Distinguishing Healthy and Carcinoma Cell Cultures Using Fluorescence Spectra Decomposition with a Genetic-Algorithm-Based Code
Abstract
:1. Introduction
2. Materials and Methods
2.1. Media
2.2. Cell lines and Culture Conditions
2.3. Cell Concentration and Viability
2.4. Intensity of Fluorescence (IF)
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rui, H.; Hongyu, S.; Dujuan, L.; Weihuang, Y.; Kai, F.; Chaoran, L.; Linxi, D.; Gaofeng, W. A Review of Biosensors for Detecting Tumor Markers in Breast Cancer. Biosensors 2022, 12, 342. [Google Scholar]
- Camarca, A.; Varriale, A.; ACapo, A.; Pennacchio, A.; Calabrese, A.; Giannattasio, C.; Almuzara, C.M.; D’Auria, S.; Staiano, M. Emergent Biosensing Technologies Based on Fluorescence Spectroscopy and Surface Plasmon Resonance. Sensors 2021, 21, 906. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.; Patel, P. Biosensors and biomarkers: Promising tools for cancer diagnosis. Int. J. Biosens. Bioelectron. 2017, 3, 313–316. [Google Scholar] [CrossRef]
- Yaku, K.; Okabe, K.; Hikosaka, K.; Nakagawa, T. NAD Metabolism in Cancer Therapeutics. Front. Oncol. 2018, 8, 622. [Google Scholar] [CrossRef]
- Navas, L.E.; Carnero, A. NAD+ metabolism, stemness, the imunne responses, and cancer. Signal Transduct. Target. Ther. 2021, 6, 2. [Google Scholar] [CrossRef]
- Croce, A.C.; Bottiroli, G. Autofluorescence spectroscopy and imaging: A tool for biomedical research and diagnosis. Eur. J. Histochem. 2014, 58, 2461. [Google Scholar] [CrossRef]
- Aubin, J.E. Autofluorescence of viable cultured mammalian cells. J. Histochem. Cyto. Chem. 1979, 27, 36–43. [Google Scholar] [CrossRef]
- Lmon, J.M.; Ohent, E.; Liet, P.; Hirshberg, J.; Wouters, A.; Ohen, H.; Thorell, B. Microspectrofluorometric Approach to the Study of Free/Bound NAD(P)H Ratio as Metabolic Indicator in Various Cell Types. Photochem. Photohiol. 1982, 36, 585–593. [Google Scholar]
- Kolenc, O.I.; Quinn, K.P. Evaluating Cell Metabolism Through Autofluorescence Imaging of NAD(P)H and FAD. Antioxid. Redox Signal. 2019, 30, 875–889. [Google Scholar] [CrossRef]
- Lakowicz, J.R.; Szmacinski, H.; Nowaczyk, K. Fluorescence lifetime imaging of free and protein bound NADH. Proc. Natl. Acad. Sci. USA 1992, 89, 1271–1275. [Google Scholar] [CrossRef]
- Schwartz, J.P.; Passonneau, J.V.; Johnson, G.S.; Pastan, I. The effect of growth conditions on NAD+. NADH ratio in normal and transformed fibroblast. J. Biol. Chem. 1974, 249, 4138–4143. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Marbois, B.N.; Faull, K.F.; Eckhert, C.D. Esterification of borate with NAD+ and NADH as studied by electrospray ionization mass spectrometry and 11B NMR spectroscopy. J. Mass Spectrom. 2003, 38, 632–640. [Google Scholar] [CrossRef]
- Chance, B.; Cohen, P.; Jobsis, F.; Schoener, B. Intracellular oxidation-reduction states in vivo. Science 1962, 137, 499–508. [Google Scholar] [CrossRef]
- Podrazky, O.; Kuncova, G. Determination of concentration of Living Immobilized Yeast Cells by Fluorescence Spectroscopy. Sens. Actuators B–Chem. 2005, 107, 126–134. [Google Scholar] [CrossRef]
- Ghukasyan, V.V.; Heikal, A.A. (Eds.) Natural Biomarkers for Cellular Metabolism Biology, Techniques, and Applications; Heikal, Series in Cellular and Clinical imaging 2015 by Taylor & Francis Group, LLC.; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar]
