Abstract
To curtail pathogens or tumors, antimicrobial or antineoplastic drugs have been developed. These drugs target microbial/cancer growth and survival, thereby improving the host’s health. In attempts to evade the detrimental effects of such drugs, these cells have evolved several mechanisms over time. Some variants of the cells have developed resistances against multiple drugs or antimicrobial agents. Such microorganisms or cancer cells are said to exhibit multidrug resistance (MDR). The drug resistance status of a cell can be determined by analyzing several genotypic and phenotypic changes, which are brought about by significant physiological and biochemical alterations. Owing to their resilient nature, treatment and management of MDR cases in clinics is arduous and requires a meticulous approach. Currently, techniques such as plating and culturing, biopsy, gene sequencing, and magnetic resonance imaging are prevalent in clinical practices for determining drug resistance status. However, the major drawbacks of using these methods lie in their time-consuming nature and the problem of translating them into point-of-care or mass-detection tools. To overcome the shortcomings of conventional techniques, biosensors with a low detection limit have been engineered to provide quick and reliable results conveniently. These devices are highly versatile in terms of analyte range and quantities that can be detected to report drug resistance in a given sample. A brief introduction to MDR, along with a detailed insight into recent biosensor design trends and use for identifying multidrug-resistant microorganisms and tumors, is presented in this review.
Keywords:
multidrug resistance; MDR; biosensors; sensors; drugs; drug efflux pump; antibiotics; antimicrobial; cancer; microorganisms 1. Introduction
The introduction should briefly place the study in a broad context and highlight its importance. Multidrug resistance (MDR) is a globally recognized pressing health concern that has rendered several drugs ineffective against many hitherto treatable diseases. Examples include tuberculosis, malaria, urinary tract infection, gonorrhea, pneumonia, and fever, which have increasingly become more difficult to treat due to MDR [1]. The underlying reason for the emergence of the multi-drug resistant strains of microorganisms can be attributed in part to the presence of antimicrobial (AM)/antibiotic residues in the environment owing to indiscriminate use of AM/antibiotics in human and veterinary medicines, livestock farming, and to a certain extent on personal negligence of the patients in consuming antibiotics [2,3,4]. MDR is becoming a major cause of concern because relatively fewer new antibiotics are being added to the developmental pipelines of the pharmaceutical industry while the existing ones are becoming redundant. For instance, a recent study revealed that 45 antibiotic drugs are at various stages of the clinical development pipelines currently [5]. However, around 26% of Escherichia coli clinical isolates from the neonatal wards of hospitals exhibited MDR [6], suggesting a disparity in the rates of newer antibiotic discovery versus the emergence of MDR.
MDR has also been recognized as a stumbling block in the effective treatment of cancers. MDR in cancers is the primary reason for tumor recurrence and refractory cancers [7,8]. While a definite underlying theory for the development of MDR in cancers is still elusive, initially, MDR exhibited by cancer cells was approached by borrowing insights from microbial MDR [9]. As with microbial MDR, most drug-resistant cancerous cells can evade the effects of various prescribed anticancer agents. However, unlike microbial MDR, these cells can develop cross-resistance to drugs that may be structurally and functionally unrelated [8]. Nevertheless, with the rampant increase in the number and types of cancers, the effects of MDR inevitably impact the treatment regime and the quality of life of patients adversely, as it limits the efficacious and extended use of drugs [10].
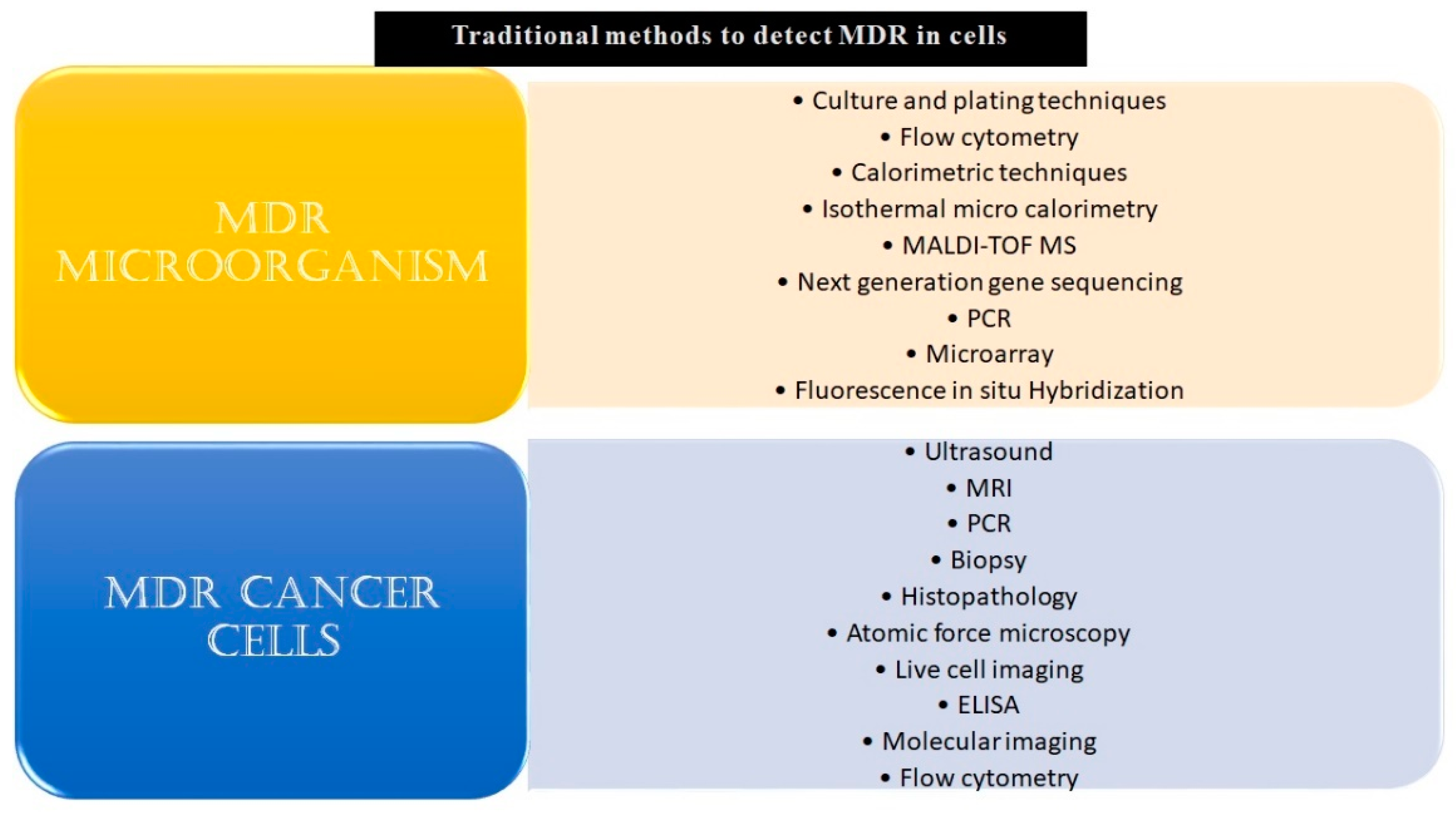
Given the challenges MDR poses by burdening healthcare systems, it is highly pertinent to develop technologies that can opportunistically screen/identify MDR strains, especially during the early stages of their onset, and monitor them. The existing methods to detect microbial and cancer MDR include culturing and plating techniques, ELIZA, biopsy, MRI, PCR, histopathology, next-generation sequencing techniques, and others, as listed in Figure 1.
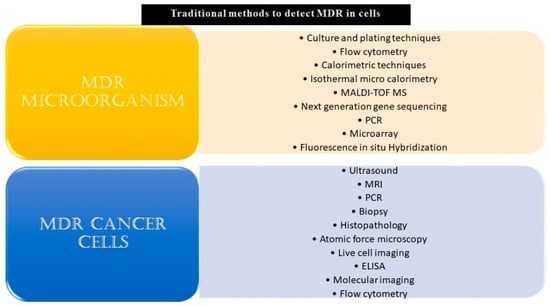
Figure 1.
Conventional methods to detect MDR in microbial or malignant systems. The conventional and modern methods which have been used in the detection of MDR specimens have been listed above. Some of these techniques, for example, culturing and plating techniques and biopsy, are considered gold standards for confirmation of drug resistance in a given sample. Though these reliably yield results, a major drawback of most of these methods is that they are time-consuming and fail to give real-time data. Other drawbacks include the requirement of skilled labor, laboratory setup, cumbersome procedures, and an inability to carry out large-scale testing.
A major shortcoming of these techniques is that they are time-intensive and hence cannot be used for real-time analysis and monitoring [11,12,13]. Other drawbacks include invasiveness, low sensitivities and selectivities, a high cost of sample processing, and the requirement of sophisticated lab infrastructure and skilled professionals to run the tests [14,15,16,17]. Similarly, most conventional techniques that are used in MDR cancer detection focus on phenotypic characteristics rather than detecting subtle but significant changes at the genetic and epigenetic levels, rendering them ineffective for the early detection of MDR [18,19,20,21].
Biosensors have emerged as an attractive option for yielding specific and selective diagnoses for MDR-related diseases [17]. Biosensor devices capitalize on the underlying physicochemical aspects of biological systems and are developed by integrating engineering, chemistry, and electronics concepts. The concept of biosensor devices is aptly reflected in the IUPAC’s definition of biosensors, which are described as devices that use specific biochemical reactions performed by biological entities like isolated enzymes, immune systems, tissues, organelles, or whole cells to detect chemical compounds, usually by electrical, thermal, or optical signals [22]. In essence, these devices identify the analyte of interest, that may be a part, or a product, of a biological system or reaction. With the help of a suitable signal processing methodology, biosensors convert the events of recognition of these moieties to measurable signal outputs that are further processed and amplified [23].
The current paradigm in biosensor development focuses on improving measurable attributes such as maximizing selectivity sensitivity, signal-to-noise ratio, reproducibility, and multiplexing and detecting an array of phenotypic traits/genotypic elements that contribute to the prevalence of MDR [15,24]. Biosensors are poised to serve as an excellent point of care diagnosis of MDR-related diseases [25,26].
In this review, we highlight various aspects of biosensor development that have helped in the detection/identification of MDR pathogens or cancer cells. Further, the concepts and the origins of MDR, and factors contributing to the emergence of these variants, are comprehensively discussed. Moreover, the application of biosensors to address the issue of microbial and cancer MDR is elaborately discussed. The present paper is expected to be of interest to those working in academia and industry on MDR mechanisms and the development of bioanalytical technologies for their detection.
2. Multidrug Resistance in Microorganisms and Biosensors
2.1. MDR Glossary and Origin of Concept
Following the landmark serendipitous discovery of penicillin, many classes of AMs against many pathogenic organisms like bacteria, protozoa, fungi, helminths, and viruses were discovered [27]. Upon the acknowledgment of drug-resistant variants, terms were coined to describe the resistance of pathogens toward various AM agents. Though there are no hard and fast lines segregating these microbial groups, the Centre for Disease Control & Prevention (CDC) and the European Centre for Disease Control (ECDC) have put forward guidelines to define these terms [28].
As per the guidelines, antimicrobial categories were designed uniquely for the epidemiologically important microorganisms Enterobacteriaceae, Staphylococcus aureus, Enterococcus spp., Acinetobacter, and Pseudomonas aeruginosa considering the drugs classes that are pertinent to each microbe, and each AM category consists of one or more drug/AM agent [29]. For example, 17 categories were made for S. aureus, containing aminoglycosides, anti-MRSA (Methicillin Resistant S. aureus) cephalosporins, antistaphylococcal β-lactams, fluoroquinolones, macrolides, oxazolidinones, phenicols, phosphoric acid, streptogramins, folate pathway inhibitors, fucidanes, glycopeptides, glycylcyclines, lipopeptides, lincosamides, and tetracyclines [29].
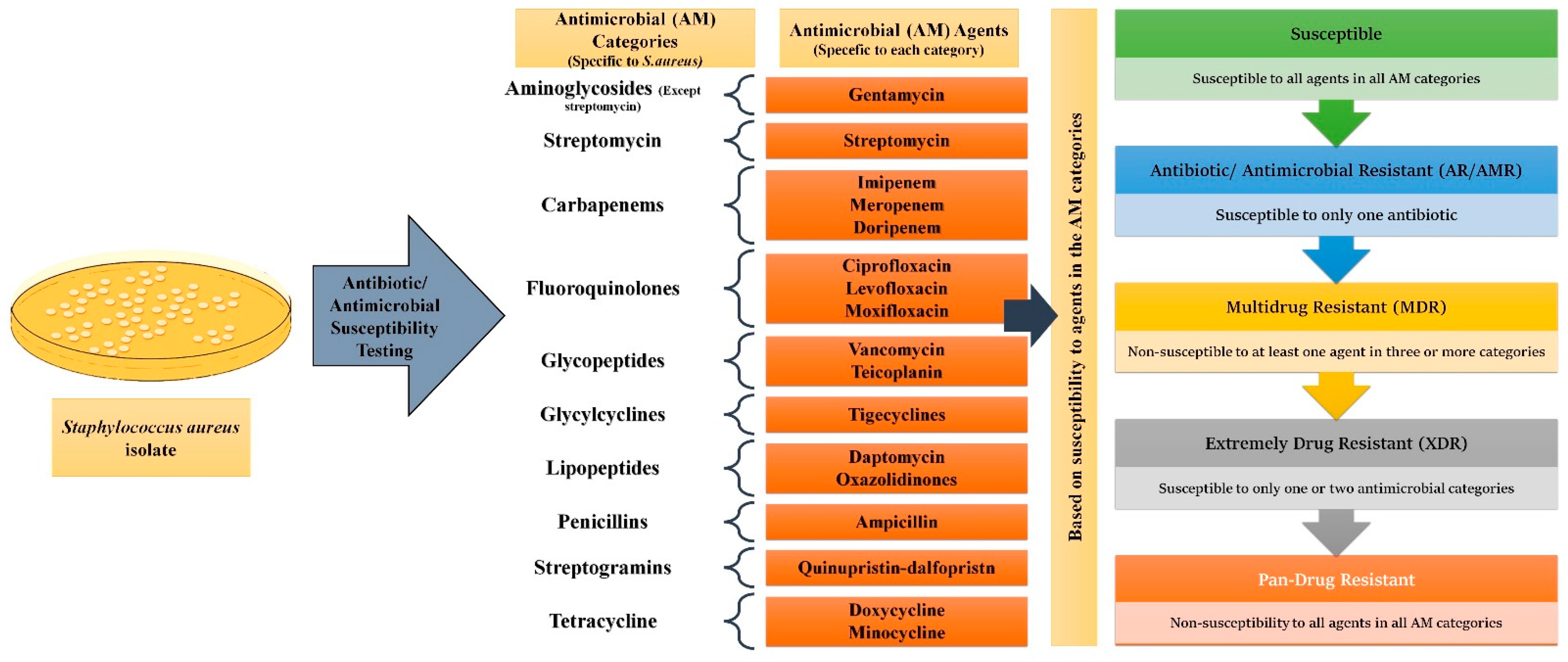
While antimicrobial resistance/antibiotic resistance (AMR/AR) is the ability of microorganisms to remain unaffected by a single AM or an antibiotic, MDR is the phenomenon where a microbe acquires non-susceptibility to at least one AM agent in a minimum of three categories [28]. XDR (Extremely Drug Resistant) pathogens remain susceptible to only one or two AM categories, whereas PDR (Pan Drug Resistant) expressing microbes show non-susceptibility to all agents in all AM categories [28]. Using S. aureus as an example, Figure 2 elaborates on how these distinctions are made.
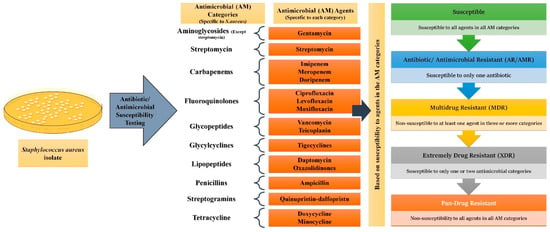
Figure 2.
Classification of microorganisms based on drug resistance to antimicrobial (AM) agents: For all epidemiologically important microbes, AM categories of pertinence have been made. Each AM category contains one or more AM agents. For Staphylococcus aureus 17 categories have been made. Depending upon the response the particular microbe shows to a different agent in the different categories, they can be divided into AR/AMR, MDR, XDR, and PDR strains.
2.2. Foundation of the Emergence of MDR
The emergence of MDR in microbial species is attributed to AM/antibiotic resistance genes in their genome or plasmids of some microbes [30]. These genes have either been inherently present in the wild-type strain or may have developed by mutation. The lateral transfer of genes from some other microorganism or the host (i.e., from a producer organism) has also been proposed as a potential mechanism for acquiring such genes [30].
Exposure to AMs imposes a selection pressure on a heterogeneous population of microbes under which the survival of the fittest prevails [31]. This selection mechanism is also influenced by various parameters, such as the drug used on the microorganism (certain classes of drugs show more propensity at promoting AM resistance development than others), the dosage of the drug, the pharmacokinetic profile of the drug, the physiological state of the host, the fitness cost of resistance incurred by the pathogen, and the influence of non-resistant therapeutics [30]. The clustering of such genes in response to a series of selection pressures over time facilitates the emergence of MDR strains. Evidence suggests that MDR is preferred over AMR/AR because of the lower fitness cost [30,31,32].
The above discussion establishes that MDR is a natural phenomenon guided by evolutionary pressures. However, the central point of contention and concern is its rapid dissemination and increasing prevalence. This necessitates the development of appropriate tools for its rapid and accurate identification.
2.3. Biochemical Basis of MDR in Microbes
The biochemical basis of microbial defenses against multiple drugs can be achieved by (a) alteration in the plasma membrane (PM) profile, (b) overcoming the drug action, and (c) alteration in the physiological state. It should be noted that these biochemical alterations may confer resistance to a single drug or more than one drug [33,34]
2.3.1. Alteration in the PM Profile
For a microbe, the PM is a barrier serving as a selectively permeable interface to the external environment. This selective barrier profoundly impacts the exchange fluxes between the cell and its environment. Pathogens demonstrating MDR may have significantly altered the structure and composition of the PM. For instance, the additional mycolic acid present in the PM of XDR Mycobacterium tuberculosis renders it impervious to a plethora of drugs [35].These structural and compositional changes hinder the drugs from reaching their targets or achieving the optimal intracellular concentration.
