Applications and Tuning Strategies for Transcription Factor-Based Metabolite Biosensors
Abstract
:1. Introduction
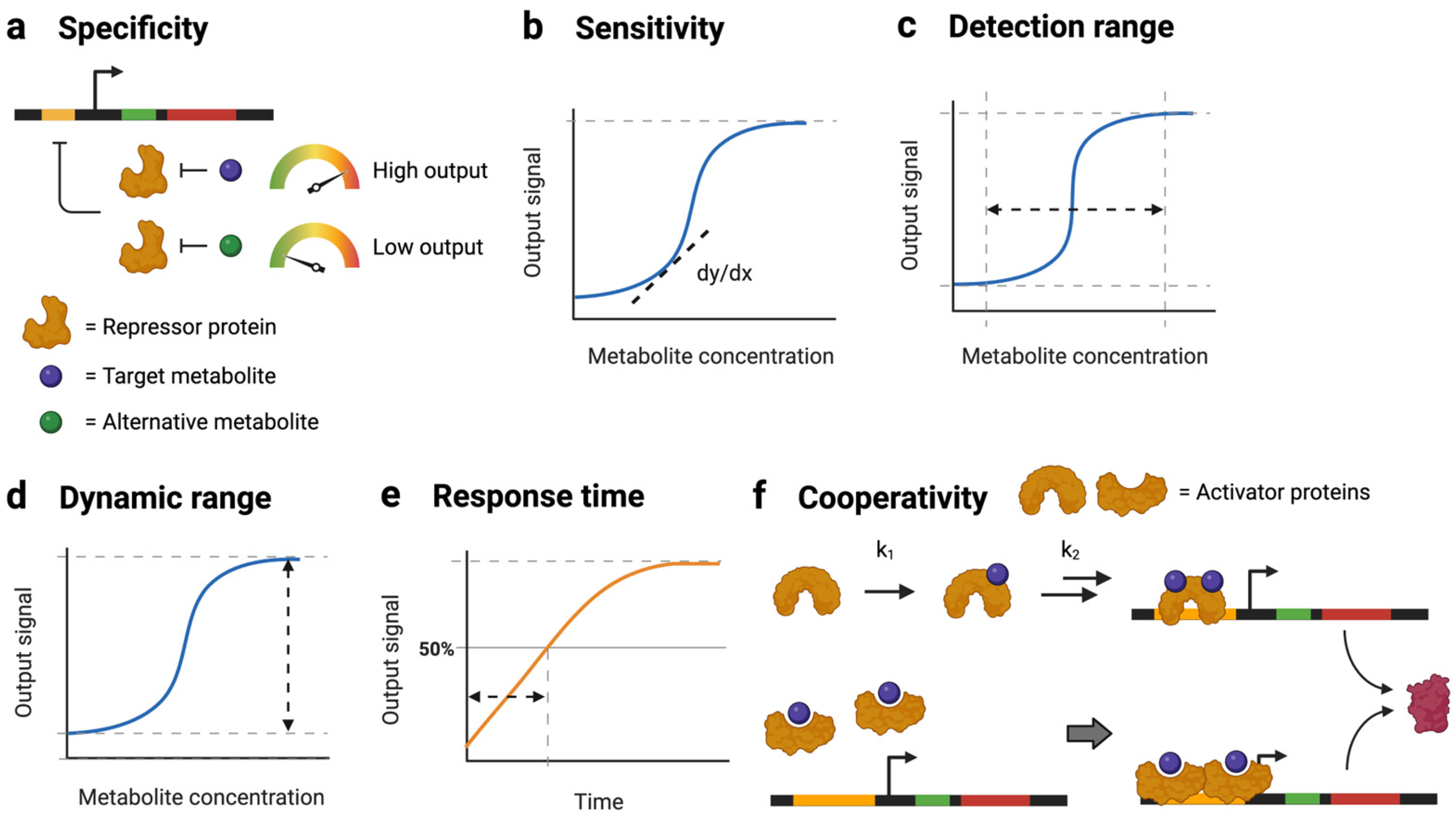
2. Tuning Strategies
2.1. Engineering TFs for Optimal Sensor Performance
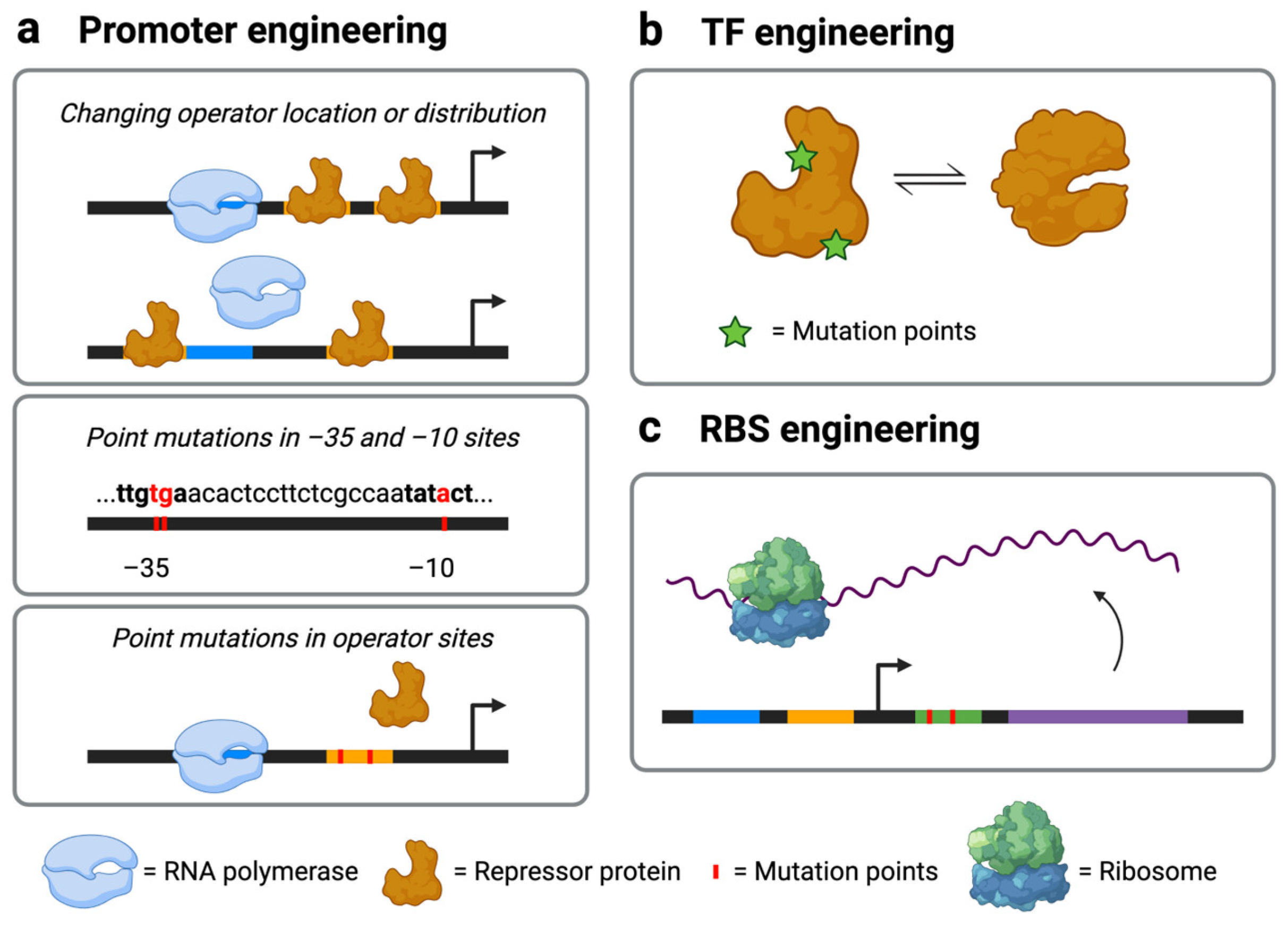
2.2. Engineering Promoters for Optimal Sensor Performance
2.3. RBS Engineering (Translational Control)
2.4. Additional Design Considerations
2.4.1. Growth-Dependent Sensor Performance
2.4.2. Tradeoffs between Sensor Performance Parameters
3. Applications
3.1. High-Throughput Screening
3.2. Metabolite Dynamics and Dynamic Metabolic Regulation
3.3. Illuminating and Engineering Metabolic Heterogeneity
4. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, D.; Evans, T.; Zhang, F. Applications and Advances of Metabolite Biosensors for Metabolic Engineering. Metab. Eng. 2015, 31, 35–43. [Google Scholar] [CrossRef] [Green Version]
- Huang, P.-J.J.; Liu, J. Simultaneous Detection of L-Lactate and D-Glucose Using DNA Aptamers in Human Blood Serum. Angew. Chem. Int. Ed. 2023, 62, e202212879. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, G.M.; Akinyemi, O.; Malik, J.; Focht, C.M.; Pritchett, E.M.; Baker, C.D.; McSally, J.P.; Jenkins, J.L.; Mathews, D.H.; Wedekind, J.E. A Riboswitch Separated from Its Ribosome-Binding Site Still Regulates Translation. Nucleic Acids Res. 2023, 51, 2464–2484. [Google Scholar] [CrossRef] [PubMed]
- Mottillo, E.P.; Mladenovic-Lucas, L.; Zhang, H.; Zhou, L.; Kelly, C.V.; Ortiz, P.A.; Granneman, J.G. A FRET Sensor for the Real-Time Detection of Long Chain Acyl-CoAs and Synthetic ABHD5 Ligands. Cell Rep. Methods 2023, 3, 100394. [Google Scholar] [CrossRef] [PubMed]
- Nuñez, S.; Barra, M.; Garrido, D. Developing a Fluorescent Inducible System for Free Fucose Quantification in Escherichia coli. Biosensors 2023, 13, 388. [Google Scholar] [CrossRef]
- Guo, X.; Wen, F.; Zheng, N.; Saive, M.; Fauconnier, M.-L.; Wang, J. Aptamer-Based Biosensor for Detection of Mycotoxins. Front. Chem. 2020, 8, 195. [Google Scholar] [CrossRef] [Green Version]
- Chinnappan, R.; AlZabn, R.; Abu-Salah, K.M.; Zourob, M. An Aptamer Based Fluorometric Microcystin-LR Assay Using DNA Strand-Based Competitive Displacement. Microchim. Acta 2019, 186, 435. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, F.; Mayer, G. Selection and Biosensor Application of Aptamers for Small Molecules. Front. Chem. 2016, 4, 25. [Google Scholar] [CrossRef] [Green Version]
- Breaker, R.R. Riboswitches and Translation Control. Cold Spring Harb. Perspect. Biol. 2018, 10, a032797. [Google Scholar] [CrossRef]
- Yaginuma, H.; Kawai, S.; Tabata, K.V.; Tomiyama, K.; Kakizuka, A.; Komatsuzaki, T.; Noji, H.; Imamura, H. Diversity in ATP Concentrations in a Single Bacterial Cell Population Revealed by Quantitative Single-Cell Imaging. Sci. Rep. 2014, 4, 6522. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.-H.; Jacobs-Wagner, C. Connecting Single-Cell ATP Dynamics to Overflow Metabolism, Cell Growth, and the Cell Cycle in Escherichia coli. Curr. Biol. 2022, 32, 3911–3924. [Google Scholar] [CrossRef] [PubMed]
- Imani, M.; Mohajeri, N.; Rastegar, M.; Zarghami, N. Recent Advances in FRET-Based Biosensors for Biomedical Applications. Anal. Biochem. 2021, 630, 114323. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; He, F.; Yu, Y.; Wang, Y. Application of FRET Biosensors in Mechanobiology and Mechanopharmacological Screening. Front. Bioeng. Biotechnol. 2020, 8, 595497. [Google Scholar] [CrossRef]