- Marose, S.; Lindemann, C.; Scheper, T. Two-Dimensional Fluorescence Spectroscopy: A New Tool for On-Line Bioprocess Monitoring. Biotechnol. Prog. 1998, 14, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Ranzan, L.; Trierweiler, L.F.; Hitzmann, B.; Trierweiler, J.O. Avoiding misleading predictions in fluorescence-based soft sensors using autoencoders. Chemom. Intell. Lab. Syst. 2022, 223, 104527. [Google Scholar] [CrossRef]
- Blacker, T.S.; Duchen, M.R. Investigating mitochondrial redox state using NADH and NADPH autofluorescence Free Radical. Biol. Med. 2016, 100, 53–65. [Google Scholar]
- Sharick, J.T.; Favreau, P.F.; Gillette, A.A.; MSdao, S.M.; Merrins, M.J.; Skala, M.C. Protein-bound NAD(P)H Lifetime is Sensitive to Multiple Fates of Glucose Carbon. Sci. Rep. 2018, 8, 5456. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Battistuz, T.; Bhat, T.N.; Bluhm, W.F.; Bourne, P.E.; Burkhardt, K.; Feng, Z.; Gilliland, G.L.; Iype, L.; Jain, S.; et al. The Protein Data Bank. Nucleic Acids 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Schaefer, P.M.; Kalinina, S.; Rueck, A.; von Arnim CA, F.; von Einem, B. NADH Autofluorescence—A Marker on its Way to Boost Bioenergetic Research. Cytom. Part A 2019, 95, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.U.; Answer, A.G.; Gosnell, M.E.; Mahbub, S.B.; Liu, G.; Goldys, E.M. Fluorescence quenching of free and bound NADH in HeLa cells determined by hyperspectral imaging and unmixing of cell autofluorescence. Biomed. Opt. Express 2017, 8, 1488–1498. [Google Scholar] [CrossRef]
- Piersma, S.R.; Visser AJ, W.G.; de Vries, S.; Duine, J.A. Optical spectro;scopy of nicotinoprotein alcohol dehydrogenase from Amycolatopsis methanolica: A comparison with horse liver alcohol dehydrogenase and UDP-galactose epimerase. Biochemistry 1998, 37, 3068–3077. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, A.; Pal, P.; Durocher, G.; Villeneuve, L.; Balassy, A.; Babai, F.; Gaboury, L.; Blanchard, L. Steady state and time-resolved fluorescence properties of metastatic and non-metastatic malignant cells from different species. J. Photochem. Photobiol. B 1995, 31, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Heikal, A. Two-photon autofluorescence dynamics imaging reveals sensitivity of intracellular NADH concentration and conformation to cell physiology at the single-cell level. J. Photochem. Photobiol. B Biol. 2005, 95, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.C.; Zheng, W.; Qu, J.Y. Sensing cell metabolism by time-resolved auto-fuorescence. Opt. Lett. 2006, 31, 3122–3124. [Google Scholar] [CrossRef]
- Lin, L.S.; Liu, L.N.; Huang, H.F.; Chen, Y.Z.; Li, B.H.; Huang, Z. Characterizing Fluorescence Lifetime of NAD(P)H in Human Leukemic Myeloid Cells and Mononuclear Cells. J. Innov. Opt. Health Sci. 2013, 6, 1350042. [Google Scholar] [CrossRef]
- Villette, S.; Pigaglio-Deshayes, S.; Vever-Bizet, C.; PValidireb, P.; Bourg-Heckly, G. Ultraviolet-induced autofluorescence characterization of normal and tumoral esophageal epithelium cells with quantitation of NAD(P)H. Photochem. Photobiol. Sci. 2006, 5, 483–492. [Google Scholar] [CrossRef] [PubMed]
