, J.M.R.; Luz, L.F.G.; Soares, M.M.C.N.; Madurro, J.M.; Brito-Madurro, A.G. Carbon ink-based electrodes modified with nanocomposite as a platform for electrochemical detection of HIV RNA. Microchem. J. 2021, 170, 106739. [Google Scholar] [CrossRef]
- Najjar, D.; Rainbow, J.; Timilsina, S.S.; Jolly, P.; de Puig, H.; Yafia, M.; Durr, N.; Sallum, H.; Alter, G.; Li, J.Z.; et al. A lab-on-a-chip for the concurrent electrochemical detection of SARS-CoV-2 RNA and anti-SARS-CoV-2 antibodies in saliva and plasma. Nat. Biomed. Eng. 2022, 6, 968–978. [Google Scholar] [CrossRef]
- Lee, Y.; Choi, J.; Han, H.-K.; Park, S.; Park, S.Y.; Park, C.; Baek, C.; Lee, T.; Min, J. Fabrication of ultrasensitive electrochemical biosensor for dengue fever viral RNA Based on CRISPR/Cpf1 reaction. Sens. Actuators B Chem. 2021, 326, 128677. [Google Scholar] [CrossRef]
- Heo, W.; Lee, K.; Park, S.; Hyun, K.-A.; Jung, H.-I. Electrochemical biosensor for nucleic acid amplification-free and sensitive detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA via CRISPR/Cas13a trans-cleavage reaction. Biosens. Bioelectron. 2022, 201, 113960. [Google Scholar] [CrossRef]
- Bukkitgar, S.D.; Shetti, N.P.; Aminabhavi, T.M. Electrochemical investigations for COVID-19 detection-A comparison with other viral detection methods. Chem. Eng. J. 2021, 420, 127575. [Google Scholar] [CrossRef] [PubMed]
- Tanwar, S.; Paidi, S.K.; Prasad, R.; Pandey, R.; Barman, I. Advancing Raman spectroscopy from research to clinic: Translational potential and challenges. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 260, 119957. [Google Scholar] [CrossRef]
- Serebrennikova, K.V.; Berlina, A.N.; Sotnikov, D.V.; Zherdev, A.V.; Dzantiev, B.B. Raman Scattering-Based Biosensing: New Prospects and Opportunities. Biosensors 2021, 11, 512. [Google Scholar] [CrossRef] [PubMed]
- Shipp, D.W.; Sinjab, F.; Notingher, I. Raman spectroscopy: Techniques and applications in the life sciences. Adv. Opt. Photon. 2017, 9, 315–428. [Google Scholar] [CrossRef]
- Wu, L.; Dias, A.; Diéguez, L. Surface enhanced Raman spectroscopy for tumor nucleic acid: Towards cancer diagnosis and precision medicine. Biosens. Bioelectron. 2022, 204, 114075. [Google Scholar] [CrossRef]
- Cao, Y.; Sun, M. Tip-enhanced Raman spectroscopy. Rev. Phys. 2022, 8, 100067. [Google Scholar] [CrossRef]
- Kneipp, K.; Wang, Y.; Kneipp, H.; Perelman, L.T.; Itzkan, I.; Dasari, R.R.; Feld, M.S. Single Molecule Detection Using Surface-Enhanced Raman Scattering (SERS). Phys. Rev. Lett. 1997, 78, 1667–1670. [Google Scholar] [CrossRef]
- Zhang, Y.-J.; Chen, S.; Radjenovic, P.; Bodappa, N.; Zhang, H.; Yang, Z.-L.; Tian, Z.-Q.; Li, J.-F. Probing the Location of 3D Hot Spots in Gold Nanoparticle Films Using Surface-Enhanced Raman Spectroscopy. Anal. Chem. 2019, 91, 5316–5322. [Google Scholar] [CrossRef]