- Ding, N.; Zhou, S.; Deng, Y. Transcription-Factor-Based Biosensor Engineering for Applications in Synthetic Biology. ACS Synth. Biol. 2021, 10, 911–922. [Google Scholar] [CrossRef]
- Li, C.; Wang, C.; Zhu, J.; Xue, F.; Sun, X.; Gu, Y. Advances and Prospects of Transcription-Factor-Based Biosensors in High-Throughput Screening for Cell Factories Construction. Food Bioeng. 2022, 1, 135–147. [Google Scholar] [CrossRef]
- Ramirez, J.C.; Grajales García, D.; Maldonado, J.; Fernández-Gavela, A. Current Trends in Photonic Biosensors: Advances towards Multiplexed Integration. Chemosensors 2022, 10, 398. [Google Scholar] [CrossRef]
- Velu, K.; Shrestha, R.G.; Shrestha, L.K.; Ariga, K. Recent Advancements in Novel Sensing Systems through Nanoarchitectonics. Biosensors 2023, 13, 286. [Google Scholar] [CrossRef]
- Ugarte-Orozco, M.J.; López-Muñoz, G.A.; Antonio-Pérez, A.; Esquivel-Ortiz, K.M.; Ramón-Azcón, J. High-Throughput Biointerfaces for Direct, Label-Free, and Multiplexed Metaplasmonic Biosensing. Curr. Res. Biotechnol. 2023, 5, 100119. [Google Scholar] [CrossRef]
- Ravikumar, S.; Baylon, M.G.; Park, S.J.; Choi, J. Engineered Microbial Biosensors Based on Bacterial Two-Component Systems as Synthetic Biotechnology Platforms in Bioremediation and Biorefinery. Microb. Cell Factories 2017, 16, 62. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Yang, X.; Lu, Y.; Shu, Q.; Zhou, S.; Deng, Y. Engineering a Fumaric Acid-Responsive Two-Component Biosensor for Dynamic Range Improvement in Escherichia coli. Syst. Microbiol. Biomanuf. 2022, 2, 533–541. [Google Scholar] [CrossRef]
- Landry, B.P.; Palanki, R.; Dyulgyarov, N.; Hartsough, L.A.; Tabor, J.J. Phosphatase Activity Tunes Two-Component System Sensor Detection Threshold. Nat. Commun. 2018, 9, 1433. [Google Scholar] [CrossRef]
- Feng, J.; Jester, B.W.; Tinberg, C.E.; Mandell, D.J.; Antunes, M.S.; Chari, R.; Morey, K.J.; Rios, X.; Medford, J.I.; Church, G.M.; et al. A General Strategy to Construct Small Molecule Biosensors in Eukaryotes. eLife 2015, 4, e10606. [Google Scholar] [CrossRef] [PubMed]
- Gheorghiu, M. A Short Review on Cell-Based Biosensing: Challenges and Breakthroughs in Biomedical Analysis. J. Biomed. Res. 2021, 35, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Tric, M.; Lederle, M.; Neuner, L.; Dolgowjasow, I.; Wiedemann, P.; Wölfl, S.; Werner, T. Optical Biosensor Optimized for Continuous In-Line Glucose Monitoring in Animal Cell Culture. Anal. Bioanal. Chem. 2017, 409, 5711–5721. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Marsafari, M.; Xu, P. Engineering Metabolite-Responsive Transcriptional Factors to Sense Small Molecules in Eukaryotes: Current State and Perspectives. Microb. Cell Factories 2019, 18, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dabirian, Y.; Li, X.; Chen, Y.; David, F.; Nielsen, J.; Siewers, V. Expanding the Dynamic Range of a Transcription Factor-Based Biosensor in Saccharomyces Cerevisiae. ACS Synth. Biol. 2019, 8, 1968–1975. [Google Scholar] [CrossRef]
- De Paepe, B.; Peters, G.; Coussement, P.; Maertens, J.; De Mey, M. Tailor-Made Transcriptional Biosensors for Optimizing Microbial Cell Factories. J. Ind. Microbiol. Biotechnol. 2017, 44, 623–645. [Google Scholar] [CrossRef] [Green Version]
- Thompson, M.G.; Costello, Z.; Hummel, N.F.C.; Cruz-Morales, P.; Blake-Hedges, J.M.; Krishna, R.N.; Skyrud, W.; Pearson, A.N.; Incha, M.R.; Shih, P.M.; et al. Robust Characterization of Two Distinct Glutarate Sensing Transcription Factors of Pseudomonas Putida L-Lysine Metabolism. ACS Synth. Biol. 2019, 8, 2385–2396. [Google Scholar] [CrossRef]