- KE Conley, K.E.; Ali, A.; Flores, B.; SJubrias, S.; Shankland, E.G. Mitochondrial NAD(P)H in vivo: Identifying natural indicators of oxidative phosphorylation in the 31P magnetic resonance spectrum. Front. Physiol. 2016, 7, 45. [Google Scholar] [CrossRef]
- Zhu, X.H.; Lu, M.; BY Lee, B.Y.; Ugurbil, K.; Chen, W. In Vivo NAD assay reveals the intracellular NAD contents and redox state in healthy human brain and their age dependences. Proc. Natl. Acad. Sci. USA 2015, 112, 2876–2881. [Google Scholar] [CrossRef]
- Ghukasyan, V.V.; Kao, F.J. Monitoring cellular metabolism with fluorescence lifetime of reduced nicotinamide adenine dinucleotide. J. Phys. Chem. C 2009, 113, 11532–11540. [Google Scholar] [CrossRef]
- Bird, D.K.; Yan, L.; Vrotsos, K.M.; Eliceiri, K.W.; Vaughan, E.M.; Keely, P.J.; White, J.G.; Ramanujam, N. Metabolic mapping of MCF10A human breast cells via multiphoton fluorescence lifetime imaging of the coenzyme NADH. Cancer Res. 2005, 65, 8766–8773. [Google Scholar] [CrossRef] [PubMed]
- Leben, R.; Köhler, M.; Radbruch, H.; Hauser, A.E.; Niesner, R.A. Systematic Enzyme Mapping of Cellular Metabolism by Phasor-Analyzed Label-Free NAD(P)H fluorescence Lifetime Imaging. Int. J. Mol. Sci. 2019, 20, 5565. [Google Scholar] [CrossRef]
- Middelburg, T.A.; Hoy, C.L.; Neumann, H.A.M.; Amelik, A.; Robinson, D.J. Correction for tissue optical properties enables quantitative skin fluorescence measurements using multi-diameter single fiber reflectance spectroscopy. J. Dermatol. Sci. 2015, 79, 64–73. [Google Scholar] [CrossRef] [PubMed]
- De Acha, N.; Socorro-Leránoz, A.B.; Elosúa, C.; Matías, I.R. Trends in the Design of Intensity-Based Optical Fiber Biosensors (2010–2020). Biosensors 2021, 11, 197. [Google Scholar] [CrossRef] [PubMed]
- Kozen, D.; Landau, S. Polynomial Decomposition Algorithms. Available online: https://www.cs.cornell.edu/~kozen/Papers/poly.pdf (accessed on 15 September 2022).
- Burstein, E.A.; Abornev, S.M.; Reshetnyak, Y.K. Decomposition of Protein Tryptophan Fluorescence Spectra into Log-Normal Components I. Decomposition Algorithms. Biophys. J. 2001, 81, 1699–1709. [Google Scholar] [CrossRef]
- Hageman, J.A.; Wehrens, R. Direct determination of molecular constants from rovibronic spektra with genetic algorithms. J. Chem. Phys. 2000, 113, 7955–7962. [Google Scholar] [CrossRef]
- Ghaheri, A.; Shoar, S.; Naderan, M.; Hoseini, S.S. The applications of genetic algorithms in medicine. Oman Med. J. 2015, 30, 406–416. [Google Scholar] [CrossRef]
- Adamek, P. Application of Genetic Algorithms to Analysis of High-Parameter Plasma. Ph.D. Thesis, Faculty of Nuclear Physics, Czech Technical University, Prague, Czech, 2003. [Google Scholar]
- Pospisilova, M.; Adamek, P.; Peterka, P.; Kubeček, V.; Kašík, I.; Matějec, V. Influence of Si-Ge-Al-Sb matrices on Tm3+ excitation levels. In Materials Science Forum; Trans Tech Publications Ltd.: Stafa-Zurich, Switzerland, 2007; p. 82. [Google Scholar]
- Sio, R.; Pospisilova, M.; Jarosikova, T. Analyses of emission spectra of biological fluorophores. In Proceedings of the Instruments and Methods for Biology and Medicine Conference 2017, FBME CTU, Kladno, Czech Republic, 5 May 2017; pp. 25–29, ISBN 978-80-01-06303-3. [Google Scholar]
- Available online: https://www.lgcstandards-atcc.org/products/all/CCL-163.aspx (accessed on 25 July 2021).
