miRNAs as Predictors of Barrier Integrity
Abstract
:1. Introduction
2. miRNAs, EVs, and Physical Barriers
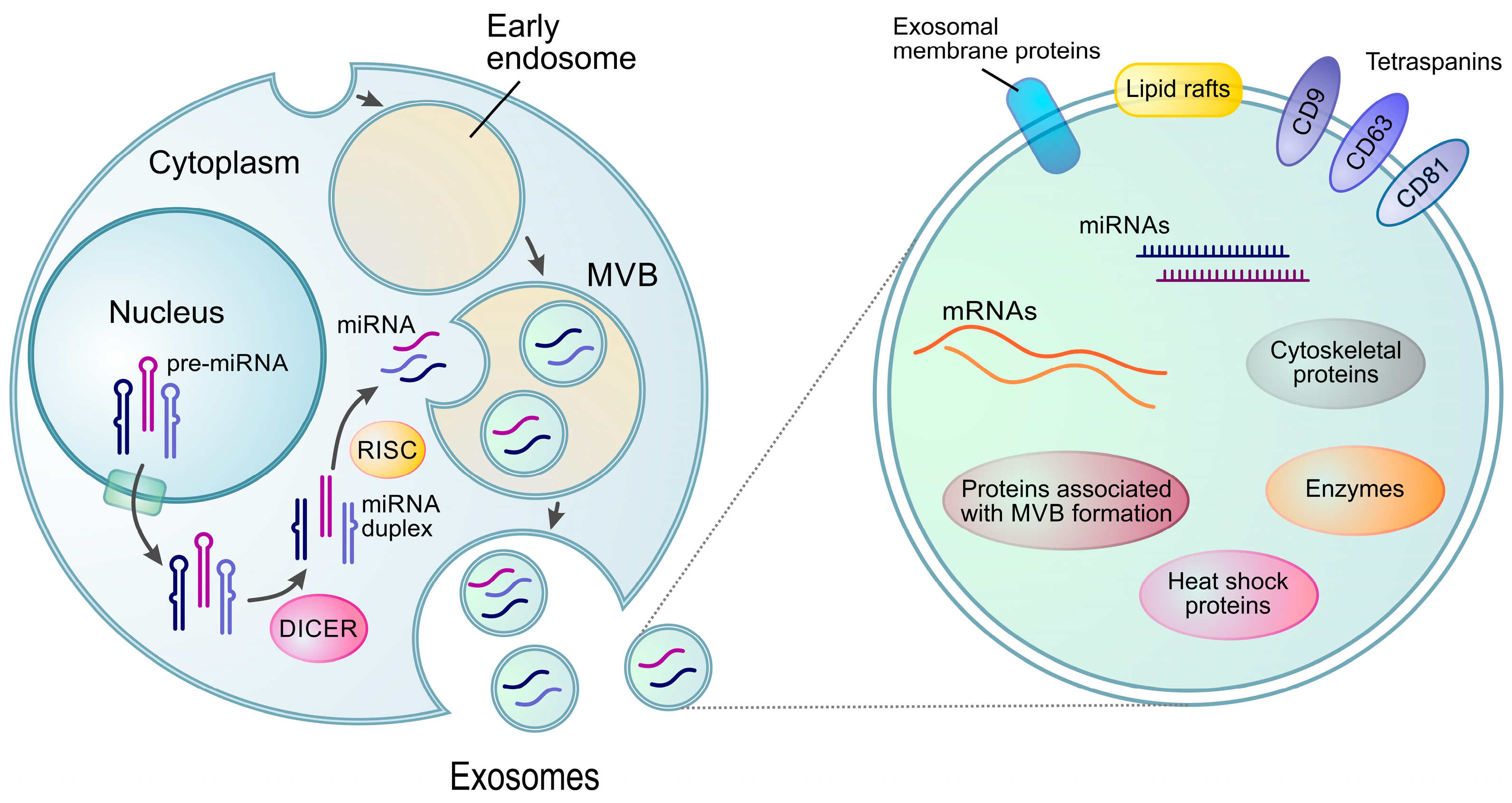
2.1. miRNA Biogenesis and Functions
2.2. miRNAs in Extracellular Vesicles (Exo-miRNAs)
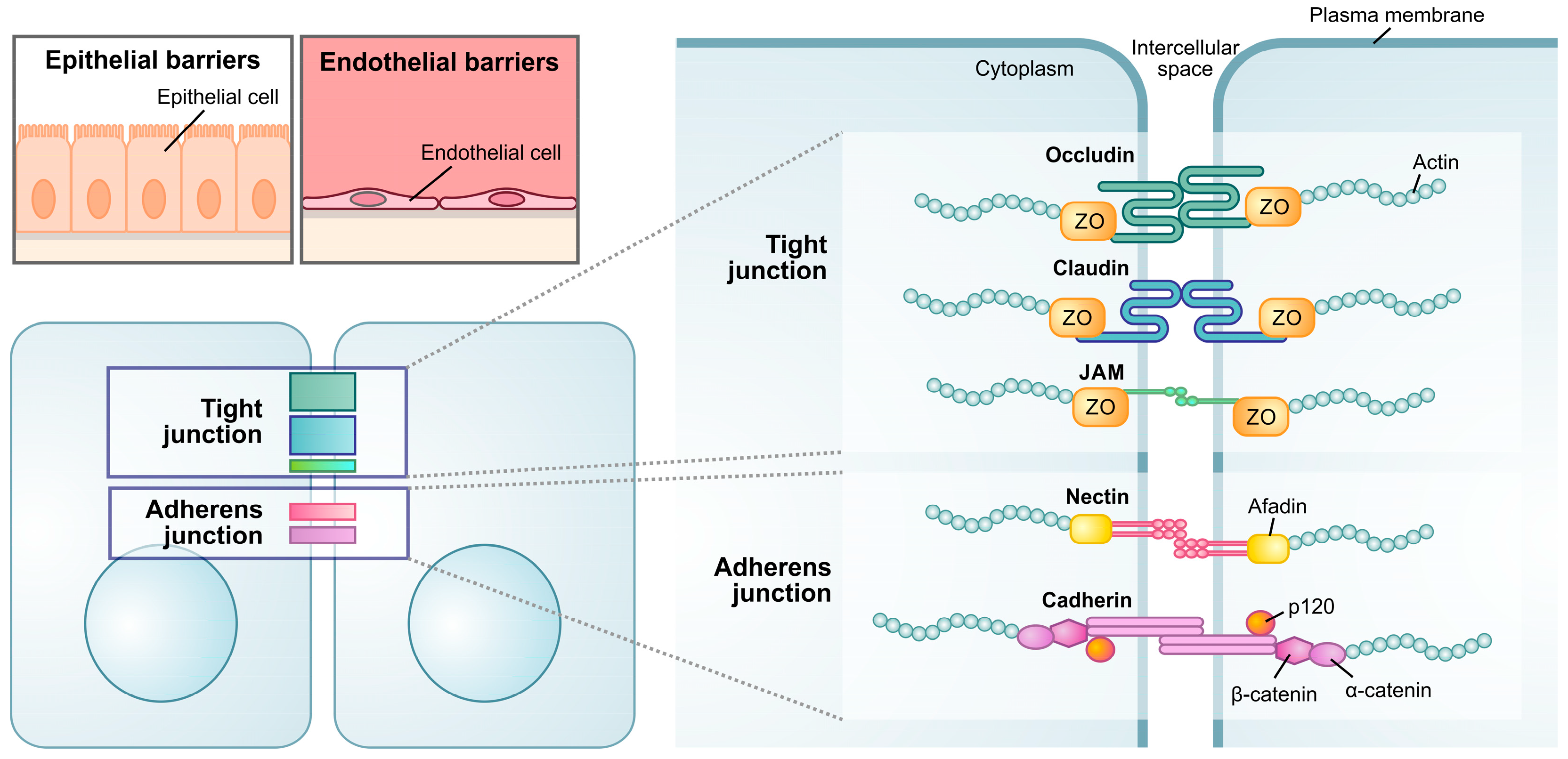
2.3. Maintenance of Physical Barriers
2.4. Maintenance of Tight Junctions
2.5. Defective Intestinal Epithelial Barrier
2.6. Defective Lung Epithelial Barrier
2.7. Defective Urinary Bladder Epithelial Barrier
2.8. Defective Blood–Brain Barrier
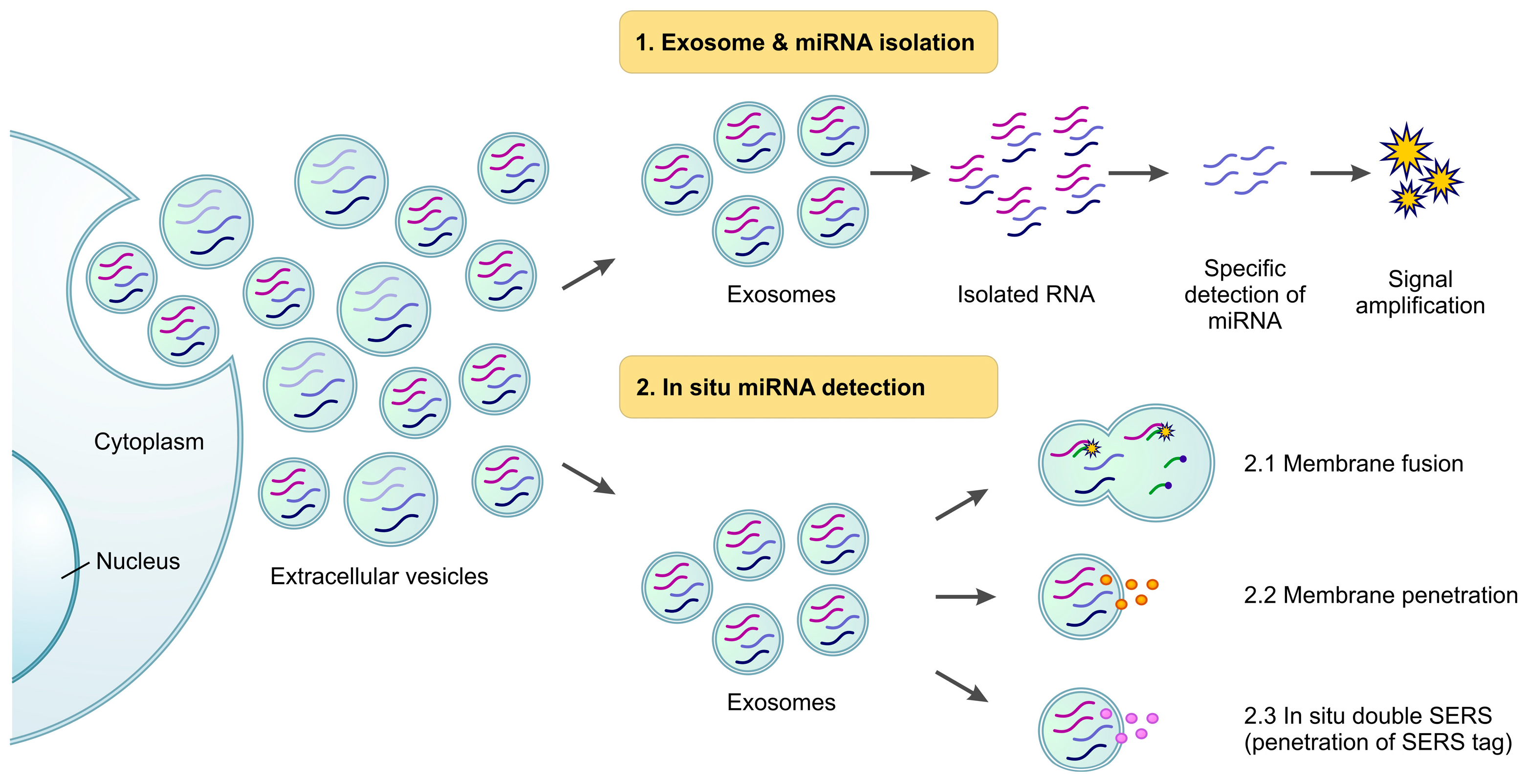
3. Biosensors and miRNA Detection
3.1. Isolation of Exosomes and Exo-miRNAs
3.1.1. Direct Detection of Isolated Exo-miRNAs [123,124] and Sequencing [95]
Direct Amplification of Exo-miRNAs
Amplification-Free Direct miRNA Detection
3.1.2. Exo-miRNA Detection Using Novel Signal Amplification Strategies
Nucleic Acid-Based Strategies
- Nucleic acid capture probes
- DNA self-assembly induced signal amplification
- Isothermal amplification
Nucleic Acid-Synergized Strategies
- Nanomaterials
- Microfluidics
Signal Sensing Strategies
- SERS (Surface-Enhanced Raman Scattering)
- SPR (Surface Plasmon Resonance)
- Electrochemical detection
3.2. In Situ Detection
3.2.1. Membrane Fusion
3.2.2. Penetration
3.2.3. In Situ Double SERS (Surface-Enhanced Raman Scattering)
4. Discussion
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002, 12, 735–739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, G.; Zhou, Y.; Chen, X. New Insight into Inter-kingdom Communication: Horizontal Transfer of Mobile Small RNAs. Front. Microbiol. 2017, 8, 768. [Google Scholar] [CrossRef] [PubMed]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Saunders, N.R.; Dziegielewska, K.M.; Møllgård, K.; Habgood, M.D. Physiology and molecular biology of barrier mechanisms in the fetal and neonatal brain. J. Physiol. 2018, 596, 5723–5756. [Google Scholar] [CrossRef] [Green Version]
- Scalise, A.A.; Kakogiannos, N.; Zanardi, F.; Iannelli, F.; Giannotta, M. The blood-brain and gut-vascular barriers: From the perspective of claudins. Tissue Barriers 2021, 9, 1926190. [Google Scholar] [CrossRef]
- Okada, C.; Yamashita, E.; Lee, S.J.; Shibata, S.; Katahira, J.; Nakagawa, A.; Yoneda, Y.; Tsukihara, T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science 2009, 326, 1275–1279. [Google Scholar] [CrossRef]
- Alarcón, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef] [Green Version]
- Denli, A.M.; Tops, B.B.; Plasterk, R.H.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. (Lausanne) 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoda, M.; Kawamata, T.; Paroo, Z.; Ye, X.; Iwasaki, S.; Liu, Q.; Tomari, Y. ATP-dependent human RISC assembly pathways. Nat. Struct. Mol. Biol. 2010, 17, 17–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic microRNA precursors that bypass Drosha processing. Nature 2007, 448, 83–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single processing center models for human Dicer and bacterial RNase III. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef] [Green Version]
- Ipsaro, J.J.; Joshua-Tor, L. From guide to target: Molecular insights into eukaryotic RNA-interference machinery. Nat. Struct. Mol. Biol. 2015, 22, 20–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef]
- Xu, K.; Lin, J.; Zandi, R.; Roth, J.A.; Ji, L. MicroRNA-mediated target mRNA cleavage and 3’-uridylation in human cells. Sci. Rep. 2016, 6, 30242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pirrò, S.; Matic, I.; Colizzi, V.; Galgani, A. The microRNA analysis portal is a next-generation tool for exploring and analyzing miRNA-focused data in the literature. Sci. Rep. 2021, 11, 9007. [Google Scholar] [CrossRef]
- Pillai, R.S. MicroRNA function: Multiple mechanisms for a tiny RNA? RNA 2005, 11, 1753–1761. [Google Scholar] [CrossRef] [Green Version]
- Sacks, D.; Baxter, B.; Campbell, B.C.V.; Carpenter, J.S.; Cognard, C.; Dippel, D.; Eesa, M.; Fischer, U.; Hausegger, K.; Hirsch, J.A.; et al. Multisociety Consensus Quality Improvement Revised Consensus Statement for Endovascular Therapy of Acute Ischemic Stroke. Int. J. Stroke 2018, 13, 612–632. [Google Scholar] [CrossRef] [Green Version]
- McCreight, J.C.; Schneider, S.E.; Wilburn, D.B.; Swanson, W.J. Evolution of microRNA in primates. PLoS ONE 2017, 12, e0176596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Connell, R.M.; Rao, D.S.; Chaudhuri, A.A.; Baltimore, D. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 2010, 10, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Binderup, H.G.; Madsen, J.S.; Heegaard, N.H.H.; Houlind, K.; Andersen, R.F.; Brasen, C.L. Quantification of microRNA levels in plasma - Impact of preanalytical and analytical conditions. PLoS ONE 2018, 13, e0201069. [Google Scholar] [CrossRef]
- Mompeón, A.; Ortega-Paz, L.; Vidal-Gómez, X.; Costa, T.J.; Pérez-Cremades, D.; Garcia-Blas, S.; Brugaletta, S.; Sanchis, J.; Sabate, M.; Novella, S.; et al. Disparate miRNA expression in serum and plasma of patients with acute myocardial infarction: A systematic and paired comparative analysis. Sci. Rep. 2020, 10, 5373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aftab, M.; Poojary, S.S.; Seshan, V.; Kumar, S.; Agarwal, P.; Tandon, S.; Zutshi, V.; Das, B.C. Urine miRNA signature as a potential non-invasive diagnostic and prognostic biomarker in cervical cancer. Sci. Rep. 2021, 11, 10323. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Lozano, E.; Sebastián-Valles, F.; Knott-Torcal, C. Circulating microRNAs in Breast Milk and Their Potential Impact on the Infant. Nutrients 2020, 12, 3066. [Google Scholar] [CrossRef]
- Rashid, H.; Hossain, B.; Siddiqua, T.; Kabir, M.; Noor, Z.; Ahmed, M.; Haque, R. Fecal MicroRNAs as Potential Biomarkers for Screening and Diagnosis of Intestinal Diseases. Front. Mol. Biosci. 2020, 7, 181. [Google Scholar] [CrossRef]
- Willms, E.; Cabañas, C.; Mäger, I.; Wood, M.J.A.; Vader, P. Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front. Immunol. 2018, 9, 738. [Google Scholar] [CrossRef] [Green Version]
- Saksena, S.; Sun, J.; Chu, T.; Emr, S.D. ESCRTing proteins in the endocytic pathway. Trends Biochem. Sci. 2007, 32, 561–573. [Google Scholar] [CrossRef]