- Hernández-Sánchez, V.; Molina, L.; Ramos, J.L.; Segura, A. New Family of Biosensors for Monitoring BTX in Aquatic and Edaphic Environments. Microb. Biotechnol. 2016, 9, 858–867. [Google Scholar] [CrossRef]
- Madar, D.; Dekel, E.; Bren, A.; Alon, U. Negative Auto-Regulation Increases the Input Dynamic-Range of the Arabinose System of Escherichia coli. BMC Syst. Biol. 2011, 5, 111. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.-Y.; Wei, W.; Yin, B.-C.; Tong, Y.; Lu, J.; Ye, B.-C. Development of a Highly Sensitive Whole-Cell Biosensor for Arsenite Detection through Engineered Promoter Modifications. ACS Synth. Biol. 2019, 8, 2295–2302. [Google Scholar] [CrossRef]
- Macazo, F.C.; Karpel, R.L.; White, R.J. Monitoring Cooperative Binding Using Electrochemical DNA-Based Sensors. Langmuir 2015, 31, 868–875. [Google Scholar] [CrossRef] [Green Version]
- Schumacher, M.A.; Miller, M.C.; Grkovic, S.; Brown, M.H.; Skurray, R.A.; Brennan, R.G. Structural Basis for Cooperative DNA Binding by Two Dimers of the Multidrug-Binding Protein QacR. EMBO J. 2002, 21, 1210–1218. [Google Scholar] [CrossRef] [PubMed]
- Tellechea-Luzardo, J.; Stiebritz, M.T.; Carbonell, P. Transcription Factor-Based Biosensors for Screening and Dynamic Regulation. Front. Bioeng. Biotechnol. 2023, 11, 1118702. [Google Scholar] [CrossRef]
- Van Brempt, M.; Peeters, A.I.; Duchi, D.; De Wannemaeker, L.; Maertens, J.; De Paepe, B.; De Mey, M. Biosensor-Driven, Model-Based Optimization of the Orthogonally Expressed Naringenin Biosynthesis Pathway. Microb. Cell Factories 2022, 21, 49. [Google Scholar] [CrossRef] [PubMed]
- d’Oelsnitz, S.; Nguyen, V.; Alper, H.S.; Ellington, A.D. Evolving a Generalist Biosensor for Bicyclic Monoterpenes. ACS Synth. Biol. 2022, 11, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ho, J.M.L.; Shis, D.L.; Gupta, C.; Long, J.; Wagner, D.S.; Ott, W.; Josić, K.; Bennett, M.R. Tuning the Dynamic Range of Bacterial Promoters Regulated by Ligand-Inducible Transcription Factors. Nat. Commun. 2018, 9, 64. [Google Scholar] [CrossRef] [Green Version]
- Peters, G.; De Paepe, B.; De Wannemaeker, L.; Duchi, D.; Maertens, J.; Lammertyn, J.; De Mey, M. Development of N-Acetylneuraminic Acid Responsive Biosensors Based on the Transcriptional Regulator NanR. Biotechnol. Bioeng. 2018, 115, 1855–1865. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; Hu, C.; Xu, X.; Lü, C.; Wang, Q.; Zhang, W.; Gao, C.; Xu, P.; Wang, X.; Ma, C. A d,l-Lactate Biosensor Based on Allosteric Transcription Factor LldR and Amplified Luminescent Proximity Homogeneous Assay. Biosens. Bioelectron. 2022, 211, 114378. [Google Scholar] [CrossRef]
- Mannan, A.A.; Liu, D.; Zhang, F.; Oyarzún, D.A. Fundamental Design Principles for Transcription-Factor-Based Metabolite Biosensors. ACS Synth. Biol. 2017, 6, 1851–1859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, Y.; Jiang, W.; Zhang, F. Developing a Genetically Encoded, Cross-Species Biosensor for Detecting Ammonium and Regulating Biosynthesis of Cyanophycin. ACS Synth. Biol. 2017, 6, 1807–1815. [Google Scholar] [CrossRef]
- Liu, D.; Xiao, Y.; Evans, B.S.; Zhang, F. Negative Feedback Regulation of Fatty Acid Production Based on a Malonyl-CoA Sensor–Actuator. ACS Synth. Biol. 2015, 4, 132–140. [Google Scholar] [CrossRef]
- Hartline, C.J.; Zhang, F. The Growth Dependent Design Constraints of Transcription-Factor-Based Metabolite Biosensors. ACS Synth. Biol. 2022, 11, 2247–2258. [Google Scholar] [CrossRef]