- Available online: https://www.lgcstandards-atcc.org/products/all/CRL-2638.aspx#culturemethod (accessed on 25 July 2021).
- Park, J.G.; Lee, J.-H.; Kang, M.-S.; Park, K.-J.; Jeon, Y.-M.; Lee, H.-J.; Kwon, H.-S.; Park, H.-S.; Yeo, K.-S.; Lee, K.-U.; et al. Characterization of cell lines established from human hepatocellular carcinoma. Int. J. Cancer 1995, 62, 276–282. [Google Scholar] [CrossRef]
- Available online: https://www.sigmaaldrich.com/technical-documents/protocols/biology/subculture-of-suspension.html (accessed on 25 July 2021).
- FluoroMax®-4 & FluoroMax®-4P with USB. In Operation Manual Part Number 810005 Version B; HORIBA: Longjumeau, France, 2005; p. 236.
- Paul, R.J.; Schneckenburger, H. Oxygen Concentration and the Oxidation-Reduction State of Yeast: Determination of Free/Bound NADH and Flavins by Time-Resolved Spectroscopy. Naturwissenschaften 1993, 83, 32–35. [Google Scholar] [CrossRef]
- Svoboda, J.; Pospisilova, M. Analyses of fluorescence spectra of healthy cells as function of carcinoma cell concentration, pH and temperature of environment. In Proceedings of the Instruments and Methods for Biology and Medicine Conference 2021, FBME CTU, Kladno, Czech Republic, 17 June 2021; pp. 22–26, ISBN 978-80-01-06917-2. [Google Scholar]
- Hahn, D.W. Light Scattering Theory Department of Mechanical and Aerospace Engineering. University of Florida. Available online: http://plaza.ufl.edu/dwhahn/Rayleigh%20and%20Mie%20Light%20Scattering.pdf (accessed on 10 December 2022).
- Shilova, O.N.; Shilov, E.S.; Deyev, S.M. The Effect of Trypan Blue Treatment on Autofluorescence of Fixed Cells. Cytom. Part A 2017, 91, 917–925. [Google Scholar] [CrossRef]
- Yuan, Y.; Yan, Z.; Miao, J.; Cai, R.; Zhang, M.; Wang, Y.; Wang, L.; Dang, W.; Wang, D.; Xiang, D.; et al. Autofluorescence of NADH is a new biomarker for sorting and characterizing cancer stem cells in human glioma. Stem Cell Res. Ther. 2019, 10, 330. [Google Scholar] [CrossRef] [PubMed]