- Termini, C.M.; Gillette, J.M. Tetraspanins Function as Regulators of Cellular Signaling. Front. Cell Dev. Biol. 2017, 5, 34. [Google Scholar] [CrossRef] [Green Version]
- Sønder, S.L.; Boye, T.L.; Tölle, R.; Dengjel, J.; Maeda, K.; Jäättelä, M.; Simonsen, A.C.; Jaiswal, J.K.; Nylandsted, J. Annexin A7 is required for ESCRT III-mediated plasma membrane repair. Sci. Rep. 2019, 9, 6726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otto, G.P.; Nichols, B.J. The roles of flotillin microdomains--endocytosis and beyond. J. Cell Sci. 2011, 124, 3933–3940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassani, S.; Cingolani, L.A. Tetraspanins: Interactions and interplay with integrins. Int. J. Biochem. Cell Biol. 2012, 44, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Keerthikumar, S.; Gangoda, L.; Liem, M.; Fonseka, P.; Atukorala, I.; Ozcitti, C.; Mechler, A.; Adda, C.G.; Ang, C.S.; Mathivanan, S. Proteogenomic analysis reveals exosomes are more oncogenic than ectosomes. Oncotarget 2015, 6, 15375–15396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsukita, S.; Furuse, M.; Itoh, M. Multifunctional strands in tight junctions. Nat. Rev. Mol. Cell Biol. 2001, 2, 285–293. [Google Scholar] [CrossRef]
- Dejana, E. Endothelial cell-cell junctions: Happy together. Nat. Rev. Mol. Cell Biol. 2004, 5, 261–270. [Google Scholar] [CrossRef]
- Schneeberger, E.E.; Lynch, R.D. The tight junction: A multifunctional complex. Am. J. Physiol. Cell Physiol. 2004, 286, C1213–C1228. [Google Scholar] [CrossRef] [Green Version]
- Jang, A.S. The apical junctional complex in respiratory diseases. Chonnam Med. J. 2014, 50, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Rios-Arce, N.D.; Collins, F.L.; Schepper, J.D.; Steury, M.D.; Raehtz, S.; Mallin, H.; Schoenherr, D.T.; Parameswaran, N.; McCabe, L.R. Epithelial Barrier Function in Gut-Bone Signaling. Adv. Exp. Med. Biol. 2017, 1033, 151–183. [Google Scholar] [CrossRef] [Green Version]
- Heinemann, U.; Schuetz, A. Structural Features of Tight-Junction Proteins. Int. J. Mol. Sci. 2019, 20, 6020. [Google Scholar] [CrossRef] [Green Version]
- Ebnet, K.; Suzuki, A.; Ohno, S.; Vestweber, D. Junctional adhesion molecules (JAMs): More molecules with dual functions? J. Cell Sci. 2004, 117, 19–29. [Google Scholar] [CrossRef] [Green Version]
- Bauer, H.; Zweimueller-Mayer, J.; Steinbacher, P.; Lametschwandtner, A.; Bauer, H.C. The dual role of zonula occludens (ZO) proteins. J. Biomed. Biotechnol. 2010, 2010, 402593. [Google Scholar] [CrossRef] [Green Version]
- Rider, L.; Tao, J.; Snyder, S.; Brinley, B.; Lu, J.; Diakonova, M. Adapter protein SH2B1beta cross-links actin filaments and regulates actin cytoskeleton. Mol. Endocrinol. 2009, 23, 1065–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortiz, M.A.; Mikhailova, T.; Li, X.; Porter, B.A.; Bah, A.; Kotula, L. Src family kinases, adaptor proteins and the actin cytoskeleton in epithelial-to-mesenchymal transition. Cell Commun. Signal. 2021, 19, 67. [Google Scholar] [CrossRef] [PubMed]
- Gumbiner, B.M. Cell adhesion: The molecular basis of tissue architecture and morphogenesis. Cell 1996, 84, 345–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Awad, W.A.; Hess, C.; Hess, M. Enteric Pathogens and Their Toxin-Induced Disruption of the Intestinal Barrier through Alteration of Tight Junctions in Chickens. Toxins 2017, 9, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eichner, M.; Protze, J.; Piontek, A.; Krause, G.; Piontek, J. Targeting and alteration of tight junctions by bacteria and their virulence factors such as Clostridium perfringens enterotoxin. Pflugers Arch. 2017, 469, 77–90. [Google Scholar] [CrossRef]
- Ding, G.; Shao, Q.; Yu, H.; Liu, J.; Li, Y.; Wang, B.; Sang, H.; Li, D.; Bing, A.; Hou, Y.; et al. Tight Junctions, the Key Factor in Virus-Related Disease. Pathogens 2022, 11, 1200. [Google Scholar] [CrossRef]
- Fu, Q.; Lin, Q.; Chen, D.; Yu, B.; Luo, Y.; Zheng, P.; Mao, X.; Huang, Z.; Yu, J.; Luo, J.; et al. β-defensin 118 attenuates inflammation and injury of intestinal epithelial cells upon enterotoxigenic Escherichia coli challenge. BMC Vet. Res. 2022, 18, 142. [Google Scholar] [CrossRef]
- Bhat, A.A.; Uppada, S.; Achkar, I.W.; Hashem, S.; Yadav, S.K.; Shanmugakonar, M.; Al-Naemi, H.A.; Haris, M.; Uddin, S. Tight Junction Proteins and Signaling Pathways in Cancer and Inflammation: A Functional Crosstalk. Front. Physiol. 2018, 9, 1942. [Google Scholar] [CrossRef] [Green Version]
- Takano, K.; Kojima, T.; Sawada, N.; Himi, T. Role of tight junctions in signal transduction: An update. EXCLI J. 2014, 13, 1145–1162. [Google Scholar] [PubMed]
- Zhuang, Y.; Peng, H.; Mastej, V.; Chen, W. MicroRNA Regulation of Endothelial Junction Proteins and Clinical Consequence. Mediators Inflamm 2016, 2016, 5078627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cichon, C.; Sabharwal, H.; Rüter, C.; Schmidt, M.A. MicroRNAs regulate tight junction proteins and modulate epithelial/endothelial barrier functions. Tissue Barriers 2014, 2, e944446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Didiano, D.; Hobert, O. Molecular architecture of a miRNA-regulated 3’ UTR. RNA 2008, 14, 1297–1317. [Google Scholar] [CrossRef] [Green Version]
- Ye, D.; Guo, S.; Al-Sadi, R.; Ma, T.Y. MicroRNA regulation of intestinal epithelial tight junction permeability. Gastroenterology 2011, 141, 1323–1333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGuckin, M.A.; Eri, R.; Simms, L.A.; Florin, T.H.; Radford-Smith, G. Intestinal barrier dysfunction in inflammatory bowel diseases. Inflamm. Bowel. Dis. 2009, 15, 100–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, T.Y.; Li, Y.Q.; Zhao, F.Q.; Sun, H.M.; Gao, Y.; Wu, B.; Yang, S.; Ji, F.Q.; Zhou, D.S. MiR-1-3p and MiR-124-3p Synergistically Damage the Intestinal Barrier in the Ageing Colon. J. Crohns Colitis 2022, 16, 656–667. [Google Scholar] [CrossRef]
- Gitter, A.H.; Wullstein, F.; Fromm, M.; Schulzke, J.D. Epithelial barrier defects in ulcerative colitis: Characterization and quantification by electrophysiological imaging. Gastroenterology 2001, 121, 1320–1328. [Google Scholar] [CrossRef]