Further, various transport proteins usually embedded in microbial species’ PM exhibit enhanced expression in MDR strains. Five major families of ABC transport proteins are associated with drug efflux through the PM of M/O. These families can be broadly divided into ATP-dependent and proton (or Na+) motive force-dependent transporters. While ABC (ATP Binding Cassette) transporters are ATP-dependent transport proteins, proton-dependent proteins include the SMR (Staphylococcal/ Small Multidrug Resistance), MF (Major Facilitator), RND (Resistance Nodulation Factor), and MATE (Multidrug and Toxic Compound Extrusion) family transporters. Any survival risk to the pathogens is averted by the active extrusion of drugs using the over-expressed families of transport proteins [35]. Apart from this mechanism, another strategy used by MDR species involves the down-regulation of porins, which are large transmembrane intrinsic proteins, restricting the passage of hydrophilic drugs like β-lactams and tetracyclines [2,31].
2.3.2. Overcoming the Drug Action
If the drug overcomes the PM barrier and enters the cell, the following mechanisms are used to achieve resistance.
Target modification: The MDR species will bring about structural or configurational changes that result from genetic or epigenetic changes, thereby manifesting the following mechanisms leading to MDR. For instance: Changes in the penicillin-binding proteins by β-lactamase producing S. aureus strains conferred their resistance to penicillin, methicillin, and other drugs [31]; changes in the sequence of glycoproteinaceous pentapeptide conferred vancomycin resistance [30]. Expression of an alternate target for the drug, exhibiting a higher affinity that preferentially allows binding of the drug to the new target, or opting for target shunting by modification of the biochemical pathway, is also a method by which the cell can evade a drug’s action [31,36]
Drug modification: By this method, drugs may be rendered ineffective by resistant strains via inactivation of drugs by structural changes catalyzed by enzymes (e.g., enzymatic inactivation of antibiotics like kanamycin and tobramycin), or by post-translational modifications like phosphorylation, and adenylation and drug sequestration [30,37].
2.3.3. Alteration in the Physiological State
The physiological state of pathogens also contributes to the emergence of MDR. A biofilm is a heterogeneous 3D structure embedded in a polymeric matrix comprising carbohydrates, nucleic acid, and proteins, commonly referred to as an extracellular polymeric matrix [38]. Biofilm formation has been associated with AM resilience [39]. This phenotype also promotes survival and persistence by providing a favorable niche protecting the microbes from external stresses like osmolarity, fluctuating pH, nutrient scarcity, mechanical forces, and even host immune cells [40,41]. MDR due to biofilm formation has been observed in pathogens like E. coli, Pseudomonas aeruginosa, and Acinetobacter baumannii. Biofilms encompass physiologically unique cells called persister cells [30]. These are metabolically inactive dormant cells tolerant to AM agents and do not undergo any genetic alteration to achieve such properties [42]. For example, the recalcitrance of chronic infections can be accredited to the presence of persister cells in biofilms. This observation is attributed to the survival of the persister cells, while most of the cells become non-viable due to antibiotics [42].
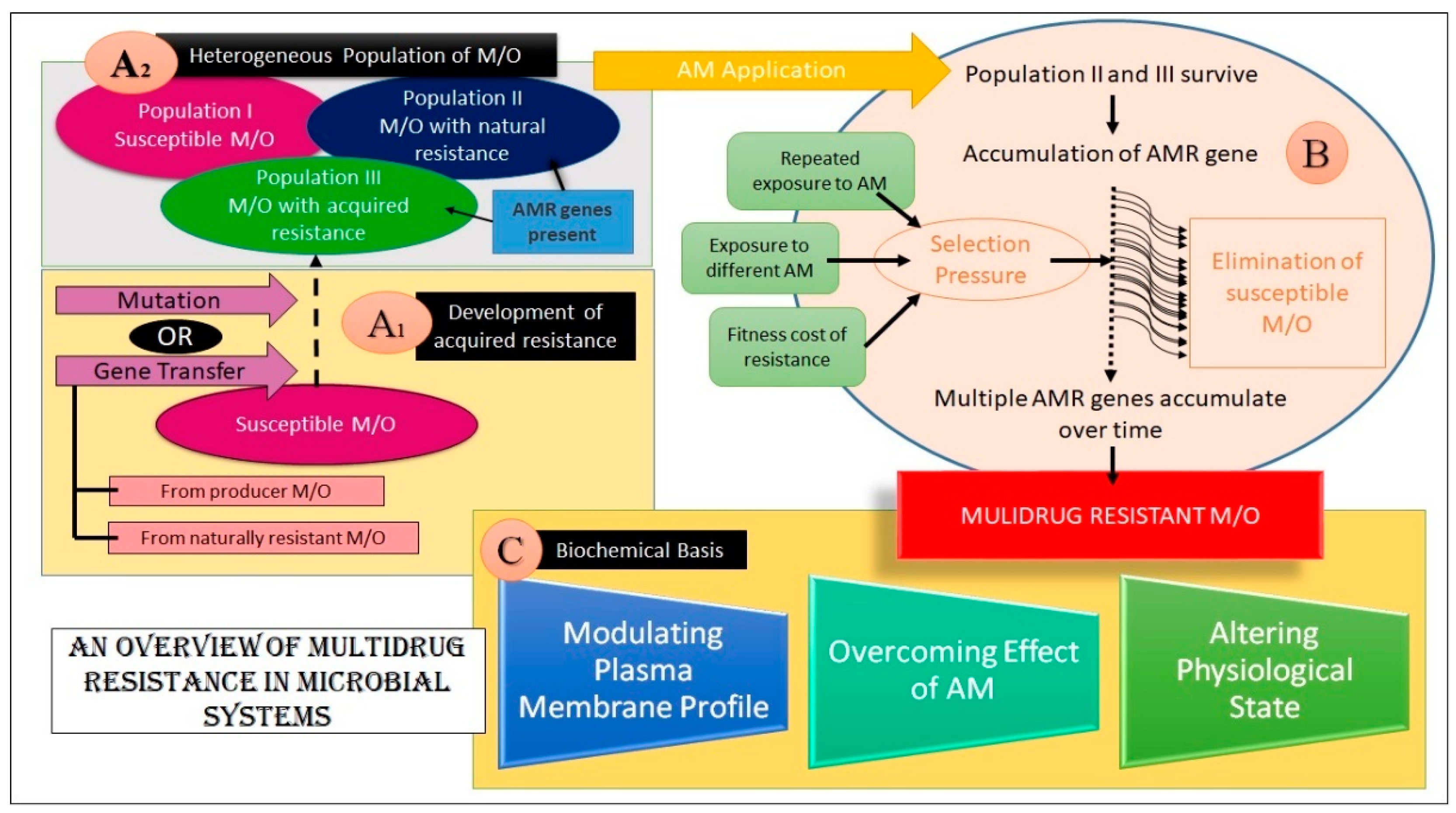
An overview of the foundation and mechanisms of MDR development in microbial systems is summarized in Figure 3.
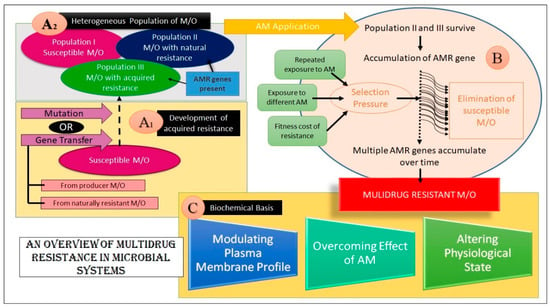
Figure 3.
An overview of MDR in microorganisms. (Key: M/O- Microorganism(s), AM- Antimicrobial, AMR- Antimicrobial Resistance) The figure aims at explaining MDR emergence in microbes. Sections A1 and A2 depict how an assortment of microbes are present in the natural pool. This heterogeneous pool harbors some microbes that are susceptible to AM action, while some of them are resistant to AM agents. This resistance may be either innate or acquired. As shown, these AMR genes may be acquired from transfer from resistant/producer M/O or may be endowed by mutation. Block B shows how the resistant group survives upon AM application and procreates, leading to the accumulation of AMR genes. Over time, selection pressure is applied wherein the susceptible microbes get killed and multiple AMR genes accumulate in the M/O, imparting MDR. These genes dictate alterations to the subject by various means, finally yielding a multidrug-resistant microbe, as shown in section C.
2.4. Use of Biosensors in Detecting MDR Microbes
The principle of biosensor design to identify MDR strains relies on detecting the genes/mutations conferring resistance. In addition, differential expression analysis of surrogate analytes that exhibit correlation with enhanced resistance or pathogenesis of the microbes is also monitored. Further, the MDR strains can be identified by analyzing altered physiological forms like biofilms. In this review, genotypic and phenotypic biosensors for detecting MDR microbial strains will be comprehensively discussed.
2.4.1. Genotypic Biosensor for MDR Microbes
The genome of an organism is the blueprint of its identity and characteristics. The emergence of MDR strains is invariably linked to alteration in the genomic configuration of an organism. This property is capitalized on to design genotypic biosensors that identify segments of DNA that impart resistance. Table 1 lists the differentially expressed or altered genes in MDR strains.
Gene sequences used to identify MDR targets may be of two types: resistance imparting genes and mutated genes. The former is absent in drug-sensitive microbes and is present in resistant variants. For example, the mecA gene is present in MRSA but absent in MSSA (Methicillin Susceptible S. aureus). Watanabe et al. designed a biosensor capable of discriminating MRSA from normal S. aureus based on the presence of the mecA gene to a limit of 10 pM. This system was suited for on-site analysis because it did not require a thermocycler for the amplification of the target DNA [43]. Similarly, a capacitive label-free DNA sensor was devised by Liu et al. to detect the ampR gene in real-time from field-sourced samples. The device exhibited a limit of detection (LOD) in the picomolar range, exhibiting high sensitivity. Additionally, it was reusable, simple, and portable based on the capacitance drop across the sensor surface due to the hybridization reaction between the target DNA and oligonucleotide probe functionalized over the gold electrode [44]. An electrochemical sensor was designed to detect the NDM 1 gene in clinical samples with multifold sensitivity (LOD: 0.042 pg/L) by Zhang et al. The biosensor developed was capable of detecting single base pair mismatches in complex bacterial clinical samples without requiring any PCR amplification [45].

Table 1.
MDR related genes in microorganisms.
Table 1.
MDR related genes in microorganisms.
| Gene | Form Associated with MDR | Function | Resistance Against | Reference |
|---|---|---|---|---|
| mecA | Normal | Codes for alternative penicillin-binding protein PBP2a | Methicillin, nafcillin, oxacillin, and cephalosporins | [46] |
| rpoB | Mutated | Codes for the β-subunit of RNA polymerase | Rifampicin and isoniazid in multidrug resistant Mycobacterium tuberculosis | [47,48,49] |
| ampR | Normal | Involved in β-lactamase transcription, a transcriptional activator of the lysR family | 3rd generation cephalosporinase | [50,51] |
| katG | Mutated | Codes for the catalase-peroxidase enzyme. | Isoniazid in M. tuberculosis when loss of function of the gene is seen | [52,53] |
| gyrA | Mutated | Codes for GyrA protein or DNA gyrase, a target of quinolones | Quinolones | [54,55] |
| inhA | Mutated | Codes for enoyl-ACP reductase of type II fatty acid synthase. which is crucial for the biosynthesis of mycolic acid (a component of the cell wall of Mycobacterium) | Isoniazid | [56,57] |
| hlyA | Normal | Codes for extracellular hyaluronate lyase Codes for α-hemolysin Shows enhanced virulence | -- | [58,59] |
| YMDD motif in reverse transcriptase | Mutated YMDD motif | Locus/motif present in RNA-dependent DNA polymerase. | Lamivudine in Hepatitis B virus | [60,61] |
| K13 gene | C580Y mutation | Codes for Kelch protein | Artemisinin in Plasmodium falciparum | [62] |
| NDM1 gene(blaNDM-1) | Normal and variants | Codes for New Delhi metallo-β-lactamase-1 (NDM-1) | Resistance to carbapeneme and β lactam antibiotics (except for azetreonam) | [45,63] |
| gliT gene | Normal | Codes for gliotoxin A virulence factor associated with invasive aspergillosis | -- | [64,65] |
The second type of target genes for MDR identification fall into the mutated category. The mutations in the genes of these species result in the expression of targets (receptors, proteins, or other biomolecules) in a form that is not recognizable by AM drugs. For example, rifampicin and isoniazid fail to act on MDR M. tuberculosis due to the presence of the mutated rpoB gene [47,48]. The unmutated rpoB codes for the β-subunit of RNA polymerase targeted by these two drugs [49]. However, the mutation in the rpoB gene brings about structural/conformational changes in the enzyme, which significantly lowers the binding affinity towards these drugs. An electrochemical sensor with LOD in the femtomolar range was designed by Haddaoui et al. to detect mutated rpoB genes in MDR-M. tuberculosis (LOD: 4 fM). This DNA sensor was based on a nanocomposite of magnetic polypyrrole/Fe3O4 tagged with naphthoquinone on PAMAM which could discriminate the wild type rpoB gene, in drop-sized samples (50 µL sample or 3 × 104 copies of DNA), from the mutated one [66]. Another gene mutation used as a target for biosensor development is the C580Y mutation in Plasmodium falciparum, which imparts resistance against choroquinone and artemisinin [62,67]. To this effect, Malpartida-Cardenas et al. developed a sensitive electrochemical sensor capable of detecting a single gene copy variation in less than 25 min. This was the first complementary metal oxide semiconductor-based lab-on-chip design biosensor for quantitative evaluation mutations in unknown P. falciparum samples [68].
Genes that impart enhanced virulence are also good targets for detecting MDR strains. For example, the hlyA gene, encoding α-hemolysin, has also been considered a target analyte, as its presence can be correlated to MDR in many pathogens [69]. An optical biosensor developed by Shi et al. provides visual detection and quantification for the hlyA gene in Listeria monocytogenes to a limit of 10 CFU/mL. The device combined loop-mediated isothermal amplification with propidium monoazide which enabled it to distinguish between viable and dead cells, and nanozyme which enabled visualization and imparted specificity and stability to the sensor [70]. Likewise, existing evidence suggests that the presence of the gliT gene correlates with invasive aspergillosis—a complication usually seen in immunocompromised patients [64,65]. A highly selective genotypic nano-biosensor was made using 1,6 hexane dithiol and chitosan stabilized AuNP for detecting the gliT gene by Bhatnagar et al., with a dynamic range of 1 × 10−14 − 1 × 10−2 M. This was the first biosensor ever developed for gliT detection sourced directly from fungal strains (LOD: 0.32 ± 0.01 × 10−14 M) [71].
There can be instances when a single analyte might not yield the required sensitivity or selectivity in detecting MDR strains. For such situations, biosensors based on multiplexing several analytes have the potential to enhance the sensitivity and selectivity limits. An example of such a biosensor developed by Dhar et. al. encompasses the simultaneous visual detection of point mutations in the rpoB, katG, and gyrA genes in MDR M. tuberculosis using split deoxyribozyme cascade probes [72]. Similarly, Bengtson et al. developed a multiplex biosensor to detect mutated rpoB, inhA, katG, and gyrA genes, and mutations in 23S rRNA, in MDR M. tuberculosis. This biosensor utilized a universal substrate significantly reducing the overall cost, had a high sensitivity compared to other devices utilizing molecular beacon probes, and could identify point mutations in both DNA and RNA [73].
Some biosensors analyze the whole DNA and/or RNA to determine the drug-resistant status of pathogens. An example of such a device is a duplex SPR-based biosensor designed to detect M. tuberculosis and pathogenic E. coli. (LOD: 5 nM). This sensor amalgamated the multiplexing ability and sensitivity of electrochemical and SPR-based sensing, with the robustness and selectivity of structure-switching nanomaterials [74]. Another electrochemical DNA-based biosensor capable of detecting point mutations to femtomolar limits in PCR samples of M. tuberculosis was developed by Bizid et al. This biosensor made use of a novel redox polymer, oligo-methoxy-phenyl-acetonitrile, deposited over a gold electrode, obtaining an LOD of 0.2 fM [75]. Tsao et al. used RNA instead of DNA to assess the MDR status of the influenza virus using peptide nucleic acid as both a sensor probe and a PCR clamp. This biosensor was sensitive to a concentration of 10 copies /mL of RNA from resistant viruses among 2 × 104 copies of RNA from non-resistant influenza viruses [76].
2.4.2. Phenotypic Biosensors for MDR Microbes
The underlying basis of phenotypic biosensors is the detection of (a) MDR microbes, (b) MDR-associated analytes, and (c) biofilms. MDR microbes exhibit pronounced changes in cell phenotype compared to the normal strains, which can be capitalized on to determine the drug resistance status of the cell. For instance, Zheng et al. developed a multiplex fluorescence carbon dot array-based biosensor that could discriminate six different bacterial strains with a high accuracy and classify them according to their gram status. The dots were functionalized with polymyxin, vancomycin, and boronic acid, allowing bacterial discrimination with 91.6% accuracy [77]. A fluorescence-based duplex biosensor was used to detect MRSA and Klebsiella pneumoniae carbapenemase 2-expressing Klebsiella pneumoniae to a limit of approximately 20 CFU/mL in real-time, using a dual platform made of a broad-spectrum fluorescent opsonin probe and specific aptamer coated magnetic beads [78]. An electrochemical sensor system developed by Gill et al. for detecting MRSA employed porous copper nanocomposites modified with vancomycin, which could be used for both detection and as a theranostic tool. This was the first time a copper nanocomposite was investigated for its potential use in the treatment of MRSA infection and it yielded satisfactory outcomes (LOD: 5 CFU/mL and MIC: 1.93 μg/mL) [79].
Another class of sensors detects the presence of analytes linked to virulence and the MDR status of the cell. Detection of the presence or activity of antibiotic-inactivating enzymes like ß-lactamases, AmpC, extended-spectrum ß-lactamases (ESBLs), carbapenemases, and cephalosporinases can also be used to confirm the drug resistance status [80,81,82]. One such example is the fluorescence-based sensor with the ability to identify ceftazidime-resistant bacteria that was developed by Thai et al. to detect various ESBL producers to a limit of 10 CFU/mL in 90 min irrespective of their genotype [83].