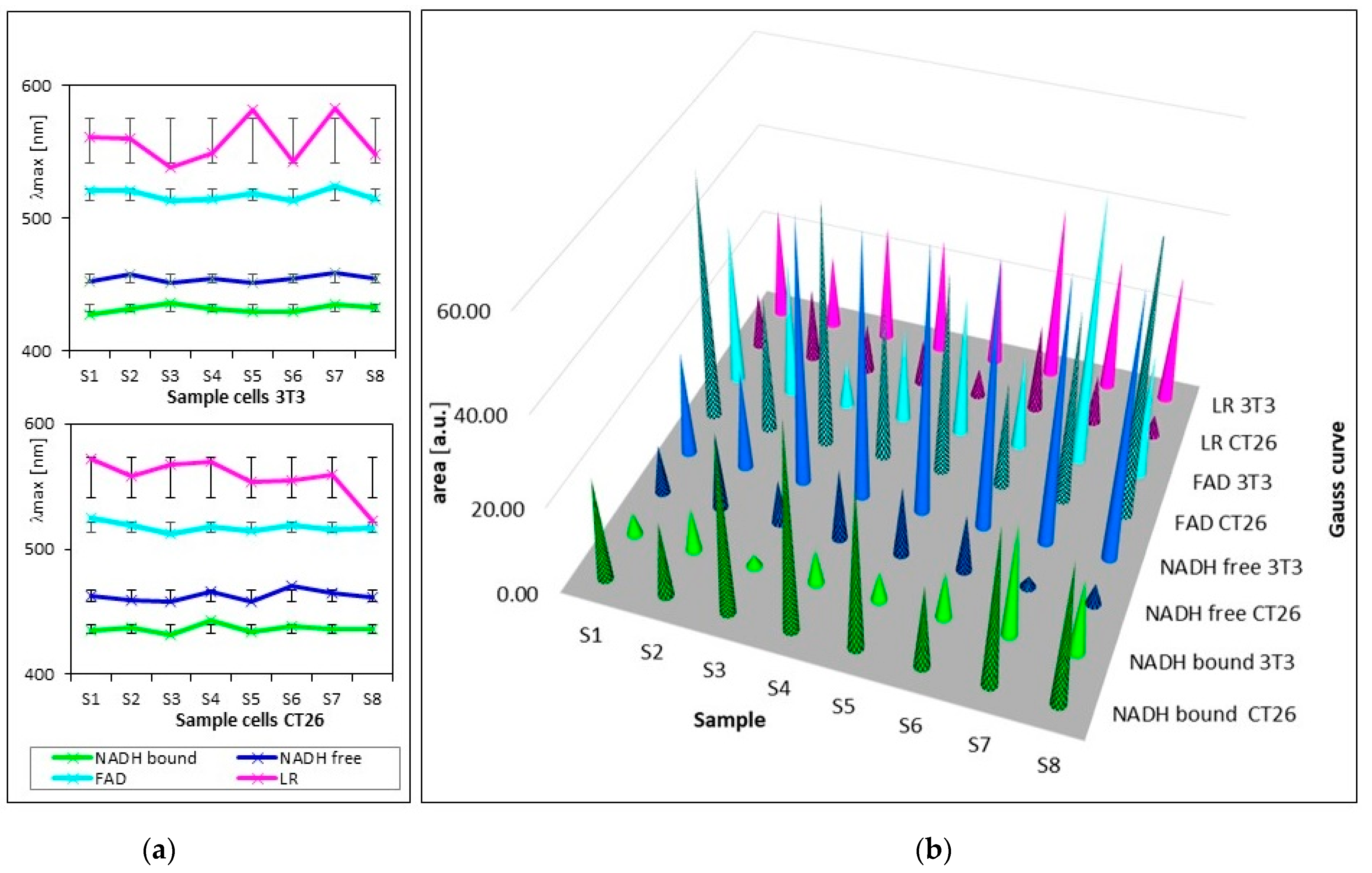
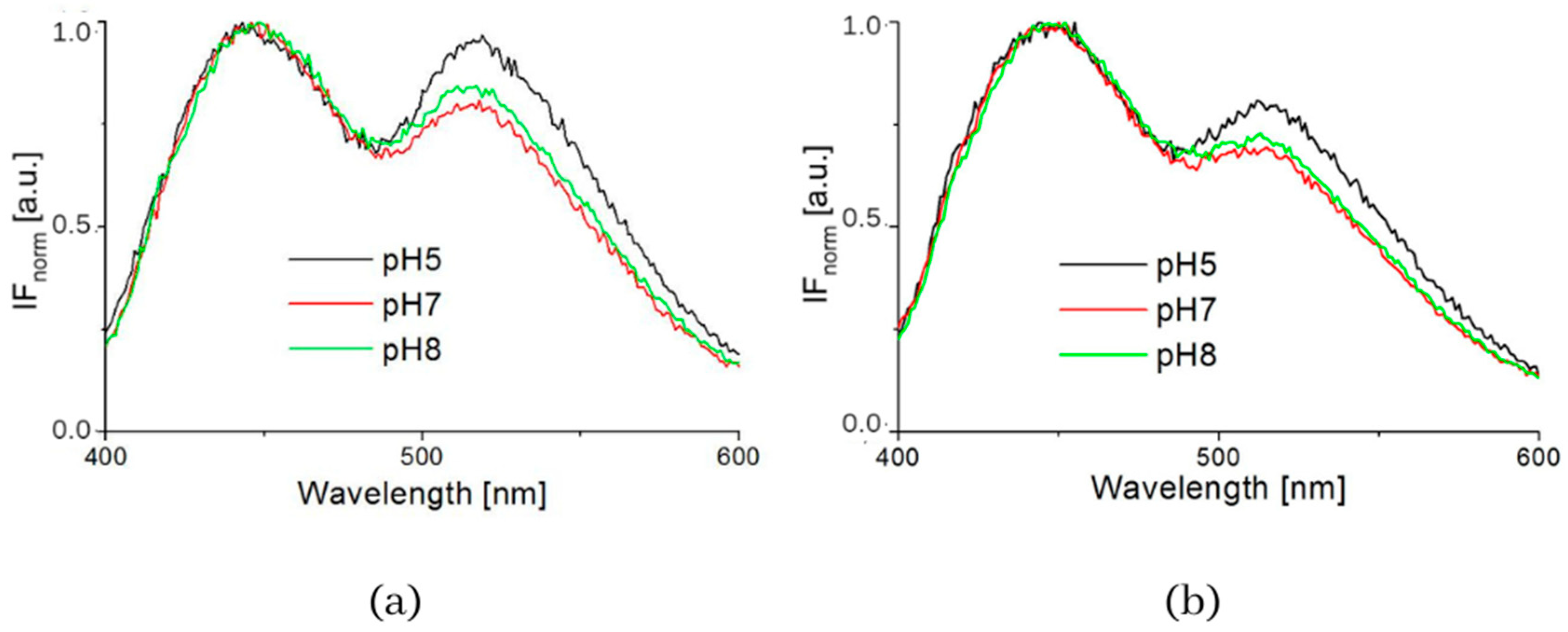
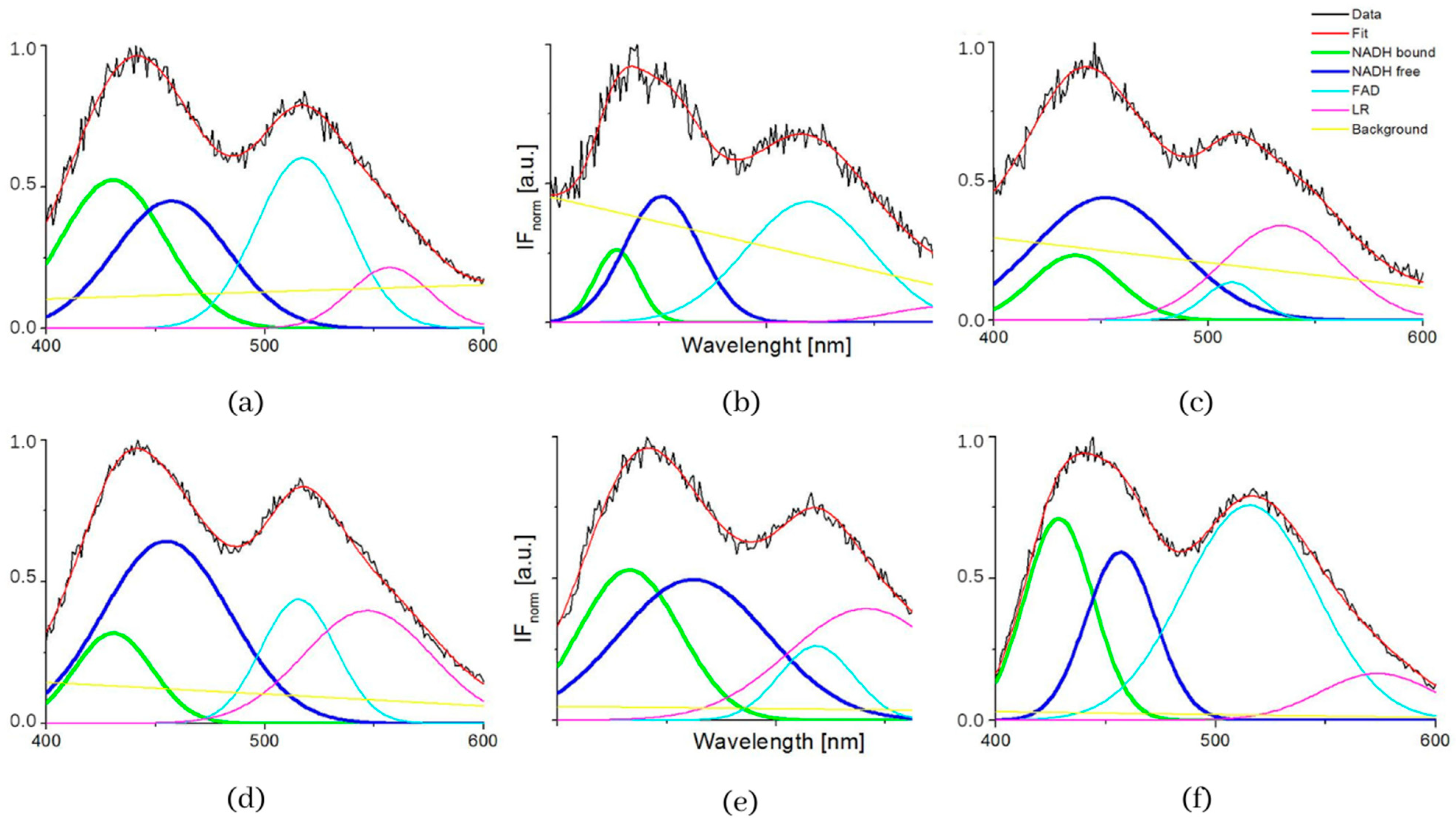


| Samples | S1 | S2 | S3 | S4 | S5 | S6 | S7 | S8 |
|---|---|---|---|---|---|---|---|---|
| Cultivation medium | M I | M II | M II | M II | M II | M II | M II | M II |
| Concentration 3T3 [cells/mL] | 5 × 104 | 1.25 × 105 | 5.9 × 104 | 1.25 × 104 | 1.17 × 105 | 5 × 104 | 2.34 × 105 | 5 × 104 |
| Concentration CT26 [cells/mL] | 5 × 104 | 1.2 × 105 | 1.33 × 105 | 1.37 × 105 | 5 × 104 | 1.25 × 104 | 5 × 104 | 1.25 × 104 |
| AF/AB | S1 | S2 | S3 | S4 | S5 | S6 | S7 | S8 | Average | S * |
|---|---|---|---|---|---|---|---|---|---|---|
| 3T3 | 4.54 | 2.32 | 25 | 2.86 | 3.03 | 2.27 | 1.2 | 2.27 | 2.6 | 7.08 |
| CT26 | 0.48 | 1 | 0.25 | 0.35 | 0.43 | 0.7 | 0.07 | 0.15 | 0.23 | 0.28 |
| Cells | 3T3 | CT26 | ||||
|---|---|---|---|---|---|---|
| pH | 5 | 7 | 8 | 5 | 7 | 8 |