- Jang, A.S.; Kumar, P.P.P.; Lim, D.-K. Attomolar Sensitive Magnetic Microparticles and a Surface-Enhanced Raman Scattering-Based Assay for Detecting SARS-CoV-2 Nucleic Acid Targets. ACS Appl. Mater. Interfaces 2022, 14, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.-H.; Shin, M.; Yang, L.; Conley, B.; Yoon, J.; Lee, S.-N.; Lee, K.-B.; Choi, J.-W. Clustered Regularly Interspaced Short Palindromic Repeats-Mediated Amplification-Free Detection of Viral DNAs Using Surface-Enhanced Raman Spectroscopy-Active Nanoarray. ACS Nano 2021, 15, 13475–13485. [Google Scholar] [CrossRef]
- Su, A.; Liu, Y.; Cao, X.; Xu, W.; Liang, C.; Xu, S. A universal CRISPR/Cas12a-mediated AuNPs aggregation-based surface-enhanced Raman scattering (CRISPR/Cas-SERS) platform for virus gene detection. Sens. Actuators B Chem. 2022, 369, 132295. [Google Scholar] [CrossRef]
- Yin, B.; Zhang, Q.; Xia, X.; Li, C.; Ho, W.K.H.; Yan, J.; Huang, Y.; Wu, H.; Wang, P.; Yi, C.; et al. A CRISPR-Cas12a integrated SERS nanoplatform with chimeric DNA/RNA hairpin guide for ultrasensitive nucleic acid detection. Theranostics 2022, 12, 5914–5930. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Miao, X.; Song, C.; Chen, N.; Xiong, J.; Gan, H.; Ni, J.; Zhu, Y.; Cheng, K.; Wang, L. Non-enzymatic signal amplification-powered point-of-care SERS sensor for rapid and ultra-sensitive assay of SARS-CoV-2 RNA. Biosens. Bioelectron. 2022, 212, 114379. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Li, Y.; Tang, B.; Zhang, C.-y. Fluorescent Biosensors Based on Single-Molecule Counting. Acc. Chem. Res. 2016, 49, 1722–1730. [Google Scholar] [CrossRef]
- Zhou, H.; Zhang, S. Recent Development of Fluorescent Light-Up RNA Aptamers. Crit. Rev. Anal. Chem. 2022, 52, 1644–1661. [Google Scholar] [CrossRef]
- Ryoo, S.-R.; Lee, J.; Yeo, J.; Na, H.-K.; Kim, Y.-K.; Jang, H.; Lee, J.H.; Han, S.W.; Lee, Y.; Kim, V.N.; et al. Quantitative and Multiplexed MicroRNA Sensing in Living Cells Based on Peptide Nucleic Acid and Nano Graphene Oxide (PANGO). ACS Nano 2013, 7, 5882–5891. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.-H.; Lim, J.; Shin, M.; Paek, S.-H.; Choi, J.-W. CRISPR-Cas12a-Based Nucleic Acid Amplification-Free DNA Biosensor via Au Nanoparticle-Assisted Metal-Enhanced Fluorescence and Colorimetric Analysis. Nano Lett. 2021, 21, 693–699. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Liu, T.; Zhang, X.-B.; Huan, S.-Y.; Wu, C.; Fu, T.; Tan, W. Multicolor Fluorescent Biosensor for Multiplexed Detection of DNA. Anal. Chim. Acta 2014, 86, 5009–5016. [Google Scholar] [CrossRef]
- Xu, Z.; Chen, D.; Li, T.; Yan, J.; He, T.; Hu, R.; Li, Y.; Yang, Y.; Liu, M. Microfluidic space coding for multiplexed nucleic acid detection via CRISPR-Cas12a and recombinase polymerase amplification. Nat. Commun. 2022, 13, 6480. [Google Scholar] [CrossRef]
- Choi, J.-H.; Ha, T.; Shin, M.; Lee, S.-N.; Choi, J.-W. Nanomaterial-Based Fluorescence Resonance Energy Transfer (FRET) and Metal-Enhanced Fluorescence (MEF) to Detect Nucleic Acid in Cancer Diagnosis. Biomedicines 2021, 9, 928. [Google Scholar] [CrossRef]