- Sun, S.; Peng, K.; Sun, S.; Wang, M.; Shao, Y.; Li, L.; Xiang, J.; Sedjoah, R.-C.A.-A.; Xin, Z. Engineering Modular and Highly Sensitive Cell-Based Biosensors for Aromatic Contaminant Monitoring and High-Throughput Enzyme Screening. ACS Synth. Biol. 2023, 12, 877–891. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Jiang, P.; Chen, W.; Xiong, D.; Huang, L.; Jia, J.; Chen, Y.; Jin, J.-M.; Tang, S.-Y. Design and Application of a Lactulose Biosensor. Sci. Rep. 2017, 7, 45994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.-F.; Xia, X.-X.; Lee, S.Y.; Qian, Z.-G. Engineering Tunable Biosensors for Monitoring Putrescine in Escherichia coli. Biotechnol. Bioeng. 2018, 115, 1014–1027. [Google Scholar] [CrossRef]
- Kasey, C.M.; Zerrad, M.; Li, Y.; Cropp, T.A.; Williams, G.J. Development of Transcription Factor-Based Designer Macrolide Biosensors for Metabolic Engineering and Synthetic Biology. ACS Synth. Biol. 2018, 7, 227–239. [Google Scholar] [CrossRef]
- Chen, C.; Liu, J.; Yao, G.; Bao, S.; Wan, X.; Wang, F.; Wang, K.; Song, T.; Han, P.; Liu, T.; et al. A Novel, Genetically Encoded Whole-Cell Biosensor for Directed Evolution of Myrcene Synthase in Escherichia coli. Biosens. Bioelectron. 2023, 228, 115176. [Google Scholar] [CrossRef]
- Ding, N.; Yuan, Z.; Zhang, X.; Chen, J.; Zhou, S.; Deng, Y. Programmable Cross-Ribosome-Binding Sites to Fine-Tune the Dynamic Range of Transcription Factor-Based Biosensor. Nucleic Acids Res. 2020, 48, 10602–10613. [Google Scholar] [CrossRef] [PubMed]
- Shilling, P.J.; Khananisho, D.; Cumming, A.J.; Söderström, B.; Daley, D.O. Signal Amplification of AraC PBAD Using a Standardized Translation Initiation Region. Synth. Biol. 2022, 7, ysac009. [Google Scholar] [CrossRef]
- Wang, B.; Kitney, R.I.; Joly, N.; Buck, M. Engineering Modular and Orthogonal Genetic Logic Gates for Robust Digital-like Synthetic Biology. Nat. Commun. 2011, 2, 508. [Google Scholar] [CrossRef] [Green Version]
- Greco, F.V.; Pandi, A.; Erb, T.J.; Grierson, C.S.; Gorochowski, T.E. Harnessing the Central Dogma for Stringent Multi-Level Control of Gene Expression. Nat. Commun. 2021, 12, 1738. [Google Scholar] [CrossRef]
- Oesterle, S.; Gerngross, D.; Schmitt, S.; Roberts, T.M.; Panke, S. Efficient Engineering of Chromosomal Ribosome Binding Site Libraries in Mismatch Repair Proficient Escherichia coli. Sci. Rep. 2017, 7, 12327. [Google Scholar] [CrossRef]
- Gong, X.; Zhang, R.; Wang, J.; Yan, Y. Engineering of a TrpR-Based Biosensor for Altered Dynamic Range and Ligand Preference. ACS Synth. Biol. 2022, 11, 2175–2183. [Google Scholar] [CrossRef]
- Collins, C.H.; Leadbetter, J.R.; Arnold, F.H. Dual Selection Enhances the Signaling Specificity of a Variant of the Quorum-Sensing Transcriptional Activator LuxR. Nat. Biotechnol. 2006, 24, 708–712. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.; Chen, Z.; Guo, S.; Zhang, C.; Huo, Y.-X. Engineering Transcription Factor BmoR Mutants for Constructing Multifunctional Alcohol Biosensors. ACS Synth. Biol. 2022, 11, 1251–1260. [Google Scholar] [CrossRef]