A significant difference in cell wall composition was observed in the susceptible and MDR variants of M. tuberculosis. An ultrastructural analysis of the cell wall of the two strains revealed that the resistant strains had a sturdier cell wall, with a higher triglyceride concentration [84]. Mannose-capped lipoarabinomannan is an amphipathic lipopolysaccharide found in all Mycobacterium species, with a critical role in its survival and pathogenesis [84]. A plasmonic fiber optic absorbance biosensor was designed to detect this particular molecule in bacterial samples to a femtogram/mL limit [85]. Endotoxins are liposaccharides associated with MDR and are present on the outer leaflet of most Gram-negative bacteria [86]. For example, L-Ara4N modified Lipid A component of gram-negative endotoxins has been known to impart polymyxin resistance [87]. Yeo et al. developed a gold electrode-based electrochemical biosensor to detect endotoxins in Escherichia coli to a limit of 0.0002 EU/mL [88]. A decreased expression of pyocyanin, a secondary metabolite and virulence factor, is seen in MDR P. aeruginosa [89,90]. An electrochemical biosensor has been developed to detect pyocyanin by Rashid et al. in matrices of varying complexities like PBS, saliva, and urine samples to micromolar concentrations, using a combination of AuNP and reduced graphene oxide [91].
The formation of biofilms is a prominent feature of various drug-resistant pathogens and chronic infections; however, their detection by conventional methods is non-trivial [92]. Specific biosensors have also been devised to detect biofilms. For instance, Kim et al. developed a surface acoustic wave-based sensor for detecting E. coli biofilms to picogram limits. The device demonstrated an amalgamation of bacterial biofilm sensing and treatment on a single chip [93]. Figure 4 gives a schematic overview of a variety of biosensors for MDR detection. The different features and specific design characteristics of many biosensors directed at detecting multidrug resistance in microbes have been extensively listed in Table 2.
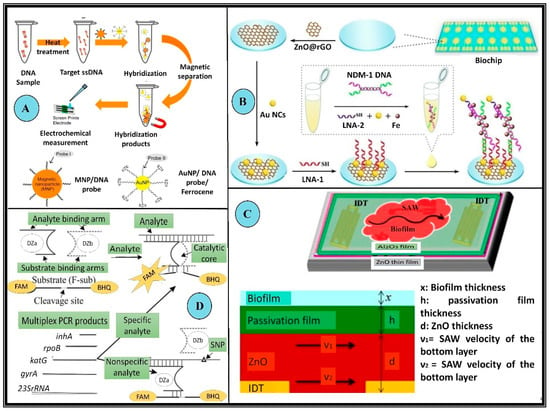
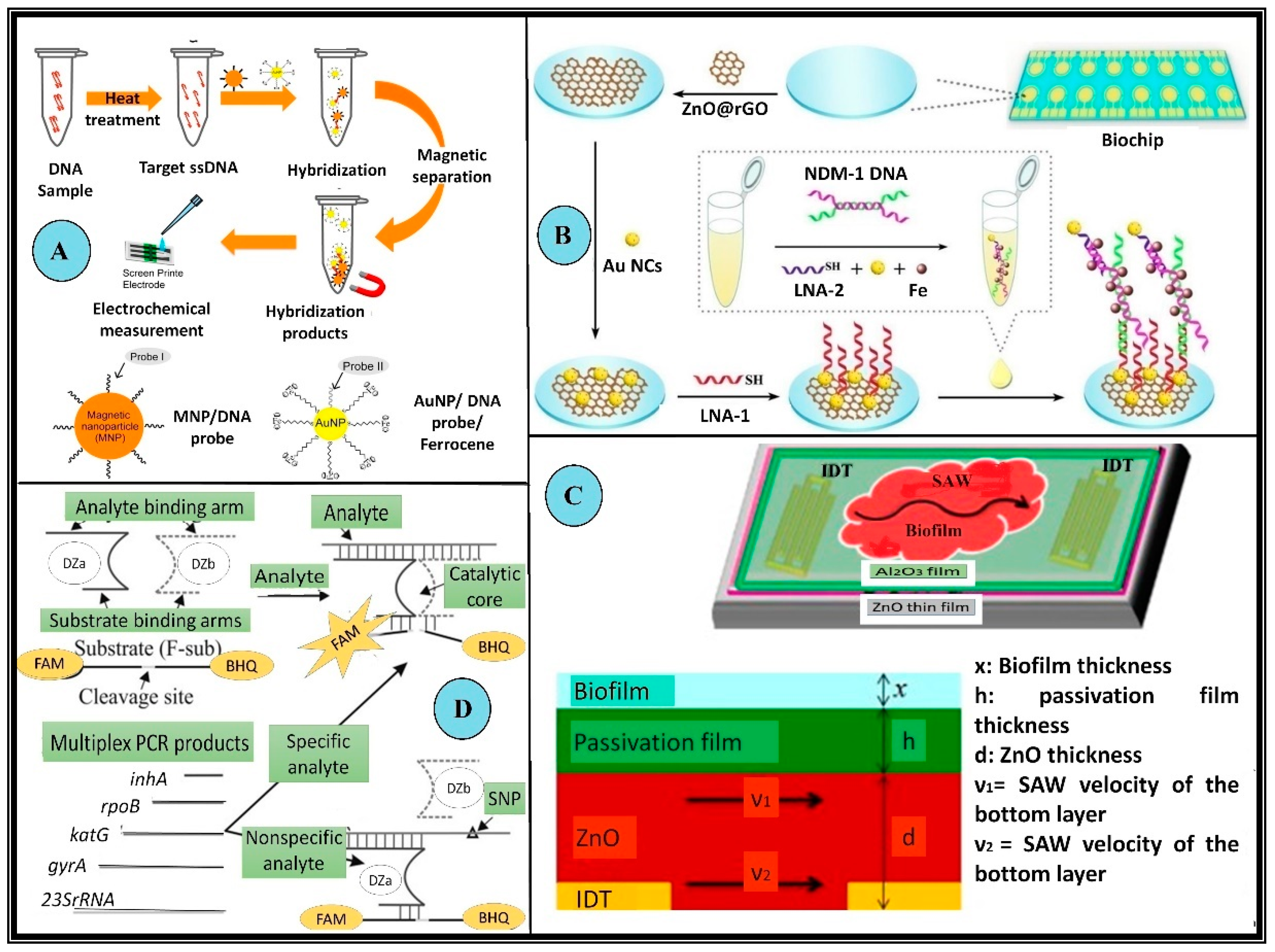
Figure 4.
Examples of biosensors employed in the detection of MDR microorganisms. (A) Schematic illustration of target DNA detection using the DNA sensing system with modified Au nanoparticles and magnetic nanoparticles to detect MRSA. These microbes possess the mecA gene which makes these variants resistant to the actions of methicillin, nafcillin, oxacillin, and cephalosporins. In this setup first, the ssDNA sample is prepared using heat treatment, which is allowed to hybridize with modified nanoparticles (Au nanoparticles and magnetic). The hybridized products are then collected via magnetic separation followed by chronoamperometric detection (B) Schematic representation of a modified 2 × 8 array biochip detecting NDM 1 resistant bacteria. These bacteria produce a special class of β-lactamases which impart enhanced resistance to the microorganism. The biochip was made using Au NC@LNA-1/NDM-1DNA/LNA-2 Au NC in a “sandwich-like” model. (C) Schematic and a cross-sectional of the inverted passivated SAW sensor for the detection of biofilms. The piezoelectric surface is decorated with an interdigitated transducer. The relative thickness of all the layers and the velocity of waves along the piezoelectric surface is crucial to the design. (D) Schematic diagram of the binary deoxyribozyme (BiDz) sensors for multiplex AMR gene detection of XDR M. tuberculosis. DNA strands Dza and Dzb reform a deoxyribozyme catalytic core by hybridizing to the adjacent regions of the analyte F-sub, a fluorescent substrate containing a 3′ black hole quencher (BHQ) and 5′ fluorescein (FAM) label. When in proximity, these emit low fluorescence signals. When the substrate is cleaved, the fluorophore is separated from the quencher allowing for an increase in fluorescence. This BiDz sensor, when paired with the specific analyte, forms the catalytic core, which cleaves F-sub and produces a fluorescent signal. Alternatively, in the presence of a mismatched (nonspecific) analyte, the catalytic core of the sensor cannot be formed and the fluorescence of F-sub remains quenched (All reproduced with permission).

Table 2.
Biosensors used to detect MDR in microbial systems.
3. Multidrug Resistance in Malignant Systems and Biosensors
3.1. Foundation and Emergence of MDR in Cancer
Though MDR in cancers is a cause for concern, the underlying principles of its development are poorly understood. Compared with MDR in microbial cells, MDR in cancers is more enigmatic owing to its highly multifactorial nature [132]. Making things worse is the fact that not all facets contributing to MDR development in cancers are known completely [133]. The significant patient-to-patient heterogeneity in characteristics of MDR tumors makes its diagnosis a difficult task.
Resistance may be innate (present before the subject is exposed to the drug) or acquired (during therapy) [133]. Innate chemoresistance results from genetic mutations, activation of pathways against xenobiotics, and tumor heterogeneity owing to pre-existing insensitive cell subpopulations [134]. Acquired chemoresistance, on the other hand, results from activating protooncogenes, altered expression of drug targets or mutations in them, or alterations in the tumor microenvironment (TME) [134]. While these factors invariably are linked with the emergence of MDR tumors, there are specific basic mechanisms to resist the actions of anticancer drugs. These include (a) preventing the drug from reaching its target, (b) effectively decreasing the drug concentration, (c) deploying compensatory mechanisms promoting survival, or (d) promoting dormancy in these cells [7,135].
3.2. Biochemical Basis of MDR in Cancer
While cancer is a widely studied disease, a complete understanding of its pathophysiology is still lacking. Cancer, when coupled with MDR, becomes even more challenging to understand. This complexity results from a cumulative effect of several mechanisms which are in constant crosstalk with one another [18]. The variability in tumor characteristics exhibiting MDR can be attributed to the differential interaction of these mechanisms across patient groups. A detailed account of all the contributing factors is beyond the scope of this review. However, the following references are excellent resources for gaining a comprehensive picture of the governing factors [2,10,30,31,36,132,134,136,137]. Figure 5 aims at summing up various factors responsible for MDR development in cancer cells.
The primary causative factor responsible for MDR in cancer cells is the differential functioning of the drug efflux pumps, belonging to the superfamily of ABC proteins. These ubiquitous transmembrane proteins catalyze the ATP-dependent transport of drugs [138]. The P-glycoprotein (Pgp) was the first efflux protein associated with MDR [10,85,139]. Later MRP1, BC RP, ABCG2/MXR, and others were recognized for their contribution to the development of MDR [84]. For a simplified overview, factors leading to MDR in neoplasm can be divided into host-associated, tumor-associated, and host tumor-interaction-associated factors [132].
3.2.1. Host-Associated Factors
These factors include genetic variations seen in the host, the differential expression of MDR-associated proteins and pumps, variation in the metabolism of drugs, the physiological state of the patient, the different drugs prescribed in due course, its pharmacological profile, among others, which heavily contribute to intrinsic drug resistance among cancer patients [9,76,78,82,132,134,137]. Genetic variations, ranging from a single point or nucleotide variation to chromosomal alteration, accounts for 20–95% of the variability observed [132]. Aneuploidy is a common phenomenon observed in cancer cells. It is hypothesized to be a prominent reason for MDR as it may lead to the deletion of essential genes which had a role in drug response [137]. Gene mutation may lead to target modification or surpassing, enhanced expression of enzymes like cytochrome P450 and glutathione-S-transferase for exogenous drug/xenobiotic metabolism [140]. Polymorphism due to ethnicity has also been acknowledged in MDR-associated genes [141]. The physiological status of the host, i.e., age, nutrition, and the presence of comorbidities, also plays a critical role in the development of MDR [140]. The drug regimen a patient is prescribed also plays a decisive role in the development of MDR because of the possibility of drug-drug interactions, as patients are generally given several drugs and supplements concomitantly [140].
3.2.2. Tumor-Associated Factors
These alterations made by tumor cells are conducive to the development of MDR. These include enhanced expression of drug efflux pumps and extracellular vesicles (EV), evasion of cell death, dysregulation of DNA damage and repair machinery, epigenetic changes, secretion of growth factors, metabolic alterations, and promotion of heterogeneity in structure [134,139,140]. Drug efflux pumps effectively decrease the intracellular drug concentration, preventing its deleterious effects [132]. EVs are specialized bodies, which serve as notable vehicles of MDR dissemination as they help in extracellular drug trafficking and sequestration [142,143]. The evasion of apoptosis by manipulating pro and anti-apoptotic genes, and avoiding anoikis and autophagy are important strategies for MDR promotion [7,9,144]. An increased DNA repair rate and secretion of growth factors such as IL-6 (a potent promoter of chronic inflammation) also accelerate MDR development [137].
3.2.3. Host-Tumor-Interaction Associated Factor
A tumor cell usually promotes the maintenance of the malignant phenotype due to intricate interactions between the host and the tumor. These interactions between the host and tumor form an ecological niche called the TME. The TME is formed by physiological and cellular (malignant and non-malignant type) constituents enabling the neoplasm to thrive and progress [145,146]. The appropriate intra-tumoral heterogeneity, and ample genetic and epigenetic diversity, allow the therapy-induced expansion of pre-existing drug-resistant cancer cell clones in the space, promoting chemoresistance [9,132,140,145]. Cancer stem cells play a crucial role in MDR development, efficient seeding of cancer upon metastasis, and enhancing the plasticity of the neoplasm [136,147,148]. This environment is notably acidic (pH 6.3–6.9); hypoxic; perennially inflamed; and filled with poorly formed vasculature, reactive oxygen species, growth factors, and signaling molecules (which promote inflammation, immunosuppression, and tumorigenesis); and has a surfeit of lactate [149,150]. These conditions produce a selective pressure which leads to the propagation of resistant cells as well as providing a haven from antineoplastic agents.
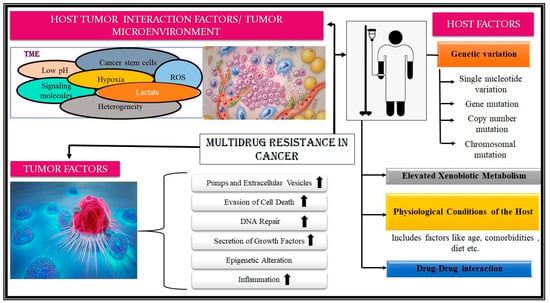
Figure 5.
Factors contributing to the development of multidrug resistance. The factors leading to the development of MDR in cancers can be grouped into ‘Host associated’, ‘Tumor associated’, and ‘Host-Tumor interaction associated’ factors. Host-associated factors are factors associated specifically with a particular patient/subject. These include genetic variation, ranging from point to chromosomal mutation; enhanced xenobiotic metabolism; drug-drug interaction; and physiological conditions. Tumor factors are the features of the tumor that facilitate the development of multidrug resistance in cancers. These include enhanced cell growth, inflammation, DNA repair, evasion of cell death, epigenetic alterations, etc. The last contributing set of factors, i.e., Host-tumor interaction-associated factors, are where the confluence of various factors produces a niche to promote MDR development. Factors like perennial inflammation of the TME, surfeit of lactate, acidic pH, lack of oxygen, etc., in the TME are some of them. (The figure has been designed taking reference from the following resources: [2,10,29,30,33,76,78,80,81,82]).
Figure 5.
Factors contributing to the development of multidrug resistance. The factors leading to the development of MDR in cancers can be grouped into ‘Host associated’, ‘Tumor associated’, and ‘Host-Tumor interaction associated’ factors. Host-associated factors are factors associated specifically with a particular patient/subject. These include genetic variation, ranging from point to chromosomal mutation; enhanced xenobiotic metabolism; drug-drug interaction; and physiological conditions. Tumor factors are the features of the tumor that facilitate the development of multidrug resistance in cancers. These include enhanced cell growth, inflammation, DNA repair, evasion of cell death, epigenetic alterations, etc. The last contributing set of factors, i.e., Host-tumor interaction-associated factors, are where the confluence of various factors produces a niche to promote MDR development. Factors like perennial inflammation of the TME, surfeit of lactate, acidic pH, lack of oxygen, etc., in the TME are some of them. (The figure has been designed taking reference from the following resources: [2,10,29,30,33,76,78,80,81,82]).
3.3. Use of Biosensors in Detecting MDR in Neoplasms
For this review, the MDR-detecting biosensors for cancerous cells have been divided into genotypic, phenotypic, and drug pharmacokinetics-based biosensors.
3.3.1. Genotypic Biosensor for MDR Cancer
Biosensors targeting genotypic traits detect any discrepancy in the cell’s genetic makeup that may result in the development of resistance. Peng et al. designed an electrochemical biosensor for detecting MDR1 using an AuNP/toluidine blue-graphene oxide-modified electrode (LOD: 3.12 fM) [151]. Further, Chen et al. made a label-free and enzyme-free biosensor with improved sensitivity to detect the same gene in clinical leukemia samples using nitrogen-doped graphene nanosheets functionalized over AuNP (LOD: 2.95 pM) [152] Graphene and related materials exhibit suitable optical, electronic, and electrochemical characteristics, making them an excellent choice for versatile fabrication [153]. Xiang et.al. developed a multiplexed fluorescence-based biosensor that could simultaneously detect seventeen different drug-sensitive and drug-resistant mutations in cell-free circulating DNA in real-time to a detection limit of 1–4 copies [154]. This biosensor identified the T790M mutation in the epidermal growth factor receptor (EGFR) gene, which makes it resistant to drugs like gefitinib and erlotinib [154,155].