- Kotla, N.G.; Isa, I.L.M.; Rasala, S.; Demir, S.; Singh, R.; Baby, B.V.; Swamy, S.K.; Dockery, P.; Jala, V.R.; Rochev, Y.; et al. Modulation of Gut Barrier Functions in Ulcerative Colitis by Hyaluronic Acid System. Adv. Sci. (Weinh) 2022, 9, e2103189. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, J.; Gao, Y.; Shen, L.; Li, S.; Chen, S. miRNA-Based Potential Biomarkers and New Molecular Insights in Ulcerative Colitis. Front. Pharmacol. 2021, 12, 707776. [Google Scholar] [CrossRef]
- Schumann, M.; Siegmund, B.; Schulzke, J.D.; Fromm, M. Celiac Disease: Role of the Epithelial Barrier. Cell. Mol. Gastroenterol. Hepatol. 2017, 3, 150–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felli, C.; Baldassarre, A.; Masotti, A. Intestinal and Circulating MicroRNAs in Coeliac Disease. Int. J. Mol. Sci. 2017, 18, 1907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, I.L.; Coutinho de Almeida, R.; Modderman, R.; Stachurska, A.; Dekens, J.; Barisani, D.; Meijer, C.R.; Roca, M.; Martinez-Ojinaga, E.; Shamir, R.; et al. Circulating miRNAs as Potential Biomarkers for Celiac Disease Development. Front. Immunol. 2021, 12, 734763. [Google Scholar] [CrossRef]
- Friebel, J.; Schinnerling, K.; Geelhaar-Karsch, A.; Allers, K.; Schneider, T.; Moos, V. Intestinal barrier dysfunction mediates Whipple’s disease immune reconstitution inflammatory syndrome (IRIS). Immun. Inflamm. Dis. 2022, 10, e622. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Winton, H.L.; Soeller, C.; Taylor, G.W.; Gruenert, D.C.; Thompson, P.J.; Cannell, M.B.; Stewart, G.A.; Garrod, D.R.; Robinson, C. The transmembrane protein occludin of epithelial tight junctions is a functional target for serine peptidases from faecal pellets of Dermatophagoides pteronyssinus. Clin. Exp. Allergy 2001, 31, 279–294. [Google Scholar] [CrossRef]
- Heijink, I.H.; Kuchibhotla, V.N.S.; Roffel, M.P.; Maes, T.; Knight, D.A.; Sayers, I.; Nawijn, M.C. Epithelial cell dysfunction, a major driver of asthma development. Allergy 2020, 75, 1902–1917. [Google Scholar] [CrossRef]
- Taka, S.; Tzani-Tzanopoulou, P.; Wanstall, H.; Papadopoulos, N.G. MicroRNAs in Asthma and Respiratory Infections: Identifying Common Pathways. Allergy Asthma Immunol. Res. 2020, 12, 4–23. [Google Scholar] [CrossRef]
- Mattila, P.; Joenväärä, S.; Renkonen, J.; Toppila-Salmi, S.; Renkonen, R. Allergy as an epithelial barrier disease. Clin. Transl. Allergy 2011, 1, 5. [Google Scholar] [CrossRef] [Green Version]
- Weidner, J.; Bartel, S.; Kılıç, A.; Zissler, U.M.; Renz, H.; Schwarze, J.; Schmidt-Weber, C.B.; Maes, T.; Rebane, A.; Krauss-Etschmann, S.; et al. Spotlight on microRNAs in allergy and asthma. Allergy 2021, 76, 1661–1678. [Google Scholar] [CrossRef]
- Hammad, N.M.; Nabil, F.; Elbehedy, E.M.; Sedeek, R.; Gouda, M.I.; Arafa, M.A.; Elalawi, S.M.; El Shahawy, A.A. Role of MicroRNA-155 as a Potential Biomarker for Allergic Rhinitis in Children. Can. Respir. J. 2021, 2021, 5554461. [Google Scholar] [CrossRef]
- Hoyer, N.; Wille, M.M.W.; Thomsen, L.H.; Wilcke, T.; Dirksen, A.; Pedersen, J.H.; Saghir, Z.; Ashraf, H.; Shaker, S.B. Interstitial lung abnormalities are associated with increased mortality in smokers. Respir. Med. 2018, 136, 77–82. [Google Scholar] [CrossRef] [Green Version]
- Ortiz-Quintero, B.; Buendía-Roldán, I.; Ramírez-Salazar, E.G.; Balderas-Martínez, Y.I.; Ramírez-Rodríguez, S.L.; Martínez-Espinosa, K.; Selman, M. Circulating microRNA Signature Associated to Interstitial Lung Abnormalities in Respiratory Asymptomatic Subjects. Cells 2020, 9, 1556. [Google Scholar] [CrossRef]
- Ali, M.S.; Singh, J.; Alam, M.T.; Chopra, A.; Arava, S.; Bhalla, A.S.; Mittal, S.; Mohan, A.; Mitra, D.K.; Hadda, V. Non-coding RNA in idiopathic interstitial pneumonia and Covid-19 pulmonary fibrosis. Mol. Biol. Rep. 2022, 49, 11535–11546. [Google Scholar] [CrossRef]
- Gonzales, J.N.; Lucas, R.; Verin, A.D. The Acute Respiratory Distress Syndrome: Mechanisms and Perspective Therapeutic Approaches. Austin J. Vasc. Med. 2015, 2, 1009. [Google Scholar]
- Lee, L.K.; Medzikovic, L.; Eghbali, M.; Eltzschig, H.K.; Yuan, X. The Role of MicroRNAs in Acute Respiratory Distress Syndrome and Sepsis, From Targets to Therapies: A Narrative Review. Anesth. Analg. 2020, 131, 1471–1484. [Google Scholar] [CrossRef]
- Lu, Q.; Yu, S.; Meng, X.; Shi, M.; Huang, S.; Li, J.; Zhang, J.; Liang, Y.; Ji, M.; Zhao, Y.; et al. MicroRNAs: Important Regulatory Molecules in Acute Lung Injury/Acute Respiratory Distress Syndrome. Int. J. Mol. Sci. 2022, 23, 5545. [Google Scholar] [CrossRef]
- Jafari, N.V.; Rohn, J.L. The urothelium: A multi-faceted barrier against a harsh environment. Mucosal. Immunol. 2022, 15, 1127–1142. [Google Scholar] [CrossRef]
- Urabe, F.; Furuta, A.; Igarashi, T.; Suzuki, Y.; Egawa, S.; Kimura, T. Urinary extracellular vesicle microRNA profiling for detection in patients with interstitial cystitis. Transl. Androl. Urol. 2022, 11, 1063–1066. [Google Scholar] [CrossRef]
- Sweeney, M.D.; Sagare, A.P.; Zlokovic, B.V. Blood-brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat. Rev. Neurol. 2018, 14, 133–150. [Google Scholar] [CrossRef]
- Bekris, L.M.; Lutz, F.; Montine, T.J.; Yu, C.E.; Tsuang, D.; Peskind, E.R.; Leverenz, J.B. MicroRNA in Alzheimer’s disease: An exploratory study in brain, cerebrospinal fluid and plasma. Biomarkers 2013, 18, 455–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hara, N.; Kikuchi, M.; Miyashita, A.; Hatsuta, H.; Saito, Y.; Kasuga, K.; Murayama, S.; Ikeuchi, T.; Kuwano, R. Serum microRNA miR-501-3p as a potential biomarker related to the progression of Alzheimer’s disease. Acta Neuropathol. Commun. 2017, 5, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balasa, R.; Barcutean, L.; Mosora, O.; Manu, D. Reviewing the Significance of Blood-Brain Barrier Disruption in Multiple Sclerosis Pathology and Treatment. Int. J. Mol. Sci. 2021, 22, 8370. [Google Scholar] [CrossRef]
- Piotrzkowska, D.; Miller, E.; Kucharska, E.; Niwald, M.; Majsterek, I. Association of miRNA and mRNA Levels of the Clinical Onset of Multiple Sclerosis Patients. Biology 2021, 10, 554. [Google Scholar] [CrossRef]