- Li, D.; Chen, H.; Gao, X.; Mei, X.; Yang, L. Development of General Methods for Detection of Virus by Engineering Fluorescent Silver Nanoclusters. ACS Sensors 2021, 6, 613–627. [Google Scholar] [CrossRef] [PubMed]
- Bardajee, G.R.; Zamani, M.; Sharifi, M. Efficient and Versatile Application of Fluorescence DNA-Conjugated CdTe Quantum Dots Nanoprobe for Detection of a Specific Target DNA of SARS Cov-2 Virus. Langmuir 2021, 37, 10223–10232. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.-C.; Chang, C.-C.; Chuang, T.-L.; Sung, K.-B.; Lin, C.-W. Rapid Detection of Virus Nucleic Acid via Isothermal Amplification on Plasmonic Enhanced Digitizing Biosensor. Biosensors 2022, 12, 75. [Google Scholar] [CrossRef] [PubMed]
- Teengam, P.; Nisab, N.; Chuaypen, N.; Tangkijvanich, P.; Vilaivan, T.; Chailapakul, O. Fluorescent paper-based DNA sensor using pyrrolidinyl peptide nucleic acids for hepatitis C virus detection. Biosens. Bioelectron. 2021, 189, 113381. [Google Scholar] [CrossRef]
- Huang, Z.; Liu, S.; Pei, X.; Li, S.; He, Y.; Tong, Y.; Liu, G. Fluorescence Signal-Readout of CRISPR/Cas Biosensors for Nucleic Acid Detection. Biosensors 2022, 12, 779. [Google Scholar] [CrossRef]
- Tao, Y.; Yi, K.; Wang, H.; Kim, H.-W.; Li, K.; Zhu, X.; Li, M. CRISPR-Cas12a-regulated DNA adsorption and metallization on MXenes as enhanced enzyme mimics for sensitive colorimetric detection of hepatitis B virus DNA. J. Colloid Interface Sci. 2022, 613, 406–414. [Google Scholar] [CrossRef]
- Zhao, X.; Tian, X.; Wang, Y.; Li, L.; Yu, Y.; Zhao, S.; Zhang, J. CRISPR-Cas12a-activated palindrome-catalytic hairpin assembly for ultrasensitive fluorescence detection of HIV-1 DNA. Anal. Chim. Acta 2022, 1227, 340303. [Google Scholar] [CrossRef]
- Tao, Y.; Yi, K.; Wang, H.; Li, K.; Li, M. Metal nanoclusters combined with CRISPR-Cas12a for hepatitis B virus DNA detection. Sens. Actuators B Chem. 2022, 361, 131711. [Google Scholar] [CrossRef]
- Alexaki, K.; Kyriazi, M.E.; Greening, J.; Taemaitree, L.; El-Sagheer, A.H.; Brown, T.; Zhang, X.; Muskens, O.L.; Kanaras, A.G. A SARS-Cov-2 sensor based on upconversion nanoparticles and graphene oxide. RSC Adv. 2022, 12, 18445–18449. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-H.; Park, Y.I.; Lin, H.-Y.; Pathania, D.; Park, K.S.; Avila-Wallace, M.; Castro, C.M.; Weissleder, R.; Lee, H. Compact and Filter-Free Luminescence Biosensor for Mobile in Vitro Diagnoses. ACS Nano 2019, 13, 11698–11706. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Su, F.; Zhang, R.; Jiang, X.; Xiao, H.; Yan, X.; Yang, C.; Fan, X.; Wu, G. Rapid point-of-care testing for SARS-CoV-2 virus nucleic acid detection by an isothermal and nonenzymatic Signal amplification system coupled with a lateral flow immunoassay strip. Sens. Actuators B Chem. 2021, 342, 129899. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-M.; Lee, C.; Lee, Y.; Lee, J.; Park, S.-J.; Park, S.; Nam, J.-M. Synthesis, Assembly, Optical Properties, and Sensing Applications of Plasmonic Gap Nanostructures. Adv. Mater. 2021, 33, 2006966. [Google Scholar] [CrossRef]