- Rogers, J.K.; Church, G.M. Genetically Encoded Sensors Enable Real-Time Observation of Metabolite Production. Proc. Natl. Acad. Sci. USA 2016, 113, 2388–2393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.; Zhang, F. Heterogeneity Coordinates Bacterial Multi-Gene Expression in Single Cells. PLoS Comput. Biol. 2020, 16, e1007643. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, F. Control Strategies to Manage Trade-Offs during Microbial Production. Curr. Opin. Biotechnol. 2020, 66, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.-J.; Zúñiga, A.; Conejero, I.; Voyvodic, P.L.; Gracy, J.; Fajardo-Ruiz, E.; Cohen-Gonsaud, M.; Cambray, G.; Pageaux, G.-P.; Meszaros, M.; et al. Programmable Receptors Enable Bacterial Biosensors to Detect Pathological Biomarkers in Clinical Samples. Nat. Commun. 2021, 12, 5216. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, V.D.; Mohan, K.; Chappell, T.C.; Mays, Z.J.S.; Nair, N.U. Cheating the Cheater: Suppressing False-Positive Enrichment during Biosensor-Guided Biocatalyst Engineering. ACS Synth. Biol. 2022, 11, 420–429. [Google Scholar] [CrossRef]
- Zhang, F.; Keasling, J. Biosensors and Their Applications in Microbial Metabolic Engineering. Trends Microbiol. 2011, 19, 323–329. [Google Scholar] [CrossRef]
- Binder, S.; Schendzielorz, G.; Stäbler, N.; Krumbach, K.; Hoffmann, K.; Bott, M.; Eggeling, L. A High-Throughput Approach to Identify Genomic Variants of Bacterial Metabolite Producers at the Single-Cell Level. Genome Biol. 2012, 13, R40. [Google Scholar] [CrossRef] [Green Version]
- Juárez, J.F.; Lecube-Azpeitia, B.; Brown, S.L.; Johnston, C.D.; Church, G.M. Biosensor Libraries Harness Large Classes of Binding Domains for Construction of Allosteric Transcriptional Regulators. Nat. Commun. 2018, 9, 3101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanko, E.K.R.; Sherlock, G.; Minton, N.P.; Malys, N. Biosensor-Informed Engineering of Cupriavidus Necator H16 for Autotrophic D-Mannitol Production. Metab. Eng. 2022, 72, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Farmer, W.R.; Liao, J.C. Improving Lycopene Production in Escherichia coli by Engineering Metabolic Control. Nat. Biotechnol. 2000, 18, 533–537. [Google Scholar] [CrossRef] [PubMed]
- Dahl, R.H.; Zhang, F.; Alonso-Gutierrez, J.; Baidoo, E.; Batth, T.S.; Redding-Johanson, A.M.; Petzold, C.J.; Mukhopadhyay, A.; Lee, T.S.; Adams, P.D.; et al. Engineering Dynamic Pathway Regulation Using Stress-Response Promoters. Nat. Biotechnol. 2013, 31, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Wang, R.; Wang, D.; Zhao, C.; Ma, Q.; Wu, H.; Xie, X. Metabolic Reprogramming and Biosensor-Assisted Mutagenesis Screening for High-Level Production of L-Arginine in Escherichia coli. Metab. Eng. 2023, 76, 146–157. [Google Scholar] [CrossRef]
- Li, J.; Nina, M.R.H.; Zhang, X.; Bai, Y. Engineering Transcription Factor XylS for Sensing Phthalic Acid and Terephthalic Acid: An Application for Enzyme Evolution. ACS Synth. Biol. 2022, 11, 1106–1113. [Google Scholar] [CrossRef] [PubMed]
- Galarion, L.H.; Mitchell, J.K.; Randall, C.P.; O’Neill, A.J. An Extensively Validated Whole-Cell Biosensor for Specific, Sensitive and High-Throughput Detection of Antibacterial Inhibitors Targeting Cell-Wall Biosynthesis. J. Antimicrob. Chemother. 2023, 78, 646–655. [Google Scholar] [CrossRef] [PubMed]
- Martin, V.J.J.; Pitera, D.J.; Withers, S.T.; Newman, J.D.; Keasling, J.D. Engineering a Mevalonate Pathway in Escherichia coli for Production of Terpenoids. Nat. Biotechnol. 2003, 21, 796–802. [Google Scholar] [CrossRef] [PubMed]