- Najafi, N.; Peymani, M. A genetic variant of pri-miR-182 may impact the risk for the onset of multiple sclerosis in the Iranian population. Am. J. Hum. Biol. 2020, 32, e23415. [Google Scholar] [CrossRef] [PubMed]
- Knowland, D.; Arac, A.; Sekiguchi, K.J.; Hsu, M.; Lutz, S.E.; Perrino, J.; Steinberg, G.K.; Barres, B.A.; Nimmerjahn, A.; Agalliu, D. Stepwise recruitment of transcellular and paracellular pathways underlies blood-brain barrier breakdown in stroke. Neuron 2014, 82, 603–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shu, B.; Zhang, X.; Du, G.; Fu, Q.; Huang, L. MicroRNA-107 prevents amyloid-β-induced neurotoxicity and memory impairment in mice. Int. J. Mol. Med. 2018, 41, 1665–1672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Vliet, E.A.; Aronica, E.; Gorter, J.A. Blood-brain barrier dysfunction, seizures and epilepsy. Semin. Cell Dev. Biol. 2015, 38, 26–34. [Google Scholar] [CrossRef]
- Marchi, N.; Granata, T.; Ghosh, C.; Janigro, D. Blood-brain barrier dysfunction and epilepsy: Pathophysiologic role and therapeutic approaches. Epilepsia 2012, 53, 1877–1886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alsharafi, W.A.; Xiao, B.; Abuhamed, M.M.; Luo, Z. miRNAs: Biological and clinical determinants in epilepsy. Front. Mol. Neurosci. 2015, 8, 59. [Google Scholar] [CrossRef] [Green Version]
- Fiorentino, M.; Sapone, A.; Senger, S.; Camhi, S.S.; Kadzielski, S.M.; Buie, T.M.; Kelly, D.L.; Cascella, N.; Fasano, A. Blood-brain barrier and intestinal epithelial barrier alterations in autism spectrum disorders. Mol. Autism. 2016, 7, 49. [Google Scholar] [CrossRef] [Green Version]
- Wong, A.; Zhou, A.; Cao, X.; Mahaganapathy, V.; Azaro, M.; Gwin, C.; Wilson, S.; Buyske, S.; Bartlett, C.W.; Flax, J.F.; et al. MicroRNA and MicroRNA-Target Variants Associated with Autism Spectrum Disorder and Related Disorders. Genes 2022, 13, 1329. [Google Scholar] [CrossRef]
- Moore, D.; Meays, B.M.; Madduri, L.S.V.; Shahjin, F.; Chand, S.; Niu, M.; Albahrani, A.; Guda, C.; Pendyala, G.; Fox, H.S.; et al. Downregulation of an Evolutionary Young miR-1290 in an iPSC-Derived Neural Stem Cell Model of Autism Spectrum Disorder. Stem. Cells Int. 2019, 2019, 8710180. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Zhu, Y.; Williams, S.; Watts, M.; Tonta, M.A.; Coleman, H.A.; Parkington, H.C.; Claudianos, C. Autism-associated miR-873 regulates ARID1B, SHANK3 and NRXN2 involved in neurodevelopment. Transl. Psychiatry 2020, 10, 418. [Google Scholar] [CrossRef]
- Ghahramani Seno, M.M.; Hu, P.; Gwadry, F.G.; Pinto, D.; Marshall, C.R.; Casallo, G.; Scherer, S.W. Gene and miRNA expression profiles in autism spectrum disorders. Brain Res. 2011, 1380, 85–97. [Google Scholar] [CrossRef]
- Jin, X.; Chen, Y.; Chen, H.; Fei, S.; Chen, D.; Cai, X.; Liu, L.; Lin, B.; Su, H.; Zhao, L.; et al. Evaluation of Tumor-Derived Exosomal miRNA as Potential Diagnostic Biomarkers for Early-Stage Non-Small Cell Lung Cancer Using Next-Generation Sequencing. Clin. Cancer Res. 2017, 23, 5311–5319. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Shen, J.; Cheng, J.; Fan, X. MicroRNA-21 regulates intestinal epithelial tight junction permeability. Cell. Biochem. Funct. 2015, 33, 235–240. [Google Scholar] [CrossRef]
- Liu, Z.; Li, C.; Chen, S.; Lin, H.; Zhao, H.; Liu, M.; Weng, J.; Liu, T.; Li, X.; Lei, C.; et al. MicroRNA-21 increases the expression level of occludin through regulating ROCK1 in prevention of intestinal barrier dysfunction. J. Cell. Biochem. 2019, 120, 4545–4554. [Google Scholar] [CrossRef]
- Smits, H.H.; van der Vlugt, L.E.; von Mutius, E.; Hiemstra, P.S. Childhood allergies and asthma: New insights on environmental exposures and local immunity at the lung barrier. Curr. Opin. Immunol. 2016, 42, 41–47. [Google Scholar] [CrossRef]
- Zhao, N.; Sun, H.; Sun, B.; Zhu, D.; Zhao, X.; Wang, Y.; Gu, Q.; Dong, X.; Liu, F.; Zhang, Y.; et al. miR-27a-3p suppresses tumor metastasis and VM by down-regulating VE-cadherin expression and inhibiting EMT: An essential role for Twist-1 in HCC. Sci. Rep. 2016, 6, 23091. [Google Scholar] [CrossRef] [Green Version]
- Harati, R.; Hammad, S.; Tlili, A.; Mahfood, M.; Mabondzo, A.; Hamoudi, R. miR-27a-3p regulates expression of intercellular junctions at the brain endothelium and controls the endothelial barrier permeability. PLoS ONE 2022, 17, e0262152. [Google Scholar] [CrossRef]
- Eyking, A.; Reis, H.; Frank, M.; Gerken, G.; Schmid, K.W.; Cario, E. MiR-205 and MiR-373 Are Associated with Aggressive Human Mucinous Colorectal Cancer. PLoS ONE 2016, 11, e0156871. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Li, X.; Zhong, Z.; Qiu, Y.; Liu, S.; Wu, H.; Tang, X.; Chen, C.; Fu, Y.; Chen, Q.; et al. 3D hESC exosomes enriched with miR-6766-3p ameliorates liver fibrosis by attenuating activated stellate cells through targeting the TGFβRII-SMADS pathway. J. Nanobiotechnol. 2021, 19, 437. [Google Scholar] [CrossRef]
- Zhu, C.S.; Zhu, L.; Tan, D.A.; Qiu, X.Y.; Liu, C.Y.; Xie, S.S.; Zhu, L.Y. Avenues Toward microRNA Detection. Comput. Struct. Biotechnol. J. 2019, 17, 904–916. [Google Scholar] [CrossRef]
- Qiu, X.Y.; Zhu, L.Y.; Zhu, C.S.; Ma, J.X.; Hou, T.; Wu, X.M.; Xie, S.S.; Min, L.; Tan, D.A.; Zhang, D.Y.; et al. Highly Effective and Low-Cost MicroRNA Detection with CRISPR-Cas9. ACS Synth. Biol. 2018, 7, 807–813. [Google Scholar] [CrossRef]
- Poujouly, C.; Le Gall, J.; Freisa, M.; Kechkeche, D.; Bouville, D.; Khemir, J.; Gonzalez-Losada, P.; Gamby, J. Microfluidic Chip for the Electrochemical Detection of MicroRNAs: Methylene Blue Increasing the Specificity of the Biosensor. Front. Chem. 2022, 10, 868909. [Google Scholar] [CrossRef]
- Kim, H.Y.; Song, J.; Park, H.G.; Kang, T. Electrochemical detection of zeptomolar miRNA using an RNA-triggered Cu 2+ reduction method. Sens. Actuators B Chem. 2022, 360, 131666. [Google Scholar] [CrossRef]
- Kaminski, M.M.; Abudayyeh, O.O.; Gootenberg, J.S.; Zhang, F.; Collins, J.J. CRISPR-based diagnostics. Nat. Biomed. Eng. 2021, 5, 643–656. [Google Scholar] [CrossRef]