- Wang, L.; Wang, X.; Wu, Y.; Guo, M.; Gu, C.; Dai, C.; Kong, D.; Wang, Y.; Zhang, C.; Qu, D.; et al. Rapid and ultrasensitive electromechanical detection of ions, biomolecules and SARS-CoV-2 RNA in unamplified samples. Nat. Biomed. Eng. 2022, 6, 276–285. [Google Scholar] [CrossRef]

| Target | Nanomaterials | Nanotechnology | Detection Method | LoD | Linear Range | Ref. |
|---|---|---|---|---|---|---|
| SARS-CoV-2 | Au nanoflower | CRISPR/Cas13a | Electrochemistry | 4.4 × 10−2 fg/mL (Orf gene) 8.1 × 10−2 fg/mL (S gene) | 1.0 × 10−1 fg/mL to 1.0 × 105 fg/mL | [94] |
| CNTs | - | Electrochemistry | 0.33 aM | 1 aM to 10 pM | [45] | |
| Au nanoneedle | 4-Wj DNA structure | Electrochemistry | 2 copies/μL (S gene) 3 copies/μL (Orf1ab gene) | 100 aM to 10 pM | [89] | |
| Au nanoporous electrode array | LIL | Electrochemistry | 1 fM | 100 pM to 1 fM | [20] | |
| PEI-Ru@Ti3C2@Au nanoparticles (Mxene) | CRISPR/Cas12a | Electrochemiluminescence | 12.8 aM | 1 aM- 500 aM | [56] | |
| CdTe QDs | FRET | Fluorescence | 2.52 × nM | - | [116] | |
| Au nanoparticles | - | SERS | 1 fM | 1 fM to 1 nM | [18] | |
| Ag nanoclusters | - | SERS | 1 fM (RdRp gene) 1 pM (E gene) | 1 fM to 1 nM (RdRp gene) 1 pM to 1 nM (E gene) | [103] | |
| Influenza | Ag nanopillars | SERS | 41.1 fM | 0 pM to 0.159 pM | [38] | |
| HPV | MoS2 GO | Nucleic aptamer | Electrochemistry | 1.75 pM | 3.5 pM to 35.3 pM | [54] |
| HBV | Mo doped ZnO nanowires | EBL | FET Electrochemistry | 1 pM | 1 pM to 10 μM | [64] |
| CuNCs | CRISPR/Cas12a | Fluorescence | 0.54 pM | 0.5 pM to 100 pM | [122] | |
| HCV | GQD/AgNC | UV-vis | 24.84 pM | 25 pM to 500 pM | [42] | |
| Ag/Zn-MOF | DNA probe | Electrochemistry | 0.64 fM | 1 fM to 100 nM | [88] | |
| HIV | CRISPR/Cas12a catalytic hairpin DNA assembly | Fluorescence | 4.2 fM | 10 fM to 100 nM | [121] | |
| HCMV | GO | Fluorescence | 68.5 pM | 61.0 pM to 488.3 pM | [48] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, H.K.; Yoon, J. Nanotechnology-Assisted Biosensors for the Detection of Viral Nucleic Acids: An Overview. Biosensors 2023, 13, 208. https://doi.org/10.3390/bios13020208
Choi HK, Yoon J. Nanotechnology-Assisted Biosensors for the Detection of Viral Nucleic Acids: An Overview. Biosensors. 2023; 13(2):208. https://doi.org/10.3390/bios13020208
Chicago/Turabian StyleChoi, Hye Kyu, and Jinho Yoon. 2023. "Nanotechnology-Assisted Biosensors for the Detection of Viral Nucleic Acids: An Overview" Biosensors 13, no. 2: 208. https://doi.org/10.3390/bios13020208
APA StyleChoi, H. K., & Yoon, J. (2023). Nanotechnology-Assisted Biosensors for the Detection of Viral Nucleic Acids: An Overview. Biosensors, 13(2), 208. https://doi.org/10.3390/bios13020208