- Hartline, C.J.; Mannan, A.A.; Liu, D.; Zhang, F.; Oyarzún, D.A. Metabolite Sequestration Enables Rapid Recovery from Fatty Acid Depletion in Escherichia coli. mBio 2020, 11, e03112–e03119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Gao, H.; Zhang, J.; Zhao, J.; Qi, Q.; Wang, Q. De Novo Design of the Global Transcriptional Factor Cra-Regulated Promoters Enables Highly Sensitive Glycolysis Flux Biosensor for Dynamic Metabolic Control. Microb. Biotechnol. 2023, 16, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Li, X.; Liu, Y.; Zhu, Y.; Li, J.; Du, G.; Chen, J.; Ledesma-Amaro, R.; Liu, L. Pyruvate-Responsive Genetic Circuits for Dynamic Control of Central Metabolism. Nat. Chem. Biol. 2020, 16, 1261–1268. [Google Scholar] [CrossRef]
- Zhou, S.; Yuan, S.-F.; Nair, P.H.; Alper, H.S.; Deng, Y.; Zhou, J. Development of a Growth Coupled and Multi-Layered Dynamic Regulation Network Balancing Malonyl-CoA Node to Enhance (2S)-Naringenin Biosynthesis in Escherichia coli. Metab. Eng. 2021, 67, 41–52. [Google Scholar] [CrossRef]
- Hartline, C.J.; Schmitz, A.C.; Han, Y.; Zhang, F. Dynamic Control in Metabolic Engineering: Theories, Tools, and Applications. Metab. Eng. 2021, 63, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Carothers, J.M.; Keasling, J.D. Design of a Dynamic Sensor-Regulator System for Production of Chemicals and Fuels Derived from Fatty Acids. Nat. Biotechnol. 2012, 30, 354–359. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, F. Metabolic Feedback Circuits Provide Rapid Control of Metabolite Dynamics. ACS Synth. Biol. 2018, 7, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Verma, B.K.; Mannan, A.A.; Zhang, F.; Oyarzún, D.A. Trade-Offs in Biosensor Optimization for Dynamic Pathway Engineering. ACS Synth. Biol. 2022, 11, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Koh, E.; Hwang, I.Y.; Lee, H.L.; De Sotto, R.; Lee, J.W.J.; Lee, Y.S.; March, J.C.; Chang, M.W. Engineering Probiotics to Inhibit Clostridioides Difficile Infection by Dynamic Regulation of Intestinal Metabolism. Nat. Commun. 2022, 13, 3834. [Google Scholar] [CrossRef]
- Zou, F.; Bai, L. Using Time-Lapse Fluorescence Microscopy to Study Gene Regulation. Methods 2019, 159–160, 138–145. [Google Scholar] [CrossRef]
- Evans, T.D.; Zhang, F. Bacterial Metabolic Heterogeneity: Origins and Applications in Engineering and Infectious Disease. Curr. Opin. Biotechnol. 2020, 64, 183–189. [Google Scholar] [CrossRef]
- Wang, T.; Dunlop, M.J. Controlling and Exploiting Cell-to-Cell Variation in Metabolic Engineering. Curr. Opin. Biotechnol. 2019, 57, 10–16. [Google Scholar] [CrossRef]
- Schmitz, A.C.; Hartline, C.J.; Zhang, F. Engineering Microbial Metabolite Dynamics and Heterogeneity. Biotechnol. J. 2017, 12, 1700422. [Google Scholar] [CrossRef]
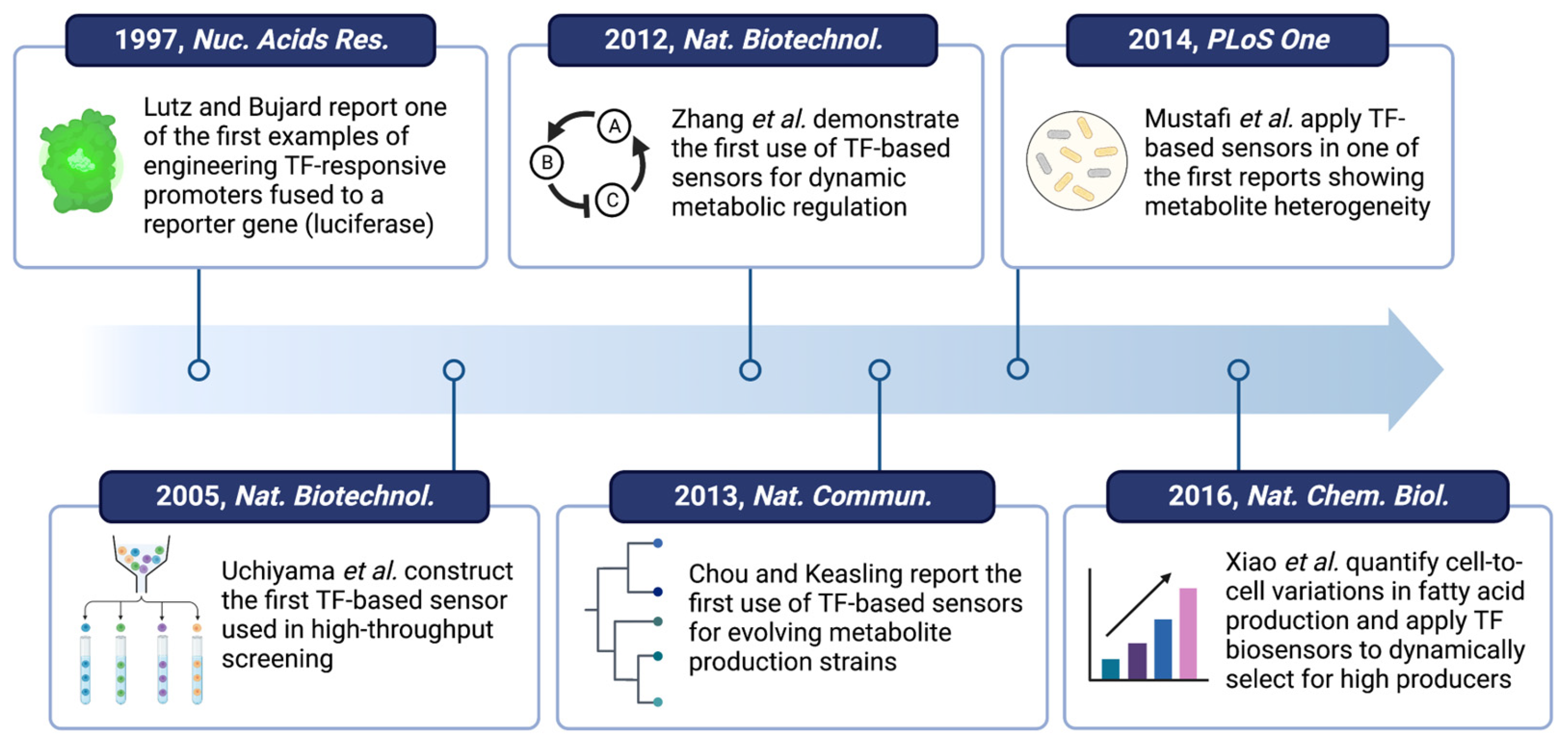
- Mustafi, N.; Grünberger, A.; Kohlheyer, D.; Bott, M.; Frunzke, J. The Development and Application of a Single-Cell Biosensor for the Detection of l-Methionine and Branched-Chain Amino Acids. Metab. Eng. 2012, 14, 449–457. [Google Scholar] [CrossRef]
- Xiao, Y.; Bowen, C.H.; Liu, D.; Zhang, F. Exploiting Nongenetic Cell-to-Cell Variation for Enhanced Biosynthesis. Nat. Chem. Biol. 2016, 12, 339–344. [Google Scholar] [CrossRef]
- Waters, J.C. Accuracy and Precision in Quantitative Fluorescence Microscopy. J. Cell Biol. 2009, 185, 1135–1148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hickey, S.M.; Ung, B.; Bader, C.; Brooks, R.; Lazniewska, J.; Johnson, I.R.D.; Sorvina, A.; Logan, J.; Martini, C.; Moore, C.R.; et al. Fluorescence Microscopy—An Outline of Hardware, Biological Handling, and Fluorophore Considerations. Cells 2021, 11, 35. [Google Scholar] [CrossRef] [PubMed]