- Dong, J.; Chen, G.; Wang, W.; Huang, X.; Peng, H.; Pu, Q.; Du, F.; Cui, X.; Deng, Y.; Tang, Z. Colorimetric PCR-Based microRNA Detection Method Based on Small Organic Dye and Single Enzyme. Anal. Chem. 2018, 90, 7107–7111. [Google Scholar] [CrossRef]
- Djebbi, K.; Shi, B.; Weng, T.; Bahri, M.; Elaguech, M.A.; Liu, J.; Tlili, C.; Wang, D. Highly Sensitive Fluorescence Assay for miRNA Detection: Investigation of the DNA Spacer Effect on the DSN Enzyme Activity toward Magnetic-Bead-Tethered Probes. ACS Omega 2022, 7, 2224–2233. [Google Scholar] [CrossRef]
- Zhu, D.; Miao, Z.Y.; Hu, Y.; Zhang, X.J. Single-step, homogeneous and sensitive detection for microRNAs with dual-recognition steps based on luminescence resonance energy transfer (LRET) using upconversion nanoparticles. Biosens. Bioelectron. 2018, 100, 475–481. [Google Scholar] [CrossRef]
- Li, D.; Xia, L.; Zhou, Q.; Wang, L.; Chen, D.; Gao, X.; Li, Y. Label-Free Detection of miRNA Using Surface-Enhanced Raman Spectroscopy. Anal. Chem. 2020, 92, 12769–12773. [Google Scholar] [CrossRef]
- Weng, S.; Lin, D.; Lai, S.; Tao, H.; Chen, T.; Peng, M.; Qiu, S.; Feng, S. Highly sensitive and reliable detection of microRNA for clinically disease surveillance using SERS biosensor integrated with catalytic hairpin assembly amplification technology. Biosens. Bioelectron. 2022, 208, 114236. [Google Scholar] [CrossRef]
- Qu, A.; Sun, M.; Xu, L.; Hao, C.; Wu, X.; Xu, C.; Kotov, N.A.; Kuang, H. Quantitative zeptomolar imaging of miRNA cancer markers with nanoparticle assemblies. Proc. Natl. Acad. Sci. USA 2019, 116, 3391–3400. [Google Scholar] [CrossRef] [Green Version]
- Hakimian, F.; Ghourchian, H.; Hashemi, A.S.; Arastoo, M.R.; Behnam Rad, M. Ultrasensitive optical biosensor for detection of miRNA-155 using positively charged Au nanoparticles. Sci. Rep. 2018, 8, 2943. [Google Scholar] [CrossRef] [Green Version]
- Lai, M.; Slaughter, G. Label-Free MicroRNA Optical Biosensors. Nanomaterials 2019, 9, 1573. [Google Scholar] [CrossRef] [Green Version]
- Zheng, W.; Yao, L.; Teng, J.; Yan, C.; Qin, P.; Liu, G.; Chen, W. Lateral Flow Test for Visual Detection of Multiple MicroRNAs. Sens. Actuators. B Chem. 2018, 264, 320–326. [Google Scholar] [CrossRef]
- Wang, N.; Zhang, J.; Xiao, B.; Sun, X.; Xie, R.; Chen, A. Recent advances in the rapid detection of microRNA with lateral flow assays. Biosens. Bioelectron. 2022, 211, 114345. [Google Scholar] [CrossRef]
- De Sousa, K.P.; Rossi, I.; Abdullahi, M.; Ramirez, M.I.; Stratton, D.; Inal, J.M. Isolation and characterization of extracellular vesicles and future directions in diagnosis and therapy. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2023, 15, e1835. [Google Scholar] [CrossRef]
- Xu, W.M.; Li, A.; Chen, J.J.; Sun, E.J. Research Development on Exosome Separation Technology. J. Membr. Biol. 2023, 256, 25–34. [Google Scholar] [CrossRef]
- Lee, K.; Fraser, K.; Ghaddar, B.; Yang, K.; Kim, E.; Balaj, L.; Chiocca, E.A.; Breakefield, X.O.; Lee, H.; Weissleder, R. Multiplexed Profiling of Single Extracellular Vesicles. ACS Nano 2018, 12, 494–503. [Google Scholar] [CrossRef]
- Morasso, C.F.; Sproviero, D.; Mimmi, M.C.; Giannini, M.; Gagliardi, S.; Vanna, R.; Diamanti, L.; Bernuzzi, S.; Piccotti, F.; Truffi, M.; et al. Raman spectroscopy reveals biochemical differences in plasma derived extracellular vesicles from sporadic Amyotrophic Lateral Sclerosis patients. Nanomedicine 2020, 29, 102249. [Google Scholar] [CrossRef]
- Butler, H.J.; Ashton, L.; Bird, B.; Cinque, G.; Curtis, K.; Dorney, J.; Esmonde-White, K.; Fullwood, N.J.; Gardner, B.; Martin-Hirsch, P.L.; et al. Using Raman spectroscopy to characterize biological materials. Nat. Protoc. 2016, 11, 664–687. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Ren, H.; Dai, B.; Li, J.; Shang, L.; Huang, J.; Shi, X. Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts. J. Exp. Clin. Cancer Res. 2018, 37, 324. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Ding, Q.; Plant, P.; Basheer, M.; Yang, C.; Tawedrous, E.; Krizova, A.; Boulos, C.; Farag, M.; Cheng, Y.; et al. Droplet digital PCR improves urinary exosomal miRNA detection compared to real-time PCR. Clin. Biochem. 2019, 67, 54–59. [Google Scholar] [CrossRef]
- Garai, K.; Adam, Z.; Herczeg, R.; Katai, E.; Nagy, T.; Pal, S.; Gyenesei, A.; Pongracz, J.E.; Wilhelm, M.; Kvell, K. Artificial Neural Network Correlation and Biostatistics Evaluation of Physiological and Molecular Parameters in Healthy Young Individuals Performing Regular Exercise. Front. Physiol. 2019, 10, 1242. [Google Scholar] [CrossRef]
- Yang, Y.; Kannisto, E.; Yu, G.; Reid, M.E.; Patnaik, S.K.; Wu, Y. An Immuno-Biochip Selectively Captures Tumor-Derived Exosomes and Detects Exosomal RNAs for Cancer Diagnosis. ACS Appl. Mater Interfaces 2018, 10, 43375–43386. [Google Scholar] [CrossRef]
- Cho, S.; Yang, H.C.; Rhee, W.J. Simultaneous multiplexed detection of exosomal microRNAs and surface proteins for prostate cancer diagnosis. Biosens. Bioelectron. 2019, 146, 111749. [Google Scholar] [CrossRef]
- Lee, J.; Kwon, M.H.; Kim, J.A.; Rhee, W.J. Detection of exosome miRNAs using molecular beacons for diagnosing prostate cancer. Artif. Cells Nanomed. Biotechnol. 2018, 46, S52–S63. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, G.P.; Zigon, E.; Rogers, G.; Davodian, D.; Lu, S.; Jovanovic-Talisman, T.; Jones, J.; Tigges, J.; Tyagi, S.; Ghiran, I.C. Detection of Extracellular Vesicle RNA Using Molecular Beacons. iScience 2020, 23, 100782. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.R.; Chen, X.; Lv, J.; Li, D.W.; Yang, C.D.; Liu, H.L.; Qian, R.C. A plasmonic nanoparticle-embedded polydopamine substrate for fluorescence detection of extracellular vesicle biomarkers in serum and urine from patients with systemic lupus erythematosus. Talanta 2022, 247, 123620. [Google Scholar] [CrossRef]
- Jang, M.; Choi, G.; Choi, Y.Y.; Lee, J.E.; Jung, J.H.; Oh, S.W.; Han, D.H.; Lee, H.; Park, J.-H.; Cheong, J.H.; et al. Extracellular vesicle (EV)-polyphenol nanoaggregates for microRNA-based cancer diagnosis. NPG Asia Mater. 2019, 11, 79. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.; Lin, Q.; Zhu, J.; Rao, Y.; Cui, L.; Bao, Y.; Ji, T. DNase I-assisted 2′-O-methyl molecular beacon for amplified detection of tumor exosomal microRNA-21. Talanta 2021, 235, 122727. [Google Scholar] [CrossRef]