Another highly sought-after method of MDR analysis is detecting and monitoring micro-RNA (miR) levels. miRs are short, non-coding, and highly conserved RNA moieties that function in post-transcriptional gene regulation via translational repression and mRNA degradation [156]. miRs may be expressed constitutively or produced only under specific circumstances differentially [157]. Fluctuations in miR levels in the case of MDR variants are described in Table 3. Hence these molecules are ideally suited to detect MDR in malignant cells. Cai et al. developed a microfluidic-based laser-induced fluorescence sensor to determine miR-21 and miR-31 levels in HeLa and A549 cell lysates in real-time, with an LOD of 0.20 fM and 0.50 fM for miR-21 and miR-31, respectively [158]. Yaman et al. developed disposable AuNP peptide nanotubes-based biosensors for detecting miR 410 in serum and prostate cancer cell lines. In minutes, these impedimetric biosensors could detect the miR to the femtomolar limit [159]. It was observed that major feats have been achieved in the development of fluorescence-based biosensors in the detection of miRs This can be attributed to such biosensors exhibiting fast response, multiplexed ability, having simple instrumentation and operation, high selectivities and specificities, a non-destructive nature, and they can be easily integrated with microfluidic platforms [21,160].

Table 3.
miRNA And Their Expression in the Case of MDR.
3.3.2. Phenotypic Biosensor for MDR Cancer
Perceptible changes in the cells accompany the MDR of the neoplasm. These changes may include some molecules being over or under-produced compared to usual, some significant changes in the TME that may be aiding in the development of resistance, an overall change in the cellular profile, and the production of anomalous molecules. Sometimes even the behavior or the overall shape becomes strikingly distinguishable, which aids in the detection of MDR cell lines. Specific biosensors are designed to detect and monitor these phenotypic changes to know about the status of the cell concerning drug resistance.
A biosensor was developed by Hu et al., which was based on the difference in electrochemical behavior as exhibited by drug-sensitive versus drug-resistant leukemia K562 cell lines to a limit of 500 cells [190]. Since the resistant and non-resistant cells have significant differences in their PM profiles, it serves as an avenue for discernment. Tao et al. developed sensing systems on Au-nanoclusters that could efficiently discern drug-resistant, cancerous, metastatic, and healthy human breast cancer cells to a limit of 200 cells, based on the unique chemistry and surface charge densities of each type of cell [191].
One of the most popular candidates for the detection of MDR is the Pgp. For instance, Chandra et al. designed an amperometric biosensor using monoclonal antibodies for Pgp as a target analyte. This sensor could detect MDR lines to an LOD of 2372 cells/mL in MDRCC cell lines [192]. Gulati et al. developed an optical biosensor with higher sensitivity, capable of detecting Pgp at concentrations as low as 27 cells/mL [193]. Another drug efflux pump that can be used to assess the MDR status is the MRP2/ABCC2 protein. Li et al. developed a microfluidics-based biosensor that was sensitive enough to detect this protein in a single cell based on hydrodynamic theory [194].
Specific proteins are overexpressed in the case of MDR cancer cell lines, to promote tumor survival (Figure 5). For example, the enhanced expression of β1 integrin, a membrane receptor, helps anchorage to the extracellular matrix and plays a role in anoikis correlated to increased resistance [195,196,197]. Jiang et al. developed an electrochemical immunosensor using a mouse anti-human monoclonal antibody functionalized over a glassy carbon electrode to detect β1 integrin to a limit of 3.5 × 103 cells/ mL [198]. Drug-resistant neoplastic cells are also known to exhibit upregulation of complement regulatory proteins. These proteins are present on the membranes of normal cells, serving as a survival strategy to prevent complement system activation. However, when expressed at abnormally high amounts in tumor cells, they lead to its evasion of attack by the complement system [199]. Choudhary et al. developed an immunosensor for early non-invasive detection of oral cancer from clinical saliva samples by detecting CD59, a complement restriction factor [200]. Brain-derived neurotrophic factor (BDNF) is a class of neurotrophins associated with decreased sensitivity to cisplatin, etoposide, and vinblastine in numerous cancers [201]. Akhtar et al. developed an ultrasensitive biosensor to detect the levels of BDNF in serum samples. This sensor was further used to study and monitor the effects of various activators of BDNF producers, like nicotine and alcohol, in the model cell lines SH-SY5Y and PC-12 [202].
Enzymes are common biomarkers that are used for the detection of MDR in cancer cells. For instance, a fluorescence-based biosensor was developed by Wang et al. to measure the real-time intracellular telomerase activity in A549, HepG2, and MCF7 cell lines [203]. Increased telomerase activity, or its upregulated expression, is a feature of cancer cells resistant to drugs like cisplatin and 5-fluorouracil, as it regulates the reactive oxygen species and imparts resistance against drugs targeting ABC transporters, DNA damage, and apoptosis [204]. Another repertoire closely related to the MDR profile is glutathione and associated enzymes. Overexpression of glutathione S-transferase has been noted in MDR cell lines. Glutathione is required as a co-factor for efflux protein ABCC1 activity [137,205,206,207,208]. A fluorescence-based biosensor was developed by Yang et al. for glutathione monitoring in chemo-resistant cell lines such as HeLa and HepG2 to a limit of 87 nM. This was the first biosensor developed for imaging glutathione levels using a two-photon nanoscale metal-organic framework [209]. DNA methyltransferases cause methylation of regions of DNA. Hypermethylation of gene segments, such as the promoter of glutathione S-transferase, is linked to higher levels of poor prognosis and MDR development [205,210]. A fluorescence-based sensor, employing a novel dumbbell-shaped DNA template for copper nanoparticles, was developed by Yin et al. to detect this enzyme to a limit of 0.16968 mU/μL [211].
The conditions in the TME promote MDR in tumors. Hypoxia and a highly acidic pH are two significant conditions prevalent in the TME. Nitro reductase is an endogenous enzyme expressed in highly hypoxic conditions [212]. Wang et al. developed a fluorescent probe for in vivo detection of the enzyme in the TME, with a dynamic range of 15–300 ng/mL and an LOD of 0.27 ng/mL [213]. To monitor the pH of the TME, a fluorescence-based biosensor was devised by Ge et al., responsive in the range of 4.2–6.4 (pKa = 5.18), capable of giving a readout in approximately 1 min [214].
3.3.3. Drug Pharmacokinetics-Based Biosensors for MDR Cancer
An indirect approach to detect MDR is by monitoring the pharmacokinetic parameters of the administered drug. The core principle of this method is based on the most common mechanisms a tumor resorts to for avoiding the adverse effects of a chemotherapeutic agent, which is the extrusion of drugs out of the cell. Monitoring the levels of the drugs in clinical samples may, therefore, serve as an indirect indicator of the MDR status of the cell. One such device was designed by Zhao et al. to detect methotrexate in serum and clinical samples. This SPR-based biosensor used folic acid functionalized AuNP and was sensitive to methotrexate to a level of 155 nM, yielding results in under 60 s [215]. A novel SER sensor design using cysteine-AuNP conjugate was developed to assess the levels of exosomes by Hunter et al. [216]. Exosomes are a subset of EVs used for transferring bioactive molecules between cancers and various destinations within and beyond the TME, helping in the development of increased chemoresistance by methods like mediating drug efflux, neutralization of antitumor antibodies, and carrying resistance imparting miRs to drug-sensitive cells [143,217,218]. This biosensor was able to detect exosomal cis-platin and exosomes to a limit of 0.17 µg/mL and 65 nM, respectively, with an accuracy of >90% in diagnosis [143,216,217,218].
Some of the examples above are illustrated in Figure 6, while Table 4 comprehensively discusses multiple biosensors which aim at detecting MDR in malignant systems.
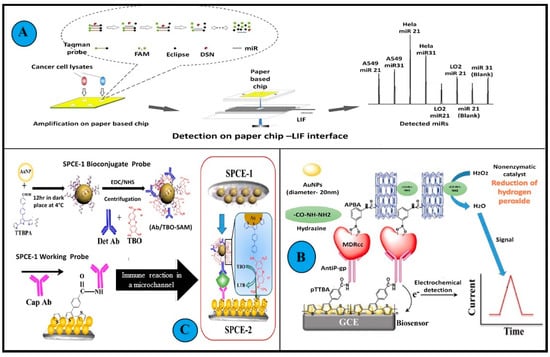
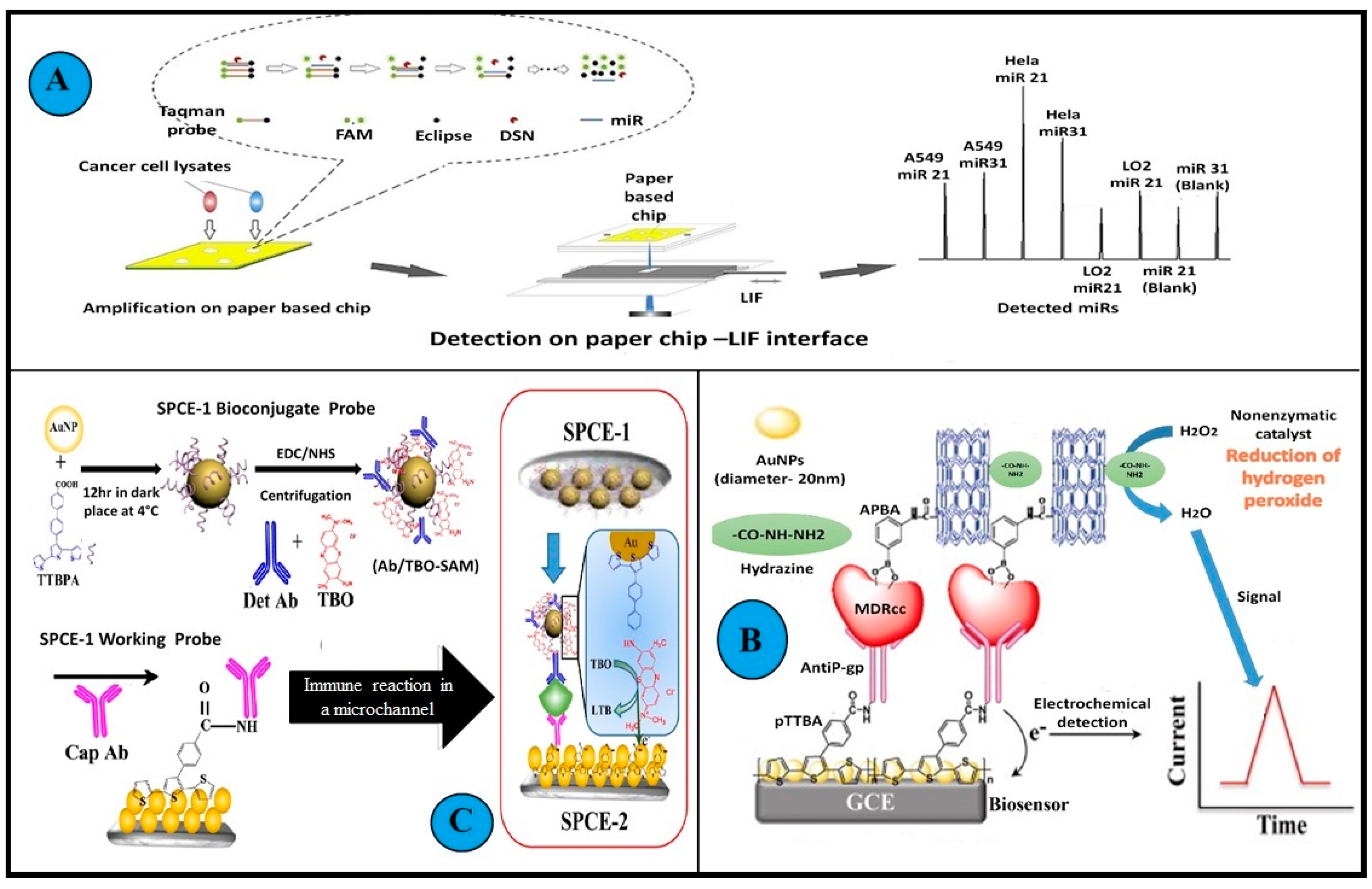
Figure 6.
Examples of biosensors used in the detection of MDR cancer cell lines. (A): A graphical abstract of amplification and subsequent detection of miRNA in cancer cells using a microfluidics paper-based fluorescent sensor. A confocal LIF detector was used for the determination of miRNAs on microfluidics paper. An interface was designed and applied to obtain a stable fluorescence signal. DSN amplification on a double-layer microfluidics paper was performed to improve the sensitivity of the system min. (B): Illustration depicts the working of immunosensors detecting BDNF. Two screen-printed carbon electrodes (SPEC) with long microfluidics channels were assembled. This sandwich immunosensing approach helped in detection by generating signals using redox indicator TBO. First an Ab-Ag complex was formed on the working probe, succeeded by the Ag and Lyophilized BDNF antibody Det Ab reacting with the biconjugate probe. (C): Schematic representation of the amperometric immunosensor fabrication and detection principle of drug-resistant cancer cells in biological matrices. The unit composition included a modified glassy carbon electrode, gold nanoparticles, polymeric TTBA (2,2′:5′, 2″-terthiophene-3′(p-benzoic acid) (TTBA)), and anti Pgp monoclonal antibodies. The MDR cancer cells are captured in between this engineered layer and APBA (amino phenyl boronic acid), multi-walled carbon nanotubes, and hydrazine conjugate. (All reproduced with permission).

Table 4.
Biosensors used in the detection of multidrug-resistant tumors.
4. Discussion and Future Perspectives
In this review, we have discussed the details of the foundation and emergence of MDR and the mechanisms by which malignant and microbial systems achieve MDR. Further, various aspects of the pathophysiology that serve as unique signatures of MDR are suitably utilized in designing different biosensors. In this section we shall discuss some pressing questions:
4.1. What Kind of Threat Does MDR Pose?
With the ever-increasing use of AM antibiotics in farming, healthcare, excessive prescription of antibiotics by clinicians, availability of antibiotics over the counter in many parts of the world, and negligence on the part of the patients coupled with globalization and population explosion; drug resistance can be perceived as the next emerging global health concern [4,233,234]. According to the latest projections, drug resistance (microbial) related deaths are projected to reach 10 million by the year 2050, amounting to a global economic burden of a staggering 100 billion USD. In 2021, the bacterial multidrug resistance market is valued at 10.359 billion USD, which is expected to reach 16.02 billion USD by 2029 [235]. For instance, the statistics of MDR TB may be considered as an example. Presently, 41% of newly reported cases of the disease are of the multidrug-resistant type [236]. In 2020, cancer accounted for nearly 10 million deaths [237]. Nevertheless, the WHO states that 30–50% of cancer cases can be managed by risk factor avoidance, early detection, and treatment strategies. Selective large-scale screening and early diagnosis are two pillars of detection [238]. Hence it becomes crucial to search for methods to tackle MDR, as cases associated with drug resistance increase exponentially with each passing year.
4.2. What Is the Current Scenario of Diagnosis?
As discussed previously, current approaches to MDR detection are primarily achieved using conventional techniques. Though these techniques offer credible results, they suffer from long turnaround times, rendering mass screenings a tedious task. Furthermore, it is difficult to scale conventional methods of MDR detection owing to the requirement of a skilled workforce for sample preparation, processing, and the interpretation of results. In addition to this, since the clinical manifestation of cancers often occur at later stages of progression, the conventional approaches fail to provide an early diagnosis of the disease as they rely on the phenotypic assessment.
4.3. How can Biosensors Be Used to Address These Problems?
Biosensors can potentially serve as a sturdy ancillary panacea to this problem. Quick turnaround times with high sensitivities, specificities, linearities, and robustness, along with ease of operation make them suitable to be used as POC devices for MDR detection [33,239,240]. These can be used in biomarker detection in a variety of sample matrices, tumor diagnosis and characterization, and medical imaging. An ideal biosensor for MDR detection is expected to differentiate resistant cell types vs. normal cells, and to give insights into the susceptibility of the cells and host immune response [33,160,240]. As is evident, most of the biosensors discussed herein are based on electrochemical or optical detection. Such biosensors offer excellent stability, reliability, robustness, compactness, and could be used for developing multiplex systems by exploiting surface chemistry, nanomaterials, and different transducing mechanisms (voltammetric, amperometric, capacitance-based, SPR, SER, fluorescence, etc.) [23]. Further, microfluidics-based sensors render the analysis of DNA damage, cytotoxicity, and detection of pathogens easier, as they can easily assimilate and define the biochemical microenvironment seamlessly both temporally and spatially [160]. These are major technological leaps that have made the development of lab-on-chip models possible [160,235,241]
The ease of reliable data generation using such sensors can add a new dimension to integrating data science into the study of MDR. Recent studies have demonstrated the use of machine learning models and algorithms to predict MDR trends in M. tuberculosis and urinary tract infections [242,243]. Some of the sensors discussed in this review have also been tested for their theragnostic abilities, hence the scope of biosensors is not limited to only diagnosis. It has been well established that a significant cornerstone for MDR prevalence is linked to the indiscriminate use of AM and its accumulation in the environment. To curtail the dissemination of MDR, many sensor systems have been designed to carefully monitor the levels of drugs and antibiotics in clinical and environmental samples. The urgency to resolve this problem has led to the development of some innovative systems that not only detect the cells but also monitor their growth.
4.4. Possible Roadblocks and Shortcomings
However, for the complete realization of biosensors’ potential, or establishing them as a valid alternative to conventional testing, several bottlenecks need to be overcome. On the designing front, work needs to be done concerning minimizing the matrix effect and system integration [239]. A disadvantage of many biosensors is the requirement of processing samples, which makes them difficult to operate [239]. A possible solution to overcome this issue would be to develop biosensors that could yield data non-invasively using saliva, sweat, and urine samples. Another focus area that needs work is the miniaturization and portability of devices and making them more user-friendly. For successful translation from a laboratory prototype to an end-user commercial product, a high capital investment is required in the research and development of such devices and for the subsequent rounds of evaluation and approval by regulators. This leads to an extremely slow rate of commercialization of biosensors [239].
5. Conclusions
MDR is a pressing scourge on the healthcare system, and the need to develop efficient solutions is imminent. While significant technological advancements have taken place in terms of the development of biosensors that can help MDR detection in complex matrices and a plethora of clinical indications, we are still far away from replacing conventional detection strategies with a biosensors-based approach. Nevertheless, with the rapid advancements in our understanding of the pathobiology of various diseases, many new biomarkers have been discovered that have huge potentials for clinical translations. This, coupled with novel biosensor systems, significantly enhances the prospect of the early diagnosis and management of MDR cancers.