- Rugbjerg, P.; Sarup-Lytzen, K.; Nagy, M.; Sommer, M.O.A. Synthetic Addiction Extends the Productive Life Time of Engineered Escherichia coli Populations. Proc. Natl. Acad. Sci. USA 2018, 115, 2347–2352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, W.; Gu, P.; Zhang, F. Steps towards ‘Drop-in’ Biofuels: Focusing on Metabolic Pathways. Curr. Opin. Biotechnol. 2018, 53, 26–32. [Google Scholar] [CrossRef]
- Bai, W.; Geng, W.; Wang, S.; Zhang, F. Biosynthesis, Regulation, and Engineering of Microbially Produced Branched Biofuels. Biotechnol. Biofuels 2019, 12, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moser, F.; Espah Borujeni, A.; Ghodasara, A.N.; Cameron, E.; Park, Y.; Voigt, C.A. Dynamic Control of Endogenous Metabolism with Combinatorial Logic Circuits. Mol. Syst. Biol. 2018, 14, e8605. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Peña, M.; Bennett, G.N. Metabolic Engineering of Escherichia coli for Quinolinic Acid Production by Assembling L-Aspartate Oxidase and Quinolinate Synthase as an Enzyme Complex. Metab. Eng. 2021, 67, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Lutz, R.; Bujard, H. Independent and Tight Regulation of Transcriptional Units in Escherichia coli Via the LacR/O, the TetR/O and AraC/I1-I2 Regulatory Elements. Nucleic Acids Res. 1997, 25, 1203–1210. [Google Scholar] [CrossRef]
- Uchiyama, T.; Abe, T.; Ikemura, T.; Watanabe, K. Substrate-Induced Gene-Expression Screening of Environmental Metagenome Libraries for Isolation of Catabolic Genes. Nat. Biotechnol. 2005, 23, 88–93. [Google Scholar] [CrossRef]
- Chou, H.H.; Keasling, J.D. Programming Adaptive Control to Evolve Increased Metabolite Production. Nat. Commun. 2013, 4, 2595. [Google Scholar] [CrossRef] [Green Version]
- Mustafi, N.; Grünberger, A.; Mahr, R.; Helfrich, S.; Nöh, K.; Blombach, B.; Kohlheyer, D.; Frunzke, J. Application of a Genetically Encoded Biosensor for Live Cell Imaging of L-Valine Production in Pyruvate Dehydrogenase Complex-Deficient Corynebacterium Glutamicum Strains. PLoS ONE 2014, 9, e85731. [Google Scholar] [CrossRef] [Green Version]
- Matilla, M.A.; Velando, F.; Martín-Mora, D.; Monteagudo-Cascales, E.; Krell, T. A Catalogue of Signal Molecules That Interact with Sensor Kinases, Chemoreceptors and Transcriptional Regulators. FEMS Microbiol. Rev. 2022, 46, fuab043. [Google Scholar] [CrossRef] [PubMed]
- Otero-Muras, I.; Mannan, A.A.; Banga, J.R.; Oyarzún, D.A. Multiobjective Optimization of Gene Circuits for Metabolic Engineering. IFAC-Pap. 2019, 52, 13–16. [Google Scholar] [CrossRef]
- Valeri, J.A.; Collins, K.M.; Ramesh, P.; Alcantar, M.A.; Lepe, B.A.; Lu, T.K.; Camacho, D.M. Sequence-to-Function Deep Learning Frameworks for Engineered Riboregulators. Nat. Commun. 2020, 11, 5058. [Google Scholar] [CrossRef]
- Nikolados, E.-M.; Wongprommoon, A.; Aodha, O.M.; Cambray, G.; Oyarzún, D.A. Accuracy and Data Efficiency in Deep Learning Models of Protein Expression. Nat. Commun. 2022, 13, 7755. [Google Scholar] [CrossRef] [PubMed]
- Höllerer, S.; Papaxanthos, L.; Gumpinger, A.C.; Fischer, K.; Beisel, C.; Borgwardt, K.; Benenson, Y.; Jeschek, M. Large-Scale DNA-Based Phenotypic Recording and Deep Learning Enable Highly Accurate Sequence-Function Mapping. Nat. Commun. 2020, 11, 3551. [Google Scholar] [CrossRef] [PubMed]
- Arano-Martinez, J.A.; Martínez-González, C.L.; Salazar, M.I.; Torres-Torres, C. A Framework for Biosensors Assisted by Multiphoton Effects and Machine Learning. Biosensors 2022, 12, 710. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Kan, S.B.J.; Lewis, R.D.; Wittmann, B.J.; Arnold, F.H. Machine Learning-Assisted Directed Protein Evolution with Combinatorial Libraries. Proc. Natl. Acad. Sci. USA 2019, 116, 8852–8858. [Google Scholar] [CrossRef] [Green Version]