- Veedu, R.N.; Wengel, J. Locked nucleic acids: Promising nucleic acid analogs for therapeutic applications. Chem. Biodivers 2010, 7, 536–542. [Google Scholar] [CrossRef]
- Saadati, A.; Hassanpour, S.; de la Guardia, M.; Mosafer, J.; Hashemzaei, M.; Mokhtarzadeh, A.; Baradaran, B. Recent advances on application of peptide nucleic acids as a bioreceptor in biosensors development. TrAC Trends Anal. Chem. 2019, 114, 56–68. [Google Scholar] [CrossRef]
- Lee, J.U.; Kim, W.H.; Lee, H.S.; Park, K.H.; Sim, S.J. Quantitative and Specific Detection of Exosomal miRNAs for Accurate Diagnosis of Breast Cancer Using a Surface-Enhanced Raman Scattering Sensor Based on Plasmonic Head-locked Gold Nanopillars. Small 2019, 15, 1804968. [Google Scholar] [CrossRef]
- Luo, L.; Wang, L.; Zeng, L.; Wang, Y.; Weng, Y.; Liao, Y.; Chen, T.; Xia, Y.; Zhang, J.; Chen, J. A ratiometric electrochemical DNA biosensor for detection of exosomal MicroRNA. Talanta 2020, 207, 120298. [Google Scholar] [CrossRef]
- Lin, M.; Wang, J.; Zhou, G.; Wu, N.; Lu, J.; Gao, J.; Chen, X.; Shi, J.; Zuo, X.; Fan, C. Programmable engineering of a biosensing interface with tetrahedral DNA nanostructures for ultrasensitive DNA detection. Angew. Chem. Int. Ed. Engl. 2015, 54, 2151–2155. [Google Scholar] [CrossRef]
- Xu, Y.; Li, X.; Niu, C.; Wu, H.; Yong, Y.; Qi, C.; Gong, W.; Bai, H.; Chen, Y.; Ding, S.; et al. Janus wireframe DNA cube-based 3D nanomachine for rapid and stable fluorescence detection of exosomal microRNA. Biosens. Bioelectron. 2022, 212, 114405. [Google Scholar] [CrossRef]
- Bi, S.; Yue, S.; Zhang, S. Hybridization chain reaction: A versatile molecular tool for biosensing, bioimaging, and biomedicine. Chem. Soc. Rev. 2017, 46, 4281–4298. [Google Scholar] [CrossRef]
- Guo, Q.; Yu, Y.; Zhang, H.; Cai, C.; Shen, Q. Electrochemical Sensing of Exosomal MicroRNA Based on Hybridization Chain Reaction Signal Amplification with Reduced False-Positive Signals. Anal. Chem. 2020, 92, 5302–5310. [Google Scholar] [CrossRef]
- Chen, X.G.; Liu, F.; Song, X.F.; Wang, Z.H.; Dong, Z.Q.; Hu, Z.Q.; Lan, R.Z.; Guan, W.; Zhou, T.G.; Xu, X.M.; et al. Rapamycin regulates Akt and ERK phosphorylation through mTORC1 and mTORC2 signaling pathways. Mol. Carcinog. 2010, 49, 603–610. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, Y.; Xie, H.; Zhao, L.; Zheng, L.; Ye, H. Applications of Catalytic Hairpin Assembly Reaction in Biosensing. Small 2019, 15, e1902989. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.Y.; Luo, S.H.; Lin, X.M.; Hu, X.M.; Zhang, Y.; Zhang, X.H.; Wu, C.M.; Zheng, L.; Wang, Q. A novel electrochemical biosensor for exosomal microRNA-181 detection based on a catalytic hairpin assembly circuit. Anal. Chim. Acta 2021, 1157, 338396. [Google Scholar] [CrossRef]
- Simmel, F.C.; Yurke, B.; Singh, H.R. Principles and Applications of Nucleic Acid Strand Displacement Reactions. Chem. Rev. 2019, 119, 6326–6369. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, X.; Situ, B.; Wu, Y.; Luo, S.; Zheng, L.; Qiu, Y. Rapid electrochemical biosensor for sensitive profiling of exosomal microRNA based on multifunctional DNA tetrahedron assisted catalytic hairpin assembly. Biosens. Bioelectron. 2021, 183, 113205. [Google Scholar] [CrossRef]
- Liu, P.; Qian, X.; Li, X.; Fan, L.; Cui, D.; Yan, Y. Enzyme-Free Electrochemical Biosensor Based on Localized DNA Cascade Displacement Reaction and Versatile DNA Nanosheets for Ultrasensitive Detection of Exosomal MicroRNA. ACS Appl. Mater. Interfaces 2020, 12, 45648–45656. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Wang, L.; Li, J.; Chen, X.; Lan, J.; Yan, A.; Lei, Y.; Yang, S.; Yang, H.; Chen, J. A Ratiometric Fluorescent Bioprobe Based on Carbon Dots and Acridone Derivate for Signal Amplification Detection Exosomal microRNA. Ana.l Chem. 2018, 90, 8969–8976. [Google Scholar] [CrossRef]
- Ali, M.M.; Li, F.; Zhang, Z.; Zhang, K.; Kang, D.K.; Ankrum, J.A.; Le, X.C.; Zhao, W. Rolling circle amplification: A versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014, 43, 3324–3341. [Google Scholar] [CrossRef]
- Al Sulaiman, D.; Juthani, N.; Doyle, P.S. Quantitative and Multiplex Detection of Extracellular Vesicle-Derived MicroRNA via Rolling Circle Amplification within Encoded Hydrogel Microparticles. Adv. Healthc. Mater. 2022, 11, e2102332. [Google Scholar] [CrossRef]
- Wright, A.V.; Nuñez, J.K.; Doudna, J.A. Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell 2016, 164, 29–44. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Zhao, X.; Chen, X.; Qiu, X.; Qing, G.; Zhang, H.; Zhang, L.; Hu, X.; He, Z.; Zhong, D.; et al. Rolling Circular Amplification (RCA)-Assisted CRISPR/Cas9 Cleavage (RACE) for Highly Specific Detection of Multiple Extracellular Vesicle MicroRNAs. Anal. Chem. 2020, 92, 2176–2185. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Zhang, Q.; Liu, M.; Wang, Y.; Lu, M. A portable system for isothermal amplification and detection of exosomal microRNAs. Biosens. Bioelectron. 2022, 196, 113707. [Google Scholar] [CrossRef]
- Maduraiveeran, G.; Sasidharan, M.; Ganesan, V. Electrochemical sensor and biosensor platforms based on advanced nanomaterials for biological and biomedical applications. Biosens. Bioelectron. 2018, 103, 113–129. [Google Scholar] [CrossRef]
- Elahi, N.; Kamali, M.; Baghersad, M.H. Recent biomedical applications of gold nanoparticles: A review. Talanta 2018, 184, 537–556. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Li, M.; Qiu, J.; Sun, Y.P. Design and fabrication of carbon dots for energy conversion and storage. Chem. Soc. Rev. 2019, 48, 2315–2337. [Google Scholar] [CrossRef]