Author Contributions
Conceptualization, N.G.J., D.S.D., S.K.S., S.J.M., N.P.S., and P.C.; resources, N.G.J., D.S.D., and S.K.S.; writing—original draft preparation, N.G.J., D.S.D., S.K.S., and P.C.; writing—review and editing, N.G.J., D.S.D., S.K.S., S.J.M., N.P.S., and P.C.; supervision, N.P.S., and P.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Pranjal Chandra and Sumit Singh acknowledge the Director of the Indian Institute of Technology (BHU) Varanasi, India, for encouragement and for providing the necessary facility for the completion of this work. Author Niharika G Jha acknowledges her fellowship provided by IIT (BHU), and Author Daphika S Dkhar (09/1217(13016)/2021-EMR-I) acknowledges CSIR for providing financial support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Centers for Disease Control and Prevention (2019). AR Threats Report. Available online: https://www.cdc.gov/drugresistance/biggest-threats.html (accessed on 24 July 2022).
- Barie, P.S. Multidrug-Resistant Organisms and Antibiotic Management. Surg. Clin. N. Am. 2012, 92, 345–391. [Google Scholar] [CrossRef]
- Menkem, E.; Lemalue, B.; Shinwin, S.; Tamunjoh, A.; Fabrice, F. Antibiotic Residues in Food Animals: Public Health Concern. Acta Ecol. Sin. 2019, 39, 411–415. [Google Scholar] [CrossRef]
- Morehead, M.S.; Scarbrough, C. Emergence of Global Antibiotic Resistance. Prim. Care—Clin. Off. Pract. 2018, 45, 467–484. [Google Scholar] [CrossRef]
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. In Current Opinion in Microbiology; Elsevier Ltd.: Amsterdam, The Netherlands, 2019; Volume 51, pp. 72–80. [Google Scholar] [CrossRef]
- Wu, D.; Ding, Y.; Yao, K.; Gao, W.; Wang, Y. Antimicrobial Resistance Analysis of Clinical Escherichia Coli Isolates in Neonatal Ward. Front. Pediatr. 2021, 9, 670470. [Google Scholar] [CrossRef]
- Li, Y.J.; Lei, Y.H.; Yao, N.; Wang, C.R.; Hu, N.; Ye, W.C.; Zhang, D.M.; Chen, Z.S. Autophagy and multidrug resistance in cancer. Chin. J. Cancer 2017, 36, 52. [Google Scholar] [CrossRef]
- Ye, Q.; Liu, K.; Shen, Q.; Li, Q.; Hao, J.; Han, F.; Jiang, R.W. Reversal of multidrug resistance in cancer by multi-functional flavonoids. Front. Oncol. 2019, 9, 487. [Google Scholar] [CrossRef]
- Baguley, B.C. Chapter 1 Multidrug Resistance in Cancer; Springer International Publishing: Berlin/Heidelberg, Germany, 2010; Volume 596. [Google Scholar] [CrossRef]
- Kunjachan, S.; Rychlik, B.; Storm, G.; Kiessling, F.; Lammers, T. Multidrug resistance: Physiological principles and nanomedical solutions. In Advanced Drug Delivery Reviews; Elsevier: Amsterdam, The Netherlands, 2013; Volume 65, pp. 1852–1865. [Google Scholar] [CrossRef]
- Mahato, K.; Maurya, P.K.; Chandra, P. Fundamentals and commercial aspects of nanobiosensors in point-of-care clinical diagnostics. 3 Biotech. 2018, 8, 149. [Google Scholar] [CrossRef]
- Cui, F.; Ye, Y.; Ping, J.; Sun, X. Biosensors and Bioelectronics Carbon dots: Current advances in pathogenic bacteria monitoring and prospect applications. Biosens. Bioelectron. 2020, 156, 112085. [Google Scholar] [CrossRef]
- Gupta, S.; Kakkar, V. Recent technological advancements in tuberculosis diagnostics—A review. Biosens. Bioelectron. 2018, 115, 14–29. [Google Scholar] [CrossRef]
- Alafeef, M.; Moitra, P.; Pan, D. Nano-enabled sensing approaches for pathogenic bacterial detection. Biosens. Bioelectron. 2020, 165, 112276. [Google Scholar] [CrossRef]
- Jayanthi, V.S.P.K.S.A.; Das, A.B.; Saxena, U. Recent advances in biosensor development for the detection of cancer biomarkers. Biosens. Bioelectron. 2017, 91, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Kaur, B.; Kumar, S.; Kaushik, B.K. Recent advancements in optical biosensors for cancer detection. Biosens. Bioelectron. 2022, 197, 113805. [Google Scholar] [CrossRef]
- Kumar, A.; Purohit, B.; Maurya, P.K.; Pandey, L.M.; Chandra, P. Engineered nanomaterial assisted signal-amplification strategies for enhancing analytical performance of electrochemical biosensors. Electroanalysis 2019, 31, 1615–1629. [Google Scholar] [CrossRef]
- Andrei, L.; Kasas, S.; Ochoa Garrido, I.; Stanković, T.; Suárez Korsnes, M.; Vaclavikova, R.; Assaraf, Y.G.; Pešić, M. Advanced Technological Tools to Study Multidrug Resistance in Cancer. Drug Resist. Updat. 2020, 48, 100658. [Google Scholar] [CrossRef]
- Zhu, Y.; Chandra, P.; Song, K.M.; Ban, C.; Shim, Y.B. Label-Free Detection of Kanamycin Based on the Aptamer-Functionalized Conducting Polymer/Gold Nanocomposite. Biosens. Bioelectron. 2012, 36, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Chandra, P.; Noh, H.B.; Shim, Y.B. Cancer Cell Detection Based on the Interaction between an Anticancer Drug and Cell Membrane Components. Chem. Commun. 2013, 49, 1900–1902. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Majdinasab, M.; Khan, R.; Li, Z.; Hayat, A.; Marty, J.L. Nanomaterials in fluorescence-based biosensors: Defining key roles. Nano-Struct. Nano-Objects 2021, 27, 100774. [Google Scholar] [CrossRef]
- Mandal, R.; Baranwal, A.; Srivastava, A.; Chandra, P. Evolving trends in bio/chemical sensor fabrication incorporating bimetallic nanoparticles. In Biosensors and Bioelectronics; Elsevier: Amsterdam, The Netherlands, 2018; Volume 117, pp. 546–561. [Google Scholar] [CrossRef]
- Purohit, B.; Vernekar, P.R.; Shetti, N.P.; Chandra, P. Biosensor Nanoengineering: Design, Operation, and Implementation for Biomolecular Analysis. Sens. Int. 2020, 1, 100040. [Google Scholar] [CrossRef]
- Gupta, N.; Renugopalakrishnan, V.; Liepmann, D.; Paulmurugan, R.; Malhotra, B.D. Cell-Based Biosensors: Recent Trends, Challenges and Future Perspectives. Biosens. Bioelectron. 2019, 141, 111435. [Google Scholar] [CrossRef]
- Mahato, K.; Kumar, A.; Maurya, P.K.; Chandra, P. Shifting Paradigm of Cancer Diagnoses in Clinically Relevant Samples Based on Miniaturized Electrochemical Nanobiosensors and Microfluidic Devices. Biosens. Bioelectron. 2018, 100, 411–428. [Google Scholar] [CrossRef]
- Tech, J.A.B.; Mahato, K.; Prasad, A.; Maurya, P.K.; Chandra, P. Nanobiosensors: Next Generation Point-of-Care Biomedical Devices for Personalized Diagnosis. J. Anal. Bioanal. Tech. 2016, 7, e125. [Google Scholar] [CrossRef]
- Ullah, H.; Ali, S. [Online] Classification of antibacterial agents and their functions. Intech 1989, 32, 137–144. Available online: https://www.intechopen.com/books/advanced-biometric-technologies/liveness-detection-in-biometrics (accessed on 8 January 2022).
- Basak, S.; Singh, P.; Rajurkar, M. Multidrug Resistant and Extensively Drug Resistant Bacteria: A Study. J. Pathog. 2016, 2016, 4065603. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Nikaido, H. Multidrug Resistance in Bacteria. Annu. Rev. Biochem. 2009, 78, 119–146. [Google Scholar] [CrossRef]
- Ray, S.M.S.; Das, S. Molecular Mechanism of Drug Resistance. In Drug Resistance in Bacteria. Fungi, Malaria, and Cancer; Springer: Cham, Switzerland, 2017; pp. 47–100. [Google Scholar] [CrossRef]
- Jacopin, E.; Lehtinen, S.; Débarre, F.; Blanquart, F. Factors Favouring the Evolution of Multidrug Resistance in Bacteria. J. R Soc. Interface 2020, 17, 20200105. [Google Scholar] [CrossRef]
- Conteduca, D.; Brunetti, G.; Dell’Olio, F.; Armenise, M.N.; Krauss, T.F.; Ciminelli, C. Monitoring of Individual Bacteria Using Electro-Photonic Traps. Biomed. Opt. Express 2019, 10, 3463. [Google Scholar] [CrossRef]
- Chang, H.-H.; Cohen, T.; Grad, Y.H.; Hanage, W.P.; O’Brien, T.F.; Lipsitch, M. Origin and Proliferation of Multiple-Drug Resistance in Bacterial Pathogens. Microbiol. Mol. Biol. Rev. 2015, 79, 101–116. [Google Scholar] [CrossRef]
- Lubelski, J.; Konings, W.N.; Driessen, A.J.M. Distribution and Physiology of ABC-Type Transporters Contributing to Multidrug Resistance in Bacteria. Microbiol. Mol. Biol. Rev. 2007, 71, 463–476. [Google Scholar] [CrossRef]
- Tanwar, J.; Das, S.; Fatima, Z.; Hameed, S. Multidrug Resistance: An Emerging Crisis. Interdiscip. Perspect. Infect. Dis. 2014, 2014, 541340. [Google Scholar] [CrossRef]
- Arora, G.; Sajid, A.; Kalia, V.C. Drug Resistance in Bacteria, Fungi, Malaria, and Cancer; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; ISBN 9783319486833. [Google Scholar] [CrossRef]
- Subhadra, B.; Hwan Oh, M.; Hee Choi, C. Quorum Sensing in Acinetobacter: With Special Emphasis on Antibiotic Resistance, Biofilm Formation and Quorum Quenching. AIMS Microbiol. 2016, 2, 27–41. [Google Scholar] [CrossRef]
- Bardbari, A.M.; Arabestani, M.R.; Karami, M.; Keramat, F.; Alikhani, M.Y.; Bagheri, K.P. Correlation between Ability of Biofilm Formation with Their Responsible Genes and MDR Patterns in Clinical and Environmental Acinetobacter Baumannii Isolates. Microb. Pathog. 2017, 108, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, F.R. The challenge of multidrug resistance: Actual strategies in the development of novel antibacterials. Appl. Microbiol. Biotechnol. 2004, 63, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilm: An emerging battleground in microbial communities. In Antimicrobial Resistance and Infection Control; BioMed Central Ltd.: London, UK, 2019; Volume 8, p. 76. [Google Scholar] [CrossRef]
- Wood, T.K.; Knabel, S.J.; Kwan, B.W. Bacterial Persister Cell Formation and Dormancy. Appl. Environ. Microbiol. 2013, 79, 7116–7121. [Google Scholar] [CrossRef]
- Watanabe, K.; Kuwata, N.; Sakamoto, H.; Amano, Y.; Satomura, T.; Suye, S.I. A Smart DNA Sensing System for Detecting Methicillin-Resistant Staphylococcus Aureus Using Modified Nanoparticle Probes. Biosens. Bioelectron. 2015, 67, 419–423. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hedström, M.; Chen, D.; Fan, X.; Mattiasson, B. A Capacitive DNA Sensor-Based Test for Simple and Sensitive Analysis of Antibiotic Resistance in Field Setting. Biosens. Bioelectron. 2015, 64, 255–259. [Google Scholar] [CrossRef]
- Zhang, L.; Liang, W.; Ran, Q.; Liu, F.; Chen, D.; Xiong, Y.; Jiang, D.; Li, Y.; Tu, D.; Luo, X.; et al. Ultrasensitive Detection of NDM-1 Resistant Bacteria Based on Signal Amplification with Sandwich-Type LNA Electrochemical Biochips. Sens. Actuators B Chem. 2020, 306, 127556. [Google Scholar] [CrossRef]
- Opal, S.M.; Pop-Vicas, A. Molecular mechanisms of antibiotic resistance in bacteria. In Eighth Edi; Elsevier, Inc.: Amsterdam, The Netherlands, 2014; Volume 1. [Google Scholar] [CrossRef]
- Sinha, P.; Srivastava, G.N.; Tripathi, R.; Mishra, M.N.; Anupurba, S. Detection of Mutations in the RpoB Gene of Rifampicin-Resistant Mycobacterium Tuberculosis Strains Inhibiting Wild Type Probe Hybridization in the MTBDR plus Assay by DNA Sequencing Directly from Clinical Specimens. BMC Microbiol. 2020, 20, 1–8. [Google Scholar] [CrossRef]
- Makadia, J.S.; Jain, A.; Patra, S.K.; Sherwal, B.L.; Khanna, A. Emerging Trend of Mutation Profile of RpoB Gene in MDR Tuberculosis, North India. Indian J. Clin. Biochem. 2012, 27, 370–374. [Google Scholar] [CrossRef]
- Adékambi, T.; Drancourt, M.; Raoult, D. The RpoB Gene as a Tool for Clinical Microbiologists. Trends Microbiol. 2009, 17, 37–45. [Google Scholar] [CrossRef]
- Lodge, J.; Busby, S.; Piddock, L. Investigation of the Pseudomonas Aeruginosa AmpR Gene and Its Role at the Chromosomal AmpC β-Lactamase Promoter. FEMS Microbiol. Lett. 1993, 111, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Sutcliffe, J.G. Nucleotide Sequence of the Ampicillin Resistance Gene of Escherichia Coli Plasmid PBR322. Proc. Natl. Acad. Sci. USA 1978, 75, 3737–3741. [Google Scholar] [CrossRef] [PubMed]
- Jaber, M.; Rattan, A.; Kumar, R. Presence of KatG Gene in Resistant Mycobacterium Tuberculosis. J. Clin. Pathol. 1996, 49, 945–947. [Google Scholar] [CrossRef] [PubMed]
- Shivekar, S.S.; Kaliaperumal, V.; Brammacharry, U.; Sakkaravarthy, A.; Raj, C.K.V.; Alagappan, C.; Muthaiah, M. Prevalence and Factors Associated with Multidrug-Resistant Tuberculosis in South India. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Jaktaji, R.P.; Mohiti, E. Study of Mutations in the DNA Gyrase GyrA Gene of Escherichia Coli. Iran. J. Pharm. Res. 2010, 9, 43–48. [Google Scholar]
- Johnning, A.; Kristiansson, E.; Fick, J.; Weijdegård, B.; Larsson, D.G.J. Resistance Mutations in GyrA and ParC Are Common in Escherichia Communities of Both Fluoroquinolone-Polluted and Uncontaminated Aquatic Environments. Front. Microbiol. 2015, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Dubnau, E.; Quemard, A.; Balasubramanian, V.; Um, K.S.; Wilson, T.; Collins, D.; de Lisle, G.; Jr, W.R.J. InhA, Encoding Target Mycobacterium. Science 1993, 263, 227–230. [Google Scholar] [CrossRef]
- Uniprot UniProtKB—P9WGR1 (INHA_MYCTU). Available online: https://www.uniprot.org/uniprot/P9WGR1 (accessed on 23 March 2022).