- Dar, D.; Dar, N.; Cai, L.; Newman, D.K. Spatial Transcriptomics of Planktonic and Sessile Bacterial Populations at Single-Cell Resolution. Science 2021, 373, eabi4882. [Google Scholar] [CrossRef]
- Zhu, Y.; Mohapatra, S.; Weisshaar, J.C. Rigidification of the Escherichia coli Cytoplasm by the Human Antimicrobial Peptide LL-37 Revealed by Superresolution Fluorescence Microscopy. Proc. Natl. Acad. Sci. USA 2019, 116, 1017–1026. [Google Scholar] [CrossRef] [Green Version]
- Tang, W.; Liu, D.R. Rewritable Multi-Event Analog Recording in Bacterial and Mammalian Cells. Science 2018, 360, eaap8992. [Google Scholar] [CrossRef] [Green Version]
- Kastberg, L.L.B.; Ard, R.; Jensen, M.K.; Workman, C.T. Burden Imposed by Heterologous Protein Production in Two Major Industrial Yeast Cell Factories: Identifying Sources and Mitigation Strategies. Front. Fungal Biol. 2022, 3, 1. [Google Scholar] [CrossRef]
- Jiang, W.; Qiao, J.B.; Bentley, G.J.; Liu, D.; Zhang, F. Modular Pathway Engineering for the Microbial Production of Branched-Chain Fatty Alcohols. Biotechnol. Biofuels 2017, 10, 244. [Google Scholar] [CrossRef]
- Bai, W.; Anthony, W.E.; Hartline, C.J.; Wang, S.; Wang, B.; Ning, J.; Hsu, F.-F.; Dantas, G.; Zhang, F. Engineering Diverse Fatty Acid Compositions of Phospholipids in Escherichia coli. Metab. Eng. 2022, 74, 11–23. [Google Scholar] [CrossRef]
- Carbonell, P.; Radivojevic, T.; García Martín, H. Opportunities at the Intersection of Synthetic Biology, Machine Learning, and Automation. ACS Synth. Biol. 2019, 8, 1474–1477. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Tuning Strategy | Advantages | Disadvantages | References |
|---|---|---|---|
| Promoter engineering | Several approaches, including changing the number or location of TF operator sites and introducing point mutations to the TF operator sites or the −35 and −10 binding sites; useful for fine-tuning sensitivity, detection range, dynamic range, cooperativity, or signal output intensity | Cannot be used to adjust sensor specificity for target metabolites | [36,37,38,39,40,41] |
| TF engineering | Can tune binding with target metabolite or DNA; useful for fine-tuning specificity, sensitivity, and dynamic range | Requires knowledge of TF structure and binding mechanism; TF expression level changes with cell growth conditions; mutations specific to one TF (e.g., specific base substitutions in one TF ligand-binding domain) may not be readily applicable to other TFs | [36,40,41,42,43,44,45,46,47,48] |
| RBS engineering | Can be used to control the rate of TF or reporter protein production; can be combined with transcriptional control for multi-layered control strategies; useful for fine-tuning the dynamic range | Cannot be used to adjust sensor specificity for target metabolites | [49,50,51,52,53] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, G.J.; Zhang, F. Applications and Tuning Strategies for Transcription Factor-Based Metabolite Biosensors. Biosensors 2023, 13, 428. https://doi.org/10.3390/bios13040428
Zhou GJ, Zhang F. Applications and Tuning Strategies for Transcription Factor-Based Metabolite Biosensors. Biosensors. 2023; 13(4):428. https://doi.org/10.3390/bios13040428
Chicago/Turabian StyleZhou, Gloria J., and Fuzhong Zhang. 2023. "Applications and Tuning Strategies for Transcription Factor-Based Metabolite Biosensors" Biosensors 13, no. 4: 428. https://doi.org/10.3390/bios13040428