| AF/AB | 0.88 | 3.12 | 3.33 | 3.33 | 0.24 | 0.84 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pospíšilová, M.; Kalábová, H.; Kuncová, G. Distinguishing Healthy and Carcinoma Cell Cultures Using Fluorescence Spectra Decomposition with a Genetic-Algorithm-Based Code. Biosensors 2023, 13, 256. https://doi.org/10.3390/bios13020256
Pospíšilová M, Kalábová H, Kuncová G. Distinguishing Healthy and Carcinoma Cell Cultures Using Fluorescence Spectra Decomposition with a Genetic-Algorithm-Based Code. Biosensors. 2023; 13(2):256. https://doi.org/10.3390/bios13020256
Chicago/Turabian StylePospíšilová, Marie, Hana Kalábová, and Gabriela Kuncová. 2023. "Distinguishing Healthy and Carcinoma Cell Cultures Using Fluorescence Spectra Decomposition with a Genetic-Algorithm-Based Code" Biosensors 13, no. 2: 256. https://doi.org/10.3390/bios13020256
APA StylePospíšilová, M., Kalábová, H., & Kuncová, G. (2023). Distinguishing Healthy and Carcinoma Cell Cultures Using Fluorescence Spectra Decomposition with a Genetic-Algorithm-Based Code. Biosensors, 13(2), 256. https://doi.org/10.3390/bios13020256