- Hynes, W.L.; Dixon, A.R.; Walton, S.L.; Aridgides, L.J. The Extracellular Hyaluronidase Gene (HylA) of Streptococcus Pyogenes. FEMS Microbiol. Lett. 2000, 184, 109–112. [Google Scholar] [CrossRef]
- Yue, S.; Zhang, Z.; Liu, Y.; Zhou, Y.; Wu, C.; Huang, W.; Chen, N.; Zhu, Z. Phenotypic and Molecular Characterizations of Multidrug-Resistant Diarrheagenic E. coli of Calf Origin. Anim. Dis. 2021, 1, 1–13. [Google Scholar] [CrossRef]
- Warner, N.; Locarnini, S. Mechanisms of Hepatitis B Virus Resistance Development. Intervirology 2014, 57, 218–224. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, X.; Wei, M.; Zhang, C.; Xu, T.; Liu, L.; Xu, Z. Potential Resistant Mutations within HBV Reverse Transcriptase Sequences in Nucleos(t)Ide Analogues-Experienced Patients with Hepatitis B Virus Infection. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zaw, M.T.; Lin, Z.; Emran, N.A. Importance of Kelch 13 C580Y Mutation in the Studies of Artemisinin Resistance in Plasmodium Falciparum in Greater Mekong Subregion. J. Microbiol. Immunol. Infect. 2020, 53, 676–681. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.U.; Maryam, L.; Zarrilli, R. Structure, Genetics and Worldwide Spread of New Delhi Metallo- β -Lactamase (NDM): A Threat to Public Health. BMC Microbiol. 2017, 17, 101. [Google Scholar] [CrossRef] [PubMed]
- Kwon-Chung, K.J.; Sugui, J.A. What Do We Know about the Role of Gliotoxin in the Pathobiology of Aspergillus Fumigatus? Med. Mycol. 2009, 47, 97–103. [Google Scholar] [CrossRef]
- Kosmidis, C.; Denning, D.W. The Clinical Spectrum of Pulmonary Aspergillosis. Thorax 2015, 70, 270–277. [Google Scholar] [CrossRef]
- Haddaoui, M.; Sola, C.; Raouafi, N.; Korri-Youssoufi, H. E-DNA Detection of RpoB Gene Resistance in Mycobacterium Tuberculosis in Real Samples Using Fe 3 O 4 /Polypyrrole Nanocomposite. Biosens. Bioelectron. 2019, 128, 76–82. [Google Scholar] [CrossRef]
- Khammanee, T.; Sawangjaroen, N.; Buncherd, H.; Tun, A.W. A LAMP-SNP Assay Detecting C580Y Mutation in Pfkelch13 Gene from Clinically Dried Blood Spot Samples. Korean J. Parasitol. 2021, 59, 15–22. [Google Scholar] [CrossRef]
- Malpartida-Cardenas, K.; Miscourides, N.; Rodriguez-Manzano, J.; Yu, L.S.; Moser, N.; Baum, J.; Georgiou, P. Quantitative and Rapid Plasmodium Falciparum Malaria Diagnosis and Artemisinin-Resistance Detection Using a CMOS Lab-on-Chip Platform. Biosens. Bioelectron. 2019, 145, 111678. [Google Scholar] [CrossRef]
- Heliyaneh Moeinizadeh, M.S. Frequency of hlyA, hlyB, hlyC and hlyD genes in uropathogenic Escherichia coli isolated from UTI patients in Shiraz. GMS Hyg. Infect. Control 2021, 16, 396. [Google Scholar] [CrossRef]
- Shi, D.; Shi, H. Combining Loop-Mediated Isothermal Amplification and Nanozyme-Strip for Ultrasensitive and Rapid Detection of Viable Listeria Monocytogenes Cells and Biofilms. LWT 2022, 154, 112641. [Google Scholar] [CrossRef]
- Bhatnagar, I.; Mahato, K.; Ealla, K.K.R.; Asthana, A.; Chandra, P. Chitosan Stabilized Gold Nanoparticle Mediated Self-Assembled GliP Nanobiosensor for Diagnosis of Invasive Aspergillosis. Int. J. Biol. Macromol. 2018, 110, 449–456. [Google Scholar] [CrossRef]
- Dhar, B.C.; Reed, A.J.; Mitra, S.; Rodriguez Sanchez, P.; Nedorezova, D.D.; Connelly, R.P.; Rohde, K.H.; Gerasimova, Y.v. Cascade of Deoxyribozymes for the Colorimetric Analysis of Drug Resistance in Mycobacterium Tuberculosis. Biosens. Bioelectron. 2020, 165, 112385. [Google Scholar] [CrossRef] [PubMed]
- Bengtson, H.N.; Homolka, S.; Niemann, S.; Reis, A.J.; da Silva, P.E.; Gerasimova, Y.v.; Kolpashchikov, D.M.; Rohde, K.H. Multiplex Detection of Extensively Drug Resistant Tuberculosis Using Binary Deoxyribozyme Sensors. Biosens. Bioelectron. 2017, 94, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Dallaire, A.M.; Patskovsky, S.; Vallée-Bélisle, A.; Meunier, M. Electrochemical Plasmonic Sensing System for Highly Selective Multiplexed Detection of Biomolecules Based on Redox Nanoswitches. Biosens. Bioelectron. 2015, 71, 75–81. [Google Scholar] [CrossRef]
- Bizid, S.; Blili, S.; Mlika, R.; Haj Said, A.; Korri-Youssoufi, H. Direct Electrochemical DNA Sensor Based on a New Redox Oligomer Modified with Ferrocene and Carboxylic Acid: Application to the Detection of Mycobacterium Tuberculosis Mutant Strain. Anal. Chim. Acta 2017, 994, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Tsao, K.C.; Chiou, C.C.; Chen, T.L.; Huang, C.G.; Hsieh, E.F.; Shih, S.R. Detection of Low Copies of Drug-Resistant Influenza Viral Gene by a Single-Tube Reaction Using Peptide Nucleic Acid as Both PCR Clamp and Sensor Probe. J. Microbiol. Immunol. Infect. 2014, 47, 254–256. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Qi, P.; Zhang, D. Identification of Bacteria by a Fluorescence Sensor Array Based on Three Kinds of Receptors Functionalized Carbon Dots. Sens. Actuators B Chem. 2019, 286, 206–213. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, L.; Xu, Y.; Xue, Y.; Qiao, L.; Ding, C.; Ling, L.; Yu, S. A Platform for Specific and Sensitive Detection of Target Bacteria by Selective Magnetic Enrichment and a Broad-Spectrum Fluorescent Probe. Sens. Actuators B Chem. 2021, 349, 130762. [Google Scholar] [CrossRef]
- Gill, A.A.S.; Singh, S.; Nate, Z.; Chauhan, R.; Thapliyal, N.B.; Karpoormath, R.; Maru, S.M.; Reddy, T.M. A Novel Copper-Based 3D Porous Nanocomposite for Electrochemical Detection and Inactivation of Pathogenic Bacteria. Sens. Actuators B Chem. 2020, 321, 128449. [Google Scholar] [CrossRef]
- Rawat, D.; Nair, D. Extended-Spectrum ß-Lactamases in Gram Negative Bacteria. J. Glob. Infect. Dis. 2010, 2, 263–274. [Google Scholar] [CrossRef]
- Queenan, A.M.; Bush, K. Carbapenemases: The versatile β-lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, G.A. AmpC Β-lactamases. Clin. Microbiol. Rev. 2009, 22, 161–182. [Google Scholar] [CrossRef] [PubMed]
- Thai, H.B.D.; Yu, J.K.; Park, B.S.; Park, Y.J.; Min, S.J.; Ahn, D.R. A Fluorogenic Substrate of Beta-Lactamases and Its Potential as a Probe to Detect the Bacteria Resistant to the Third-Generation Oxyimino-Cephalosporins. Biosens. Bioelectron. 2016, 77, 1026–1031. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.; Torrelles, J.B. Mannose-Capped Lipoarabinomannan in Mycobacterium Tuberculosis Pathogenesis. Pathog. Dis. 2018, 76, 1–15. [Google Scholar] [CrossRef]
- Divagar, M.; Bandaru, R.; Janakiraman, V.; Sai, V.V.R. A plasmonic fiberoptic absorbance biosensor for mannose-capped lipoarabinomannan based tuberculosis diagnosis. Biosens. Bioelectron. 2020, 167, 112488. [Google Scholar] [CrossRef]
- Lüderitz, O.; Freudenberg, M.A.; Galanos, C.; Lehmann, V.; Rietschel, E.T.; Shaw, D.H. Lipopolysaccharides of Gram-Negative Bacteria. Curr. Top. Membr. Transp. 1982, 17, 79–151. [Google Scholar] [CrossRef]
- Raetz, C.R.H.; Whitfield, C. Lipopolysaccharide Endotoxins Endotoxins as Activators of Innate Immunity. Annu. Rev. Biochem. 2008, 71, 635–700. [Google Scholar] [CrossRef]
- Yeo, T.Y.; Choi, J.S.; Lee, B.K.; Kim, B.S.; Yoon, H.I.; Lee, H.Y.; Cho, Y.W. Electrochemical Endotoxin Sensors Based on TLR4/MD-2 Complexes Immobilized on Gold Electrodes. Biosens. Bioelectron. 2011, 28, 139–145. [Google Scholar] [CrossRef]
- Lau, G.W.; Hassett, D.J.; Ran, H.; Kong, F. The Role of Pyocyanin in Pseudomonas Aeruginosa Infection. Trends Mol. Med. 2004, 10, 599–606. [Google Scholar] [CrossRef]
- Fuse, K.; Fujimura, S.; Kikuchi, T.; Gomi, K.; Iida, Y.; Nukiwa, T.; Watanabe, A. Reduction of Virulence Factor Pyocyanin Production in Multidrug-Resistant Pseudomonas Aeruginosa. J. Infect. Chemother. 2013, 19, 82–88. [Google Scholar] [CrossRef]
- Rashid, J.I.A.; Kannan, V.; Ahmad, M.H.; Mon, A.A.; Taufik, S.; Miskon, A.; Ong, K.K.; Yusof, N.A. An Electrochemical Sensor Based on Gold Nanoparticles-Functionalized Reduced Graphene Oxide Screen Printed Electrode for the Detection of Pyocyanin Biomarker in Pseudomonas Aeruginosa Infection. Mater. Sci. Eng. C 2021, 120, 111625. [Google Scholar] [CrossRef]
- Xu, Y.; Dhaouadi, Y.; Stoodley, P.; Ren, D. Sensing the Unreachable: Challenges and Opportunities in Biofilm Detection. Curr. Opin. Biotechnol. 2020, 64, 79–84. [Google Scholar] [CrossRef]
- Kim, Y.W.; Sardari, S.E.; Meyer, M.T.; Iliadis, A.A.; Wu, H.C.; Bentley, W.E.; Ghodssi, R. An ALD Aluminum Oxide Passivated Surface Acoustic Wave Sensor for Early Biofilm Detection. Sens. Actuators B Chem. 2012, 163, 136–145. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, Z.; Li, Y.; Xie, G. Amplified Electrochemical Detection of MecA Gene in Methicillin-Resistant Staphylococcus Aureus Based on Target Recycling Amplification and Isothermal Strand-Displacement Polymerization Reaction. Sens. Actuators B Chem. 2015, 221, 148–154. [Google Scholar] [CrossRef]
- Trinh, K.T.L.; Stabler, R.A.; Lee, N.Y. Fabrication of a Foldable All-in-One Point-of-Care Molecular Diagnostic Microdevice for the Facile Identification of Multiple Pathogens. Sens. Actuators B Chem. 2020, 314, 128057. [Google Scholar] [CrossRef]
- Pedrosa, P.; Veigas, B.; Machado, D.; Couto, I.; Viveiros, M.; Baptista, P.v. Gold Nanoprobes for Multi Loci Assessment of Multi-Drug Resistant Tuberculosis. Tuberculosis 2014, 94, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Matsishin, M.; Rachkov, A.; Errachid, A.; Dzyadevych, S.; Soldatkin, A. Development of Impedimetric DNA Biosensor for Selective Detection and Discrimination of Oligonucleotide Sequences of the RpoB Gene of Mycobacterium Tuberculosis. Sens. Actuators B Chem. 2016, 222, 1152–1158. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, Y.; Zhang, L.; Yang, Y.; Yang, S.; Yang, L.; Chen, H.; Liu, C.; Li, J.; Xie, G. Integration of multiplex PCR and CRISPR-Cas allows highly specific detection of multidrug-resistant Acinetobacter baumannii. Sens. Actuators Part B 2021, 334, 129600. [Google Scholar] [CrossRef]
- Hu, C.; Kalsi, S.; Zeimpekis, I.; Sun, K.; Ashburn, P.; Turner, C.; Sutton, J.M.; Morgan, H. Ultra-Fast Electronic Detection of Antimicrobial Resistance Genes Using Isothermal Amplification and Thin Film Transistor Sensors. Biosens. Bioelectron. 2017, 96, 281–287. [Google Scholar] [CrossRef]
- Maldonado, J.; González-Guerrero, A.B.; Fernández-Gavela, A.; González-López, J.J.; Lechuga, L.M. Ultrasensitive label-free detection of unamplified multidrug-resistance bacteria genes with a bimodal waveguide interferometric biosensor. Diagnostics 2020, 10, 845. [Google Scholar] [CrossRef]
- Shin, H.H.; Lee, H.J.; Hwang, M.J.; Kim, J.; Kim, H.; Nam, S.H.; Park, J.S.; Hwang, J.E.; Kim, E.S.; Park, Y.S.; et al. Gold-Silver Core-Shell Nanodumbbells in Solution State as a Highly Sensitive and Reproducible Assay Platform for Bacterial Genome Detection. Sens. Actuators B Chem. 2021, 349, 130784. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, J.; Chen, P.; Zhou, X.; Yang, Y.; Wu, S.; Niu, L.; Han, Y.; Wang, L.; Chen, P.; et al. Label-Free, Electrochemical Detection of Methicillin-Resistant Staphylococcus Aureus DNA with Reduced Graphene Oxide-Modified Electrodes. Biosens. Bioelectron. 2011, 26, 3881–3886. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Deng, K.; Xia, H.; Yao, C.; Chen, Q.; Zhang, L.; Liu, Z.; Fu, W. Isothermal Detection of Multiple Point Mutations by a Surface Plasmon Resonance Biosensor with Au Nanoparticles Enhanced Surface-Anchored Rolling Circle Amplification. Biosens. Bioelectron. 2013, 49, 442–449. [Google Scholar] [CrossRef]
- Sun, S.C.; Dou, H.Y.; Chuang, M.C.; Kolpashchikov, D.M. Multi-Labeled Electrochemical Sensor for Cost-Efficient Detection of Single Nucleotide Substitutions in Folded Nucleic Acids. Sens. Actuators B Chem. 2019, 287, 569–575. [Google Scholar] [CrossRef]
- Trinh, T.N.D.; Thai, D.A.; Lee, N.Y. Pop-up Paper-Based and Fully Integrated Microdevice for Point-of-Care Testing of Vancomycin-Resistant Enterococcus. Sens. Actuators B Chem. 2021, 345, 130362. [Google Scholar] [CrossRef]
- Tawil, N.; Mouawad, F.; Lévesque, S.; Sacher, E.; Mandeville, R.; Meunier, M. The Differential Detection of Methicillin-Resistant, Methicillin-Susceptible and Borderline Oxacillin-Resistant Staphylococcus Aureus by Surface Plasmon Resonance. Biosens. Bioelectron. 2013, 49, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Liu, Q.; Wu, Z.; Lu, Z. Detection of hlyA Gene of Listeria Monocytogenes with Electrochemical DNA Biosensor. In Proceedings of the 2nd International Conference on Bioinformatics and Biomedical Engineering, Shanghai, China, 16–18 May 2008; pp. 375–378. [Google Scholar] [CrossRef]
- Shin, Y.; Perera, A.P.; Tang, W.Y.; Fu, D.L.; Liu, Q.; Sheng, J.K.; Gu, Z.; Lee, T.Y.; Barkham, T.; Kyoung Park, M. A Rapid Amplification/Detection Assay for Analysis of Mycobacterium Tuberculosis Using an Isothermal and Silicon Bio-Photonic Sensor Complex. Biosens. Bioelectron. 2015, 68, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wei, L.; Chen, L. Application of Metal-Organic Framework (MOF)-Based Enzymatic Amplification Strategy for the Sensitive Electrochemical Detection of Tuberculosis. Sens. Actuators B Chem. 2020, 324, 128724. [Google Scholar] [CrossRef]
- Tiwari, I.; Singh, M.; Pandey, C.M.; Sumana, G. RSC Advances Electrochemical Detection of a Pathogenic Escherichia Coli Speci Fi c DNA Sequence Based on a Graphene Oxide—Chitosan Composite Decorated with Nickel Ferrite Nanoparticles. RSC Adv. 2015, 5, 67115–67124. [Google Scholar] [CrossRef]
- Wang, J.C.; Tung, Y.C.; Ichiki, K.; Sakamoto, H.; Yang, T.H.; ichiro Suye, S.; Chuang, H.S. Culture-Free Detection of Methicillin-Resistant Staphylococcus Aureus by Using Self-Driving Diffusometric DNA Nanosensors. Biosens. Bioelectron. 2020, 148, 111817. [Google Scholar] [CrossRef]
- Ahangar, L.E.; Mehrgardi, M.A. Amplified Detection of Hepatitis B Virus Using an Electrochemical DNA Biosensor on a Nanoporous Gold Platform. Bioelectrochemistry 2017, 117, 83–88. [Google Scholar] [CrossRef]
- Chandra, S.; Mahto, T.K.; Chowdhuri, A.R.; Das, B.; Sahu, S. kumar One Step Synthesis of Functionalized Carbon Dots for the Ultrasensitive Detection of Escherichia Coli and Iron (III). Sens. Actuators B Chem. 2017, 245, 835–844. [Google Scholar] [CrossRef]
- Safardoust-Hojaghan, H.; Salavati-Niasari, M.; Amiri, O.; Hassanpour, M. Preparation of Highly Luminescent Nitrogen Doped Graphene Quantum Dots and Their Application as a Probe for Detection of Staphylococcus Aureus and E. Coli. J. Mol. Liq 2017, 241, 1114–1119. [Google Scholar] [CrossRef]
- Wang, N.; Wang, Y.; Guo, T.; Yang, T.; Chen, M.; Wang, J. Green Preparation of Carbon Dots with Papaya as Carbon Source for Effective Fluorescent Sensing of Iron (III) and Escherichia Coli. Biosens. Bioelectron. 2016, 85, 68–75. [Google Scholar] [CrossRef]
- Chen, K.H.; Lee, S.H.; Kok, L.C.; Ishdorj, T.O.; Chang, H.Y.; Tseng, F.G. A 3D-ACEK/SERS System for Highly Efficient and Selectable Electrokinetic Bacteria Concentration/Detection/ Antibiotic-Susceptibility-Test on Whole Blood. Biosens. Bioelectron. 2022, 197, 113740. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zheng, Y.; Liu, Y.; Feng, X.; Dong, Q.; Li, J.; Wang, J.; Zhao, C. A Concise Detection Strategy of Staphylococcus Aureus Using N-Succinyl-Chitosan-Dopped Bacteria-Imprinted Composite Film and AIE Fluorescence Sensor. J. Hazard. Mater. 2022, 423, 126934. [Google Scholar] [CrossRef] [PubMed]