- Li, T.; Liang, Y.; Li, J.; Yu, Y.; Xiao, M.M.; Ni, W.; Zhang, Z.; Zhang, G.J. Carbon Nanotube Field-Effect Transistor Biosensor for Ultrasensitive and Label-Free Detection of Breast Cancer Exosomal miRNA21. Anal. Chem. 2021, 93, 15501–15507. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, D.; Zhao, M.; Wu, H.; Shen, B.; Liu, P.; Ding, S. Electrochemical biosensor for ultrasensitive exosomal miRNA analysis by cascade primer exchange reaction and MOF@Pt@MOF nanozyme. Biosens. Bioelectron. 2020, 168, 112554. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Xianyu, Y.; Jiang, X. Point-of-care biochemical assays using gold nanoparticle-implemented microfluidics. Chem. Soc. Rev. 2014, 43, 6239–6253. [Google Scholar] [CrossRef]
- Ramshani, Z.; Zhang, C.; Richards, K.; Chen, L.; Xu, G.; Stiles, B.L.; Hill, R.; Senapati, S.; Go, D.B.; Chang, H.C. Extracellular vesicle microRNA quantification from plasma using an integrated microfluidic device. Commun. Biol. 2019, 2, 189. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Han, J.; Park, J.S.; Kim, J.H.; Lee, E.S.; Cha, B.S.; Park, K.S. DNA barcode-based detection of exosomal microRNAs using nucleic acid lateral flow assays for the diagnosis of colorectal cancer. Talanta 2022, 242, 123306. [Google Scholar] [CrossRef]
- Langer, J.; Jimenez de Aberasturi, D.; Aizpurua, J.; Alvarez-Puebla, R.A.; Auguié, B.; Baumberg, J.J.; Bazan, G.C.; Bell, S.E.J.; Boisen, A.; Brolo, A.G.; et al. Present and Future of Surface-Enhanced Raman Scattering. ACS Nano 2020, 14, 28–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, D.; Huang, C.; Zheng, J.; Tang, J.; Li, J.; Yang, J.; Yang, R. Quantitative detection of exosomal microRNA extracted from human blood based on surface-enhanced Raman scattering. Biosens. Bioelectron. 2018, 101, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.H.; Lee, J.U.; Jeon, M.J.; Park, K.H.; Sim, S.J. Three-dimensional hierarchical plasmonic nano-architecture based label-free surface-enhanced Raman spectroscopy detection of urinary exosomal miRNA for clinical diagnosis of prostate cancer. Biosens. Bioelectron. 2022, 205, 114116. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.H.; Park, J.; Kang, S.; Kim, M. Surface plasmon resonance: A versatile technique for biosensor applications. Sensors 2015, 15, 10481–10510. [Google Scholar] [CrossRef] [Green Version]
- Song, S.; Lee, J.U.; Jeon, M.J.; Kim, S.; Sim, S.J. Detection of multiplex exosomal miRNAs for clinically accurate diagnosis of Alzheimer’s disease using label-free plasmonic biosensor based on DNA-Assembled advanced plasmonic architecture. Biosens. Bioelectron. 2022, 199, 113864. [Google Scholar] [CrossRef]
- Boriachek, K.; Umer, M.; Islam, M.N.; Gopalan, V.; Lam, A.K.; Nguyen, N.T.; Shiddiky, M.J.A. An amplification-free electrochemical detection of exosomal miRNA-21 in serum samples. Analyst 2018, 143, 1662–1669. [Google Scholar] [CrossRef]
- Cheng, W.; Ma, J.; Cao, P.; Zhang, Y.; Xu, C.; Yi, Y.; Li, J. Enzyme-free electrochemical biosensor based on double signal amplification strategy for the ultra-sensitive detection of exosomal microRNAs in biological samples. Talanta 2020, 219, 121242. [Google Scholar] [CrossRef]
- Saha, S.; Allelein, S.; Pandey, R.; Medina-Perez, P.; Osman, E.; Kuhlmeier, D.; Soleymani, L. Two-Step Competitive Hybridization Assay: A Method for Analyzing Cancer-Related microRNA Embedded in Extracellular Vesicles. Anal. Chem. 2021, 93, 15913–15921. [Google Scholar] [CrossRef]
- Gao, X.; Li, S.; Ding, F.; Fan, H.; Shi, L.; Zhu, L.; Li, J.; Feng, J.; Zhu, X.; Zhang, C. Rapid Detection of Exosomal MicroRNAs Using Virus-Mimicking Fusogenic Vesicles. Angew. Chem. Int. Ed. Engl. 2019, 58, 8719–8723. [Google Scholar] [CrossRef]
- Zhou, S.; Hu, T.; Han, G.; Wu, Y.; Hua, X.; Su, J.; Jin, W.; Mou, Y.; Mou, X.; Li, Q.; et al. Accurate Cancer Diagnosis and Stage Monitoring Enabled by Comprehensive Profiling of Different Types of Exosomal Biomarkers: Surface Proteins and miRNAs. Small 2020, 16, e2004492. [Google Scholar] [CrossRef]
- Cao, Y.; Yu, X.; Zeng, T.; Fu, Z.; Zhao, Y.; Nie, B.; Zhao, J.; Yin, Y.; Li, G. Molecular Characterization of Exosomes for Subtype-Based Diagnosis of Breast Cancer. J. Am. Chem. Soc. 2022, 144, 13475–13486. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xie, M.; Shi, M.; Yuan, K.; Wu, Y.; Meng, H.M.; Qu, L.; Li, Z. Spatial Confinement-Derived Double-Accelerated DNA Cascade Reaction for Ultrafast and Highly Sensitive. Anal. Chem. 2022, 94, 2227–2235. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Liu, C.; Li, Y.; Ma, Y.; Deng, J.; Li, L.; Sun, J. Thermophoretic Detection of Exosomal microRNAs by Nanoflares. J. Am. Chem. Soc. 2020, 142, 4996–5001. [Google Scholar] [CrossRef]
- Jiang, S.; Li, Q.; Wang, C.; Pang, Y.; Sun, Z.; Xiao, R. In Situ Exosomal MicroRNA Determination by Target-Triggered SERS and Fe. ACS Sens. 2021, 6, 852–862. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.Y.; Li, M.X.; Pan, W.L.; Chen, Y.; Li, M.M.; Pang, J.X.; Zheng, L.; Chen, J.X.; Duan, W.J. In Situ Detection of Plasma Exosomal MicroRNA-1246 for Breast Cancer Diagnostics by a Au Nanoflare Probe. ACS Appl. Mater. Interfaces 2018, 10, 39478–39486. [Google Scholar] [CrossRef]
- Finn, N.A.; Searles, C.D. Intracellular and Extracellular miRNAs in Regulation of Angiogenesis Signaling. Curr. Angiogenes. 2012, 4, 299–307. [Google Scholar] [CrossRef] [Green Version]
- Bueno Marinas, M.; Celeghin, R.; Cason, M.; Bariani, R.; Frigo, A.C.; Jager, J.; Syrris, P.; Elliott, P.M.; Bauce, B.; Thiene, G.; et al. A microRNA Expression Profile as Non-Invasive Biomarker in a Large Arrhythmogenic Cardiomyopathy Cohort. Int. J. Mol. Sci. 2020, 21, 1536. [Google Scholar] [CrossRef] [Green Version]
- Song, R.; Ro, S.; Yan, W. In situ hybridization detection of microRNAs. Methods Mol. Biol. 2010, 629, 287–294. [Google Scholar] [CrossRef] [Green Version]
- Sun, B.; Liang, Z.; Xie, B.P.; Li, R.T.; Li, L.Z.; Jiang, Z.H.; Bai, L.P.; Chen, J.X. Fluorescence sensing platform based on ruthenium(II) complexes as high 3S (sensitivity, specificity, speed) and "on-off-on" sensors for the miR-185 detection. Talanta 2018, 179, 658–667. [Google Scholar] [CrossRef]
- Le, B.H.; Seo, Y.J. Highly sensitive MicroRNA 146a detection using a gold nanoparticle-based CTG repeat probing system and isothermal amplification. Anal Chim Acta 2018, 999, 155–160. [Google Scholar] [CrossRef]

| Type and Size of EV | Molecular Markers | Contents |
|---|---|---|