- Hiremath, S.D.; Bhosle, A.A.; Nayse, A.; Biswas, S.; Biswas, M.; Bhasikuttan, A.C.; Banerjee, M.; Chatterjee, A. A Redox-Coupled Carbon Dots-MnO2 Nanosheets Based Sensory Platform for Label-Free and Sensitive Detection of E. Coli. Sens. Actuators B Chem. 2021, 339, 129918. [Google Scholar] [CrossRef]
- Wang, H.Y.; Chen, C.Y.; Chu, P.Y.; Zhu, Y.X.; Hsieh, C.H.; Lu, J.J.; Wu, M.H. Application of an Optically Induced Dielectrophoresis (ODEP)-Based Microfluidic System for the Detection and Isolation of Bacteria with Heterogeneity of Antibiotic Susceptibility. Sens. Actuators B Chem. 2020, 307, 127540. [Google Scholar] [CrossRef]
- Xia, H.; Huang, J.; Lu, X.; Wang, G.; Zhang, Z.; Yue, J.; Li, Q.; Wang, S.; Yan, J.; Deng, L.; et al. Autocatalytic MNAzyme-Integrated Surface Plasmon Resonance Biosensor for Simultaneous Detection of Bacteria from Nosocomial Bloodstream Infection Specimens. Sens. Actuators B Chem. 2021, 330, 129255. [Google Scholar] [CrossRef]
- Wang, H.; Chi, Z.; Cong, Y.; Wang, Z.; Jiang, F.; Geng, J.; Zhang, P.; Ju, P.; Dong, Q.; Liu, C. Development of a Fluorescence Assay for Highly Sensitive Detection of: Pseudomonas Aeruginosa Based on an Aptamer-Carbon Dots/Graphene Oxide System. RSC Adv. 2018, 8, 32454–32460. [Google Scholar] [CrossRef]
- Villamizar, R.A.; Maroto, A.; Rius, F.X.; Inza, I.; Figueras, M.J. Fast Detection of Salmonella Infantis with Carbon Nanotube Field Effect Transistors. Biosens. Bioelectron. 2008, 24, 279–283. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, X.; Jiang, H. Label-Free Detection of Acinetobacter Baumannii through the Induced Fluorescence Quenching of Thiolated AuAg Nanoclusters. Sens. Actuators B Chem. 2018, 277, 388–393. [Google Scholar] [CrossRef]
- Kumar, N.; Wang, W.; Ortiz-Marquez, J.C.; Catalano, M.; Gray, M.; Biglari, N.; Hikari, K.; Ling, X.; Gao, J.; van Opijnen, T.; et al. Dielectrophoresis Assisted Rapid, Selective and Single Cell Detection of Antibiotic Resistant Bacteria with G-FETs. Biosens. Bioelectron. 2020, 156, 112123. [Google Scholar] [CrossRef]
- Quero, G.; Zuppolini, S.; Consales, M.; Diodato, L.; Vaiano, P.; Venturelli, A.; Santucci, M.; Spyrakis, F.; Costi, M.P.; Giordano, M.; et al. Long Period Fiber Grating Working in Reflection Mode as Valuable Biosensing Platform for the Detection of Drug Resistant Bacteria. Sens. Actuators B Chem. 2016, 230, 510–520. [Google Scholar] [CrossRef]
- Atchudan, R.; Edison, T.N.J.I.; Chakradhar, D.; Perumal, S.; Shim, J.J.; Lee, Y.R. Facile Green Synthesis of Nitrogen-Doped Carbon Dots Using Chionanthus Retusus Fruit Extract and Investigation of Their Suitability for Metal Ion Sensing and Biological Applications. Sens. Actuators B Chem. 2017, 246, 497–509. [Google Scholar] [CrossRef]
- Sypabekova, M.; Jolly, P.; Estrela, P.; Kanayeva, D. Electrochemical Aptasensor Using Optimized Surface Chemistry for the Detection of Mycobacterium Tuberculosis Secreted Protein MPT64 in Human Serum. Biosens. Bioelectron. 2019, 123, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.Z.; Yu, H.W.; Wang, N.; Zhang, Z.; Wan, G.C.; Liu, H.; Guan, X.; Chang, D. Electrochemical DNA Biosensor Based on a Glassy Carbon Electrode Modified with Gold Nanoparticles and Graphene for Sensitive Determination of Klebsiella Pneumoniae Carbapenemase. J. Biotechnol 2015, 214, 133–138. [Google Scholar] [CrossRef]
- Hilton, S.H.; Hall, C.; Nguyen, H.T.; Everitt, M.L.; DeShong, P.; White, I.M. Phenotypically Distinguishing ESBL-Producing Pathogens Using Paper-Based Surface Enhanced Raman Sensors. Anal. Chim. Acta 2020, 1127, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Andersson, T.; Bläckberg, A.; Lood, R.; Bergdahl, G.E. Development of a Molecular Imprinting-Based Surface Plasmon Resonance Biosensor for Rapid and Sensitive Detection of Staphylococcus Aureus Alpha Hemolysin From Human Serum. Front. Cell. Infect. Microbiol. 2020, 10, 571578. [Google Scholar] [CrossRef]
- Poma, N.; Vivaldi, F.; Bonini, A.; Salvo, P.; Kirchhain, A.; Melai, B.; Bottai, D.; Tavanti, A.; di Francesco, F. A Graphenic and Potentiometric Sensor for Monitoring the Growth of Bacterial Biofilms. Sens. Actuators B Chem. 2020, 323, 128662. [Google Scholar] [CrossRef]
- Assaraf, Y.G.; Brozovic, A.; Gonçalves, A.C.; Jurkovicova, D.; Linē, A.; Machuqueiro, M.; Saponara, S.; Sarmento-Ribeiro, A.B.; Xavier, C.P.R.; Vasconcelos, M.H. The Multi-Factorial Nature of Clinical Multidrug Resistance in Cancer. Drug Resist. Updat. 2019, 46, 100645. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Feng, S.S.; Guo, Y. Nanomedicine against Multidrug Resistance in Cancer Treatment. Nanomedicine 2012, 7, 465–468. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Chen, X. Drug resistance and combating drug resistance in cancer. In Cancer Drug Resistance; OAE Publishing, Inc.: Alhambra, CA, USA, 2019; Volume 2, pp. 141–160. [Google Scholar] [CrossRef]
- Szakács, G.; Hall, M.D.; Gottesman, M.M.; Boumendjel, A.; Kachadourian, R.; Day, B.J.; Baubichon-Cortay, H.; di Pietro, A. Targeting the Achilles Heel of Multidrug-Resistant Cancer by Exploiting the Fitness Cost of Resistance. Chem Rev. 2014, 114, 5753–5774. [Google Scholar] [CrossRef] [PubMed]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The different mechanisms of cancer drug resistance: A brief review. In Advanced Pharmaceutical Bulletin; Tabriz University of Medical Sciences: Tabriz, Iran, 2017; Volume 7, pp. 339–348. [Google Scholar] [CrossRef]
- Bukowski, K.; Kciuk, M.; Kontek, R. Mechanisms of multidrug resistance in cancer chemotherapy. Int. J. Mol. Sci. 2020, 21, 3233. [Google Scholar] [CrossRef] [PubMed]
- van Veen, H.W.; Konings, W.N. The ABC Family of Multidrug Transporters in Microorganisms. Biochim. Biophys. Acta Bioenerg. 1998, 1365, 31–36. [Google Scholar] [CrossRef]
- Robey, R.W.; Pluchino, K.M.; Hall, M.D.; Fojo, A.T.; Bates, S.E.; Gottesman, M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. In Nature Reviews. Cancer; Nature Publishing Group: Berlin, Germany, 2018; Volume 18, pp. 452–464. [Google Scholar] [CrossRef]
- Gillet, J.P.; Gottesman, M.M. Mechanisms of Multidrug Resistance in Cancer. Methods Mol. Biol. 2010, 596, 47–76. [Google Scholar] [CrossRef]
- Kimchi-Sarfaty, C.; Marple, A.H.; Shinar, S.; Kimchi, A.M.; Scavo, D.; Roma, M.I.; Kim, I.W.; Jones, A.; Arora, M.; Gribar, J.; et al. Ethnicity-related polymorphisms and haplotypes in the human ABCB1 gene. Pharmacogenomics 2007, 8, 29–39. [Google Scholar] [CrossRef]
- Wen-Hsuan, C.; Cerione Richard, A.; Antonyak Mark, A. Cancer cell signaling methods and protocols third edition. In Cancer Cell Signaling; Springer: Berlin, Germany, 2021; Available online: springer.com/book/10.1007/978-1-0716-0759-6 (accessed on 26 January 2022).
- Momen-Heravi, F.; Bala, S. The miRNA and Extracellular Vesicles in Alcoholic Liver Disease; Elsevier, Inc.: Amsterdam, The Netherlands, 2016; Volume 2000, ISBN 9780128007730. [Google Scholar] [CrossRef]
- Paoli, P.; Giannoni, E.; Chiarugi, P. Anoikis Molecular Pathways and Its Role in Cancer Progression. Biochim. Biophys. Acta Mol. Cell Res. 2013, 1833, 3481–3498. [Google Scholar] [CrossRef]
- Arneth, B. Tumor microenvironment. Medicina 2019, 56, 15. [Google Scholar] [CrossRef]
- Kim, J.; Bae, J.S.; Tam, M. Tumor-associated macrophages and neutrophils in tumor microenvironment. Mediat. Inflamm. 2016, 2016, 6058147. [Google Scholar] [CrossRef]
- Phi, L.T.H.; Sari, I.N.; Yang, Y.G.; Lee, S.H.; Jun, N.; Kim, K.S.; Lee, Y.K.; Kwon, H.Y. Cancer stem cells (CSCs) in drug resistance and their therapeutic implications in cancer treatment. In Stem Cells International; Hindawi Publishing Limited: London, UK, 2018; Volume 2018, p. 5416923. [Google Scholar] [CrossRef]
- Robles-Flores. Cancer Cell Signaling Methods and Protocols Third Edition; Fighting Cancer Resistance. 2021. Available online: www.springer.com/series/7651 (accessed on 20 July 2022).
- Ricardo, P. Lactate in the Tumor Microenvironment: An Essential; MDPI: Basel, Switzerland, 2020; pp. 1–29. [Google Scholar]
- Pérez-Tomás, R.; Pérez-Guillén, I. Lactate in the tumor microenvironment: An essential molecule in cancer progression and treatment. Cancers 2020, 12, 3244. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.P.; Hu, Y.; Liu, P.; Deng, Y.N.; Wang, P.; Chen, W.; Liu, A.L.; Chen, Y.Z.; Lin, X.H. Label-Free Electrochemical DNA Biosensor for Rapid Detection of Mutidrug Resistance Gene Based on Au Nanoparticles/Toluidine Blue-Graphene Oxide Nanocomposites. Sens. Actuators B Chem. 2015, 207, 269–276. [Google Scholar] [CrossRef]
- Chen, M.; Hou, C.; Huo, D.; Bao, J.; Fa, H.; Shen, C. An Electrochemical DNA Biosensor Based on Nitrogen-Doped Graphene/Au Nanoparticles for Human Multidrug Resistance Gene Detection. Biosens. Bioelectron. 2016, 85, 684–691. [Google Scholar] [CrossRef]
- Prattis, I.; Hui, E.; Gubeljak, P.; Kaminski Schierle, G.S.; Lombardo, A.; Luigi, G. Occhipinti, Graphene for Biosensing Applications in Point-of-Care Testing. Trends Biotechnol. 2021, 39, 1065–1077. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Z.; Zou, B.; Zhang, L.; Ma, X.; Qi, X.; Wei, W.; Song, Q.; Zhou, G. Ultra-Sensitive and Multiplex Digital-PCR for Quantifying the Mutants in Cell Free DNA by Employing Invasive Reaction as Identifier. Sens. Actuators B Chem. 2020, 320, 128362. [Google Scholar] [CrossRef]
- Ma, C.; Wei, S.; Song, Y. T790M and Acquired Resistance of EGFR TKI: A Literature Review of Clinical Reports; Journal for Thoracic Diseases: Hong Kong, China, 2010; Volume 3. [Google Scholar] [CrossRef]
- Brien, J.O.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, L.A.; Murphy, P.R. MicroRNA: Biogenesis, function and role in cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef]
- Cai, X.; Zhang, H.; Yu, X.; Wang, W. A Microfluidic Paper-Based Laser-Induced Fluorescence Sensor Based on Duplex-Specific Nuclease Amplification for Selective and Sensitive Detection of MiRNAs in Cancer Cells. Talanta 2020, 216, 120996. [Google Scholar] [CrossRef] [PubMed]
- Yaman, Y.T.; Vural, O.A.; Bolat, G.; Abaci, S. One-Pot Synthesized Gold Nanoparticle-Peptide Nanotube Modified Disposable Sensor for Impedimetric Recognition of MiRNA 410. Sens. Actuators B Chem. 2020, 320, 128343. [Google Scholar] [CrossRef]
- Qu, H.; Fan, C.; Chen, M.; Zhang, X.; Yan, Q.; Wang, Y.; Zhang, S.; Gong, Z.; Shi, L.; Li, X.; et al. Recent Advances of Fluorescent Biosensors Based on Cyclic Signal Amplification Technology in Biomedical Detection. J. Nanobiotechnol. 2021, 19, 403. [Google Scholar] [CrossRef]
- Dong, Z.; Ren, L.; Lin, L.; Li, J.; Huang, Y.; Li, J. Effect of MicroRNA-21 on Multidrug Resistance Reversal in A549/DDP Human Lung Cancer Cells. Mol. Med. Rep. 2015, 11, 682–690. [Google Scholar] [CrossRef]
- Feng, Y.H.; Tsao, C.J. Emerging Role of MicroRNA-21 in Cancer (Review). Biomed. Rep. 2016, 5, 395–402. [Google Scholar] [CrossRef]
- National Library of Medicine MIR155 MicroRNA 155 [Homo Sapiens (Human)]. Available online: https://www.ncbi.nlm.nih.gov/gene/406947#summary (accessed on 26 July 2022).
- Bayraktar, R.; van Roosbroeck, K. miR-155 in cancer drug resistance and as target for miRNA-based therapeutics. Cancer Metastasis Rev. 2018, 37, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Qiu, Y.; Yagüe, E.; Ji, W.; Liu, J.; Zhang, J. MiRNA-205 Targets VEGFA and FGF2 and Regulates Resistance to Chemotherapeutics in Breast Cancer. Cell Death Dis. 2016, 7, e2291. [Google Scholar] [CrossRef] [PubMed]
- Jopling, C.L. Liver-specific microRNA-122. RNA Biol. 2012, 9, 137–142. [Google Scholar] [CrossRef]
- Yahya, S.M.M.; Fathy, S.A. Possible Role of MicroRNA-122 in Modulating Multidrug Resistance of Hepatocellular Carcinoma. Indian J. Clin. Biochem. 2018, 33, 21–30. [Google Scholar] [CrossRef]
- Dong, J.; Liu, Y.; Liao, W.; Liu, R.; Shi, P.; Wang, L. MiRNA-223 Is a Potential Diagnostic and Prognostic Marker for Osteosarcoma. J. Bone Oncol. 2016, 5, 74–79. [Google Scholar] [CrossRef]
- Wang, H.; Chen, J.; Zhang, S.; Zheng, X.; Xie, S.; Mao, J.; Cai, Y.; Lu, X.; Hu, L.; Shen, J.; et al. MiR—223 regulates autophagy associated with cisplatin resistance by targeting FBXW7 in human non—Small cell lung cancer. Cancer Cell Int. 2020, 20, 258. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Zheng, Z.M.; Li, X.N.; Li, Z.F.; Wang, Y.; Geng, Y.F.; Bai, L.; Zhang, X.B. MiR-223 modulates multidrug resistance via downregulation of ABCB1 in hepatocellular carcinoma cells. Exp. Biol. Med. 2013, 238, 1024–1032. [Google Scholar] [CrossRef]
- Stepicheva, N.A.; Song, J.I.A.L. Function and Regulation of MicroRNA-31 in Development and Disease. Mol. Reprod. Dev. 2018, 83, 654–674. [Google Scholar] [CrossRef]
- Humphries, B.; Yang, C. The microRNA-200 family: Small molecules with novel roles in cancer development, progression and therapy. Oncotarget 2015, 6, 6472–6498. [Google Scholar] [CrossRef]
- Lee, H.; Kim, C.; Kang, H.; Tak, H.; Ahn, S.; Yoon, S.K.; Kuh, H.J.; Kim, W.; Lee, E.K. microRNA-200a-3p increases 5- fluorouracil resistance by regulating dual speci fi city phosphatase 6 expression. Exp. Mol. Med. 2017, 49, e327. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liao, Y.; Tang, L. MicroRNA-34 family: A potential tumor suppressor and therapeutic candidate in cancer. J. Exp. Clin. Cancer Res. 2019, 5, 53. [Google Scholar] [CrossRef]
- Physiology, C. MicroRNA-34a Attenuates Metastasis and Chemoresistance of Bladder Cancer Cells by Targeting the TCF1/LEF1 Axis. Cell. Physiol. Biochem. 2018, 400037, 87–98. [Google Scholar] [CrossRef]
- Schneider, J.W.; Dittmer, D.P. Diagnosis and Treatment of Kaposi Sarcoma Johann. Am. J. Clin. Dermatol. 2018, 18, 529–539. [Google Scholar] [CrossRef]
- Zhang, J.; Pu, X.; Xiong, Y. kshv-mir-k12-1-5p Promotes Cell Growth and Metastasis by Targeting SOCS6 in Kaposi’s Sarcoma Cells; Dove Medical Press: Macclesfield, UK, 2019; pp. 4985–4995. [Google Scholar]
- Rta, O.R.F.; Lin, X.; Liang, D.; He, Z.; Deng, Q.; Robertson, E.S.; Lan, K. MiR-K12-7-5p Encoded by Kaposi’ s Sarcoma-Associated Herpesvirus Stabilizes the Latent State by Targeting Viral. PLoS ONE 2011, 6, e0016224. [Google Scholar] [CrossRef]
- Wang, K. Role of microRNA—410 in molecular oncology: A double edged sword. J. Cell. Biochem. 2018, 119, 8737–8742. [Google Scholar] [CrossRef]
- Chen, R.E.N.; Li, X.; He, B.I.N.; Hu, W.E.I. MicroRNA-410 Regulates Autophagy-Related Gene ATG16L1 Expression and Enhances Chemosensitivity via Autophagy Inhibition in Osteosarcoma. Mol. Med. Rep. 2017, 15, 1326–1334. [Google Scholar] [CrossRef]
- Milevskiy, M.J.G.; Gujral, U.; Del, C.; Marques, L.; Stone, A.; Northwood, K.; Burke, L.J.; Gee, J.M.W.; Nephew, K.; Clark, S.; et al. MicroRNA-196a Is Regulated by ER and Is a Prognostic Biomarker in ER + Breast Cancer. Br. J. Cancer 2019, 120, 621–632. [Google Scholar] [CrossRef]