| Exosome (30–100 nm) | ALIX, TSG101, CD63 | Nucleic acids (DNA, mRNA, miRNA), lipids, proteins, metabolites |
| Ectosome (microvesicle) (100–1000 nm) | ARF6, VCAMP3 | Nucleic acids (DNA, mRNA, miRNA), lipids, proteins, metabolites |
| Oncosome (Apoptotic body) (1–5 µm) | Cytokeratin-18 | Proteins, metabolites |
| Junctional Complexes | Function | Main Junctional Proteins |
|---|---|---|
| Tight junction (TJ) | Maintain cellular polarity and establish distinct fluid compartments | Zonula occludens (ZO-1, ZO-2, and ZO-3) bound to occludin or claudins and actin |
| Adherens junction (AJ) | Initiation and stabilization of cell–cell adhesion, regulation of the actin cytoskeleton, intracellular signaling and transcriptional regulation | E-cadherin linked to actin cytoskeleton, VE-cadherin, catenins, and signaling elements via membrane proteins |
| Organ | Disease | Cell Type | miRNA |
|---|---|---|---|
| Intestine | Inflammatory bowel diseases Crohn’s Disease [56] | epithelial | miR-1-3p and miR-124-3p [57] |
| Ulcerative colitis [58,59] | epithelial | miR-21, miR-122a, miR-124, miR-191a, miR-212, miR-675, miR-874, miR-93, miR-200b, miR-20a, miR-30c miR-93, miR-106b, miR-122, miR-130a, miR-132, miR-2, miR-142-3p, miR-146b, miR-155, miR-192, miR-196, miR-320, miR-10a, miR-141, miR-320, miR-346, miR-665 [60] | |
| Celiac disease [61] | epithelial | miR-31-5p, miR-192, miR-194, miR-449a, and miR-638 [62] miR-29c and miR-224 [63] | |
| Infectious diarrheal syndromes [64] | epithelial | ||
| Lung | Asthma [65,66] | epithelial | Let-7a [67], miR-let-7f, miR-181c, miR-487b, miR-34b-5p, miR-34c-5p, miR-449a, miR-449b-5p, miR-155, miR-203, miR-221, miR-3065 |
| Pollen allergy [68] | epithelial | [69] miR-21, miR-146a miR-155 [70], miR-151a, miR-143, miR-124, miR-223, miR-10a, miR-572, miR-1228-, miR-483, miR-1908, miR-126, miR-92a, miR-125a, miR-19a, miR-26a, miR-106a, miR-181c, -3177 | |
| Interstitial pneumonia [71] | epithelial | miR-193a-5p, miR-502-3p, miR-200c-3p, miR-16-5p, miR-21-5p, miR-126-3p, miR-34a-5p [72] miR-375, miR-193a, miR-106b, miR-18a, miR-15a, and miR-374a [73] | |
| Acute respiratory distress syndrome [74] and acute lung injury | endothelial | miR-155, miR-223, miR-146a, miR-27a/b [75], miR-34a, miR-132, miR-15a, miR-21 [76] | |
| Urinary bladder- Urothelial Barrier | Interstitial cystitis/bladder pain syndrome [77] | epithelial | miR-373-5p, miR-6766-5p [78] |
| Blood-brain barrier | Alzheimer’s disease (AD) [79] | endothelial | miR-15a/16-1 [80] |
| miR-501-3p [81] | |||
| Multiple sclerosis (MS) [82] | endothelial | miR-182 [83,84] | |
| ischemia/stroke [85] | endothelial | miR-107 [86] | |
| Epilepsy [87,88] | endothelial | miR-143, miR-27a-3p, miR-101, miR-132, miR-210, miR-125a-5p, miR-126-3p, miR-98, let-7g, miR-1303, miR-132, miR-27b, miR-150, miR-150, miR-182, miR-285, miR-181c, miR-30a [89] | |
| Autism spectrum disorder [90] | endothelial | miR-6780a-3p, miR-1225-5p, miR-2277-3p, miR-548j-5p, miR-100-5p [91], miR-1290 [92], miR-873 [93] miR-486, miR-181b [94] |
| miRNA | Targeted Barrier Protein | References |
|---|---|---|
| miR-15a, b/16-1 | Claudin-2, 5 | [95] |
| miR-16-5p | Claudin-2 | [96] |
| miR-21, -5p | Claudin-2, occludin, ZO-1 | [96,97,98] |
| miR-27a/b, a-3p | Claudin-2, 5, 8, VE-cadherin | [99,100] |
| miR-34, -a | Claudin-8, ZO-1 | [101] |
| miR-93 | Claudin-3 | [102] |
| miR-101 | VE-cadherin | [103] |
| miR-122a | Occludin, ZO-1 | [55,104] |
| miR-122 | Claudin-2, 8, occluding | [105] |
| miR-125a,b, -5p | Claudin-2 | [96,106] |
| miR-142-5p | Claudin-1 | [107] |
| miR-142-3p | Claudin-12, ZO-1, occludin | [108] |
| miR-144 | Claudin-1, ZO-1 | [108] |
| miR-155 | Claudin-1, 8, ZO-1, occludin | [106,109] |
| miR-182 | Claudin-5 | [110] |
| miR-195-5p | Claudin-2 | [111] |
| miR-200a | ZO-1 | [112] |
| miR-200b | Claudin-8, ZO-1, occludin | [113] |
| miR-200c | Claudin-8, ZO-1, occludin | [114] |
| miR-200 | ZO-1 | [115] |
| miR-210 | Occludin | [116] |
| miR-212 | ZO-1, occluding | [117] |
| miR-373-5p | ZO-1 | [95] |
| miR-429 | Claudin-8, ZO-1, occludin | [118] |
| miR-874 | Claudin-1, occluding | [119] |
| miR-1290 | Claudin-1, 5, ZO-1 | [120,121] |
| Assays | miRNA | Sensitivity (mol/L) | Biological Recognition Elements |
|---|---|---|---|
| Electrochemical [105,106] | Let-7a, miR-21, miR-141, miR-122, miR-196a | 10−7–10−18 | ssDNA probe, ssRNA probe, hairpin-shaped probe, HCR, CHA, DSNSA, (clustered regularly interspaced short palindromic repeats) CRISPR–Cas system [104,107] |
| Colorimetric [108] | miR-221, miR-21, miR-141 Let 7a, -7b, -7d, miR-1178, miR-1248 | 10−7–10−18 | CHA, EXPAR, DSNSA, hairpin-shaped probe, |
| CRISPR–Cas system [104,107] | |||
| Fluorescent [109] | miR-19b, miR-21, miR-92a, miR-148, miR-146a, miR-185 | 10−8–10−17 | ssDNA probe, hairpin-shaped probe, HCR, CHA, DSNSA, EXPAR, RCA, CRISPR–Cas system [104,107] |
| Luminescence [110] | miR-21 | 10−8–10−12 | CRISPR–Cas system [104,107] |
| Raman scattering [111,112] | miR-21a, | ||
| 155a, 10b, 96a,125a, 34a | 10−9–10−15 | Hairpin-shaped probe | |
| Gold nanorod [113] | miR-21 | 10−17–10−17 | Hairpin-shaped probe |
| miR-200b | |||
| Optical biosensor [114,115] | miR-133a, miR-141, miR-21, miR-155 | 10−17–10−17 | Hairpin-shaped probe, CHA, ssDNA probe |
| 10−17 | ssDNA probe | ||
| Lateral flow [116,117] | miR-21, miR-224 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bovari-Biri, J.; Garai, K.; Banfai, K.; Csongei, V.; Pongracz, J.E. miRNAs as Predictors of Barrier Integrity. Biosensors 2023, 13, 422. https://doi.org/10.3390/bios13040422
Bovari-Biri J, Garai K, Banfai K, Csongei V, Pongracz JE. miRNAs as Predictors of Barrier Integrity. Biosensors. 2023; 13(4):422. https://doi.org/10.3390/bios13040422
Chicago/Turabian StyleBovari-Biri, Judit, Kitti Garai, Krisztina Banfai, Veronika Csongei, and Judit E. Pongracz. 2023. "miRNAs as Predictors of Barrier Integrity" Biosensors 13, no. 4: 422. https://doi.org/10.3390/bios13040422
APA StyleBovari-Biri, J., Garai, K., Banfai, K., Csongei, V., & Pongracz, J. E. (2023). miRNAs as Predictors of Barrier Integrity. Biosensors, 13(4), 422. https://doi.org/10.3390/bios13040422