- Xin, H.; Wang, C.; Liu, Z. miR-196a-5p promotes metastasis of colorectal cancer via targeting I κ B α. BMC Cancer 2019, 19, 30. [Google Scholar] [CrossRef]
- Gao, Y.; Feng, B.; Han, S.; Zhang, K.; Chen, J.; Li, C.; Wang, R. The Roles of MicroRNA-141 in Human Cancers: From Diagnosis to Treatment. Cell. Physiol. Biochem. 2016, 38, 427–448. [Google Scholar] [CrossRef]
- Kinose, Y.; Sawada, K.; Nakamura, K.; Kimura, T. The role of microRNAs in ovarian cancer. BioMed Res. Int. 2014, 2014, 249393. [Google Scholar] [CrossRef] [PubMed]
- Su, J.L.; Chen, P.S.; Johansson, G.; Kuo, M.L. Function and regulation of Let-7 family microRNAs. MicroRNA 2012, 1, 34–39. [Google Scholar] [CrossRef]
- Kolenda, T.; Przybyła, W.; Teresiak, A. The Mystery of Let-7d—A Small RNA with Great Power. Contemp. Oncol./Współczesna Onkol. 2014, 18, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Yang, W.; Feng, W.; Cao, L.; Wang, X.; Niu, L.; Li, Y.; Zhou, W.; Zhang, Y.; Liu, J.; et al. Molecular Mechanisms and Clinical Implications of MiRNAs in Drug Resistance of Colorectal Cancer. Ther. Adv. Med. Oncol. 2020, 12, 1758835920947342. [Google Scholar] [CrossRef]
- Garg, H.; Suri, P.; Gupta, J.C.; Talwar, G.P.; Dubey, S. Survivin: A Unique Target for Tumor Therapy. Cancer Cell Int. 2016, 16, 49. [Google Scholar] [CrossRef] [PubMed]
- Lei, W.; Gui-mei, Z.; Zuo-Hua, F. Down-Regulation of Survivin Expression Reversed Multidrug Resistance in Adriamycin- Resistant HL-60/ADR Cell Line 1. Acta Pharmacol. Sin. 2003, 24, 1235–1240. [Google Scholar]
- Hu, H.; Jiang, H.; Wang, X.; Chen, B. Detection and Distinguishability of Leukemia Cancer Cells Based on Au Nanoparticles Modified Electrodes. Electrochem. Commun. 2008, 10, 1121–1124. [Google Scholar] [CrossRef]
- Tao, Y.; Li, M.; Auguste, D.T. Pattern-Based Sensing of Triple Negative Breast Cancer Cells with Dual-Ligand Cofunctionalized Gold Nanoclusters. Biomaterials 2017, 116, 21–33. [Google Scholar] [CrossRef]
- Chandra, P.; Noh, H.B.; Pallela, R.; Shim, Y.B. Ultrasensitive Detection of Drug Resistant Cancer Cells in Biological Matrixes Using an Amperometric Nanobiosensor. Biosens. Bioelectron. 2015, 70, 418–425. [Google Scholar] [CrossRef]
- Gulati, P.; Kaur, P.; Rajam, M.v.; Srivastava, T.; Ali, M.A.; Mishra, P.; Islam, S.S. Leukemia Biomarker Detection by Using Photoconductive Response of CNT Electrode: Analysis of Sensing Mechanism Based on Charge Transfer Induced Fermi Level Fluctuation. Sens. Actuators B Chem. 2018, 270, 45–55. [Google Scholar] [CrossRef]
- Li, Y.; Chen, D.; Zhang, Y.; Liu, C.; Chen, P.; Wang, Y.; Feng, X.; Du, W.; Liu, B.F. High-Throughput Single Cell Multidrug Resistance Analysis with Multifunctional Gradients-Customizing Microfluidic Device. Sens. Actuators B Chem. 2016, 225, 563–571. [Google Scholar] [CrossRef]
- Berrazouane, S.; Boisvert, M.; Salti, S.; Mourad, W.; Al-Daccak, R.; Barabé, F.; Aoudjit, F. Beta1 integrin blockade overcomes doxorubicin resistance in human T-cell acute lymphoblastic leukemia. Cell Death Dis. 2019, 10, 357. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.; Guo, J.; Liao, Q.; Zhao, Y. β 1 and β 3 Integrins in Breast, Prostate and Pancreatic Cancer: A Novel Implication (Review). Oncol. Lett. 2018, 15, 5412–5416. [Google Scholar] [CrossRef] [PubMed]
- Morozevich, G.E.; Kozlova, N.I.; Preobrazhenskaya, M.E.; Ushakova, N.A.; Eltsov, I.A.; Shtil, A.A.; Berman, A.E. The role of β 1 integrin subfamily in anchorage-dependent apoptosis of breast carcinoma cells differing in multidrug resistance. Biochem. Biokhimiia. 2006, 71, 489–495. [Google Scholar] [CrossRef]
- Jiang, X.; Tan, L.; Zhang, B.; Zhang, Y.; Tang, H.; Xie, Q.; Yao, S. Detection of Adherent Cells Using Electrochemical Impedance Spectroscopy Based on Molecular Recognition of Integrin Β1. Sens. Actuators B Chem. 2010, 149, 87–93. [Google Scholar] [CrossRef]
- Odening, K.E.; Li, W.; Rutz, R. Enhanced complement resistance in drug-selected P-glycoprotein expressing multi-drug-resistant ovarian carcinoma cells. Clin. Exp. Immunol. 2008, 155, 239–248. [Google Scholar] [CrossRef]
- Choudhary, M.; Yadav, P.; Singh, A.; Kaur, S.; Ramirez-Vick, J. CD 59 Targeted Ultrasensitive Electrochemical Immunosensor for Fast and Noninvasive Diagnosis of Oral Cancer. Electroanalysis 2016, 28, 2565–2574. [Google Scholar] [CrossRef]
- Meng, L.; Liu, B.; Ji, R.U.I.; Jiang, X.I.N.; Yan, X.; Xin, Y. Targeting the BDNF / TrkB Pathway for the Treatment of Tumors. Oncol. Lett. 2019, 17, 2031–2039. [Google Scholar] [CrossRef]
- Akhtar, M.H.; Hussain, K.K.; Gurudatt, N.G.; Chandra, P.; Shim, Y. Ultrasensitive Dual Probe Immunosensor for the Monitoring of Nicotine Induced-Brain Derived Neurotrophic Factor Released from Cancer Cells. Biosens. Bioelectron. 2018, 116, 108–115. [Google Scholar] [CrossRef]
- Wang, X.; Yang, D.; Liu, M.; Cao, D.; He, N.; Wang, Z. Highly Sensitive Fluorescence Biosensor for Intracellular Telomerase Detection Based on a Single Patchy Gold/Carbon Nanosphere via the Combination of Nanoflare and Hybridization Chain Reaction. Biosens. Bioelectron. 2019, 137, 110–116. [Google Scholar] [CrossRef]
- Lipinska, N.; Romaniuk, A.; Paszel-Jaworska, A.; Toton, E.; Kopczynski, P.; Rubis, B. Telomerase and drug resistance in cancer. Cell. Mol. Life Sci. 2017, 74, 4121–4132. [Google Scholar] [CrossRef]
- Gao, P.; Yang, X.; Xue, Y.W.; Zhang, X.F.; Wang, Y.; Liu, W.J.; Wu, X.J. Promoter methylation of glutathione S-transferase p 1 and multidrug resistance gene 1 in bronchioloalveolar carcinoma and its correlation with DNA methyltransferase 1 expression. Cancer 2009, 115, 3222–3232. [Google Scholar] [CrossRef] [PubMed]
- Sodani, K.; Patel, A.; Kathawala, R.J.; Chen, Z.S. Multidrug Resistance Associated Proteins in Multindrung Resistance. Chin. J. Cancer 2012, 31, 58–72. [Google Scholar] [CrossRef] [PubMed]
- Baker, E.K.; El-osta, A. The Rise of DNA Methylation and the Importance of Chromatin on Multidrug Resistance in Cancer. Exp. Cell Res. 2003, 290, 177–194. [Google Scholar] [CrossRef]
- Jones, P.M.; George, A.M. Multidrug Resistance in Parasites: ABC Transporters, P-Glycoproteins and Molecular Modelling. Int. J. Parasitol 2005, 35, 555–566. [Google Scholar] [CrossRef]
- Yang, P.; Zheng, Y.; Wang, Y.; Yue, J.; Gao, G.; Ma, Y.; Tang, B. A novel sensitive NMOF fluorescent probe for two photon imaging of glutathione in chemo-resistant cancer cells. Sens. Actuators Part B 2021, 348, 130680. [Google Scholar] [CrossRef]
- Fattahi, S.; Golpour, M.; Amjadi-Moheb, F.; Sharifi-Pasandi, M.; Khodadadi, P.; Pilehchian-Langroudi, M.; Ashrafi, G.H.; Akhavan-Niaki, H. DNA Methyltransferases and Gastric Cancer: Insight into Targeted Therapy. Epigenomics 2018, 10, 1477–1497. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Liu, F.; Fan, T.; Ren, Y.; Jiang, Y. Rapid Detection of Methyltransferases Utilizing Dumbbell DNA-Templated Copper Nanoparticles. Sens. Actuators B Chem. 2018, 276, 499–506. [Google Scholar] [CrossRef]
- Zastawna, M.; Colas, C.; Hiebel, M.; Suzenet, F. Nitro-Pyrazinotriazapentalene Scaffolds—Nitroreductase Quantification and In Vitro Fluorescence Imaging of Hypoxia; Sensors and Actuators: Amsterdam, The Netherlands, 2021; Volume 346. [Google Scholar]
- Wang, Y.; Zhang, L.; Huang, Y.; Wang, X.; Zhang, L.; Chen, L. Rational Design of a Nitroreductase-Activatable Two-Photon Fluorescent Probe for Hypoxia Imaging in Cell and in Vivo. Sens. Actuators B Chem. 2020, 310, 127755. [Google Scholar] [CrossRef]
- Ge, Y.; Wei, P.; Wang, T.; Cao, X.; Zhang, D.; Li, F. A Simple Fluorescent Probe for Monitoring PH in Cellsbased on New Fluorophorepyrido [1,2-a] Benzimidazole. Sens. Actuators B Chem. 2018, 254, 314–320. [Google Scholar] [CrossRef]
- Zhao, S.S.; Bukar, N.; Toulouse, J.L.; Pelechacz, D.; Robitaille, R.; Pelletier, J.N.; Masson, J.F. Miniature Multi-Channel SPR Instrument for Methotrexate Monitoring in Clinical Samples. Biosens. Bioelectron. 2015, 64, 664–670. [Google Scholar] [CrossRef]
- Hunter, R.A.; Asare-Werehene, M.; Mandour, A.; Tsang, B.K.; Anis, H. Determination of Chemoresistance in Ovarian Cancer by Simultaneous Quantification of Exosomes and Exosomal Cisplatin with Surface Enhanced Raman Scattering. Sens. Actuators B Chem. 2021, 354, 131237. [Google Scholar] [CrossRef]
- Kalyane, D.; Choudhary, D.; Polaka, S.; Goykar, H.; Anup, N.; Tambe, V.; Kalia, K.; Tekade, R.K. Exosomes in Multidrug-Resistant Cancer. Curr. Opin. Pharm. 2020, 54, 109–120. [Google Scholar] [CrossRef]
- Tai, Y.L.; Chen, K.C.; Hsieh, J.T.; Shen, T.L. Exosomes in Cancer Development and Clinical Applications. Cancer Sci. 2018, 109, 2364–2374. [Google Scholar] [CrossRef] [PubMed]
- Zada, S.; Lu, H.; Dai, W.; Tang, S.; Khan, S.; Yang, F.; Qiao, Y.; Fu, P.; Dong, H.; Zhang, X. Multiple Amplified MicroRNAs Monitoring in Living Cells Based on Fluorescence Quenching of Mo2B and Hybridization Chain Reaction. Biosens. Bioelectron. 2022, 197, 113815. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wang, M.; Xiong, X.; Liu, Y.; Chen, X. Simultaneous Fluorescent Detection of Multiplexed MiRNA of Liver Cancer Based on DNA Tetrahedron Nanotags. Talanta 2020, 210, 120677. [Google Scholar] [CrossRef]
- Peng, W.; Huang, Y.; Zhao, Q.; Lian, G.; Chen, M.; Piao, J.; Gong, X.; Chang, J. A Fluorescent Signal “Removal” Sensor via Duplex-Specific Nuclease-Aided Cleavage for MiRNA Detection in Flow Cytometry. Colloids Surf. B Biointerfaces 2020, 185, 110570. [Google Scholar] [CrossRef]
- Na, H.K.; Wi, J.S.; Son, H.Y.; Ok, J.G.; Huh, Y.M.; Lee, T.G. Discrimination of Single Nucleotide Mismatches Using a Scalable, Flexible, and Transparent Three-Dimensional Nanostructure-Based Plasmonic MiRNA Sensor with High Sensitivity. Biosens. Bioelectron. 2018, 113, 39–45. [Google Scholar] [CrossRef]
- Wang, B.; Chen, Z.; Ren, D.; You, Z. A Novel Dual Energy Transfer Probe for Intracellular MRNA Detection with High Robustness and Specificity. Sens. Actuators B Chem. 2019, 279, 342–350. [Google Scholar] [CrossRef]
- Li, Q.; Zhou, S.; Zhang, T.; Zheng, B.; Tang, H. Bioinspired Sensor Chip for Detection of MiRNA-21 Based on Photonic Crystals Assisted Cyclic Enzymatic Amplification Method. Biosens. Bioelectron. 2020, 150, 111866. [Google Scholar] [CrossRef]
- Isin, D.; Eksin, E.; Erdem, A. Graphene Oxide Modified Single-Use Electrodes and Their Application for Voltammetric MiRNA Analysis. Mater. Sci. Eng. C 2017, 75, 1242–1249. [Google Scholar] [CrossRef]
- Bahari, D.; Babamiri, B.; Salimi, A.; Rashidi, A. Graphdiyne/Graphene Quantum Dots for Development of FRET Ratiometric Fluorescent Assay toward Sensitive Detection of MiRNA in Human Serum and Bioimaging of Living Cancer Cells. J. Lumin 2021, 239, 118371. [Google Scholar] [CrossRef]
- Han, Y.; Qiang, L.; Gao, Y.; Gao, J.; He, Q.; Liu, H.; Han, L.; Zhang, Y. Large-Area Surface-Enhanced Raman Spectroscopy Substrate by Hybrid Porous GaN with Au/Ag for Breast Cancer MiRNA Detection. Appl Surf. Sci. 2021, 541, 148456. [Google Scholar] [CrossRef]
- Mao, Y.; Sun, Y.; Xue, J.; Lu, W.; Cao, X. Ultra-Sensitive and High Efficiency Detection of Multiple Non-Small Cell Lung Cancer-Related MiRNAs on a Single Test Line in Catalytic Hairpin Assembly-Based SERS-LFA Strip. Anal. Chim. Acta 2021, 1178, 338800. [Google Scholar] [CrossRef]
- Wang, Q.; Sun, H.; Wen, D.; Wang, L.; Li, L.; Kong, J.; Zhang, X. Ultrasensitive Electrochemical Detection of MiRNA Based on Polymerization Signal Amplification. Talanta 2021, 235, 122744. [Google Scholar] [CrossRef]
- Wu, X.; Li, Y.; Yang, M.Y.; Mao, C.B. Simultaneous ultrasensitive detection of two breast cancer microRNA biomarkers by using a dual nanoparticle/nanosheet fluorescence resonance energy transfer sensor. Mater. Today Adv. 2021, 12, 100163. [Google Scholar] [CrossRef]
- Gulati, P.; Kaur, P.; Rajam, M.v.; Srivastava, T.; Mishra, P.; Islam, S.S. Vertically Aligned Multi-Walled Carbon Nanotubes Based Flexible Immunosensor for Extreme Low-Level Detection of Multidrug Resistant Leukemia Cells. Sens. Actuators B Chem. 2019, 301, 127047. [Google Scholar] [CrossRef]
- Zhang, H.; Jiang, H.; Sun, F.; Wang, H.; Zhao, J.; Chen, B.; Wang, X. Rapid Diagnosis of Multidrug Resistance in Cancer by Electrochemical Sensor Based on Carbon Nanotubes-Drug Supramolecular Nanocomposites. Biosens. Bioelectron. 2011, 26, 3361–3366. [Google Scholar] [CrossRef]
- WHO. Antimicrobial Resistance—À Global Epidemic. 2010. Available online: https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance (accessed on 3 August 2022).
- Eduardo, C.R.; Laschi, S.; Ilaria Palchetti, E.T. Advances in Antimicrobial Resistance Monitoring Using Sensor and Biosensor: A Review; MDPI: Berlin, Germany, 2021. [Google Scholar]
- Global Multidrug Resistant Bacteria Market-Industry Trends and Forcast to 2029. Available online: https://www.databridgemarketresearch.com/reports/global-multidrug-resistant-bacteria-market (accessed on 15 August 2022).
- Monedero-Recuero, I.; Gegia, M.; Wares, D.F.; Chadha, S.S.; Mirzayev, F. Situational Analysis of 10 Countries with a High Burden of Drug-Resistant Tuberculosis 2 Years Post-UNHLM Declaration: Progress and Setbacks in a Changing Landscape. Int. J. Infect. Dis. 2021, 108, 557–567. [Google Scholar] [CrossRef]
- WHO Newsroom. Cancer. 2022. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer#:~:text=Cancer isa leading cause, and rectum and prostate cancers (accessed on 6 July 2022).
- Rebecca, A.J.; Seigel, L.; Miller, K.D.; Fuchs, H.E. Cancer Statistics; American Cancer Society: Atlanta, Georgia, 2022. [Google Scholar]
- Sin, M.L.; Mach, K.E.; Wong, P.K.; Liao, J.C. Advances and Challenges in Biosensor- Based Diagnosis of Infectious Diseases. Adolesc. Med. 2003, 14, 337–364. [Google Scholar] [CrossRef]
- Mahato, K.; Srivastava, A.; Chandra, P. Paper Based Diagnostics for Personalized Health Care: Emerging Technologies and Commercial Aspects. Biosens. Bioelectron. 2017, 96, 246–259. [Google Scholar] [CrossRef] [PubMed]
- Global Biosensors Market Size to Reach USD 50.7 Billion in 2030|Rising Need for Real-Time Monitoring Devices and Increasing Demand for Point of Care Healthcare Services Are Some Key Factors Driving Industry Demand, Says Emergen Research. Available online: https://www.prnewswire.com/news-releases/global-biosensors-market-size-to-reach-usd-50-7-billion-in-2030--rising-need-for-real-time-monitoring-devices-and-increasing-demand-for-point-of-care-healthcare-services-are-some-key-factors-driving-industry-demand (accessed on 15 August 2022).
- Mancini, A.; Vito, L.; Marcelli, E.; Piangerelli, M.; de Leone, R.; Pucciarelli, S.; Merelli, E. Machine Learning Models Predicting Multidrug Resistant Urinary Tract Infections Using “DsaaS”. BMC Bioinform. 2020, 21, 347. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.H.; Khan, D.M.; Jamal, K.; Ahmad, Z.; Manzoor, S.; Khan, Z. Prediction of Multidrug-Resistant Tuberculosis Using Machine Learning Algorithms in SWAT, Pakistan. J. Healthc Eng. 2021, 2021, 2567080. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).